Abstract
Vaccination of mice with activated autoantigen-reactive CD4+ T cells (T cell vaccination, TCV) has been shown to induce protection from the subsequent induction of a variety of experimental autoimmune diseases, including experimental allergic encephalomyelitis (EAE). Although the mechanisms involved in TCV-mediated protection are not completely known, there is some evidence that TCV induces CD8+ regulatory T cells that are specific for pathogenic CD4+ T cells. Previously, we demonstrated that, after superantigen administration in vivo, CD8+ T cells emerge that preferentially lyse and regulate activated autologous CD4+ T cells in a T cell receptor (TCR) Vβ-specific manner. This TCR Vβ-specific regulation is not observed in β2-microglobulin-deficient mice and is inhibited, in vitro, by antibody to Qa-1. We now show that similar Vβ8-specific Qa-1-restricted CD8+ T cells are also induced by TCV with activated CD4+ Vβ8+ T cells. These CD8+ T cells specifically lyse murine or human transfectants coexpressing Qa-1 and murine TCR Vβ8. Further, CD8+ T cell hybridoma clones generated from B10.PL mice vaccinated with a myelin basic protein-specific CD4+Vβ8+ T cell clone specifically recognize other CD4+ T cells and T cell tumors that express Vβ8 and the syngeneic Qa-1a but not the allogeneic Qa-1b molecule. Thus, Vβ-specific Qa-1-restricted CD8+ T cells are induced by activated CD4+ T cells. We suggest that these CD8+ T cells may function to specifically regulate activated CD4+ T cells during immune responses.
The injection of naive animals with irradiated, antigen-activated, autoimmune CD4+ T cells (T cell vaccination, TCV) was initially shown to protect rats from the subsequent induction of experimental allergic encephalomyelitis (EAE) (1). TCV has since been used with various degrees of success in a variety of animal models of autoimmunity (2) and in human autoimmune disease (3, 4). Although it has generally been thought that TCV augments the normal mechanisms employed by regulatory T cells to control the pathogenic potential and/or outgrowth of disease-causing T cells (5), the precise mechanisms by which TCV may ameliorate autoimmune manifestations are unknown.
Several lines of evidence have suggested that CD8+ T cells mediate the protective effect of TCV by down-regulating autoimmune T cells. For example, in studies of patients with multiple sclerosis, vaccination with autologous myelin basic protein (MBP)-reactive CD4+ T cells (3) was shown to induce CD8+ T cells that inhibited the antigen-induced proliferation of vaccine T cell clones and also specifically lysed the vaccine CD4+ T cell clones in vitro. Similarly, the induction of EAE in rats by the injection of an encephalitogenic CD4+ T cell clone was shown to induce CD8+ T cells that recognize the encephalitogenic T cell clone in vitro and that, upon adoptive transfer, protect naive animals from the induction of EAE in vivo (6). These regulatory CD8+ T cells preferentially recognized the vaccine T cells but not other T cells activated by different antigens. This clonotypic recognition by the CD8+ cells suggested that the T cell receptor (TCR) or peptides derived from the TCR of autoimmune T cells are part of the target structure recognized by regulatory CD8+ T cells. In this regard, TCR peptide immunization, although it may function by different mechanisms than TCV, efficiently prevents EAE in rats and mice (7–9) and thus provides additional evidence that the recognition of TCR structures is involved in the immunoregulation of EAE. However, regulatory CD8+ T cells may not necessarily be specific for unique idiotypes expressed by particular autoimmune CD4+ clones, and it is possible that CD8+ regulatory cells may recognize variable portions of the TCR common to a set of TCRs. In this regard pathogenic autoimmune T cell populations, although not clonally homogeneous, are known to be restricted in terms of their recognition of particular peptide(s) and in their TCR gene usage (10–12).
It is important to emphasize that although the above studies strongly suggest the possibility that TCV induces regulatory CD8+ T cells that recognize the TCR or TCR peptide expressed by autologous CD4+ T cells, there have been no experiments to directly demonstrate this point at a molecular level. In this regard, it is of interest that, in a different in vivo system, we have shown that the staphylococcal enterotoxin B (SEB)-induced deletion of CD4+ Vβ8+ T cells depends, in part, on CD8+ T cells. Moreover we have shown that, during the period of in vivo deletion of the CD4+ Vβ8+ cells, one can culture from the animals CD8+ cytotoxic T lymphocytes (CTL) that kill in vitro activated autologous CD4+ Vβ8+ T cells but not T cells that express other Vβ TCR. This TCR Vβ-specific killing requires β2-microglobulin (β2m)-associated class I major histocompatibility complex (MHC) molecules expressed on the target cells and is inhibited by antisera to the class I-b MHC molecule Qa-1 but not by antibodies to conventional class I-a MHC molecules (13).
Because the protective effect of TCV has been ascribed to the CD8+ T cell recognition of TCR structures expressed by CD4+ cells and because we had identified, in a different context, CD8+ T cells that in fact recognize TCR structures on CD4+ cells, we considered the possibility that TCV induces Vβ-specific Qa-1-restricted CD8+ T cells analogous to those we had identified in SEB-primed mice. To test this hypothesis we used antigen- or superantigen-activated purified CD4+ Vβ8+ T cells as vaccine T cells and assayed the specificity and Qa-1 restriction of the CD8+ T cells induced by TCV. These CD8+ T cells were found to be TCR Vβ specific and Qa-1 restricted. Moreover, CD8+ T cell hybridoma clones generated from B10.PL mice vaccinated with a MBP-specific CD4+Vβ8+ T cell clone displayed the same Vβ specificity and Qa-1 restriction. Thus, clones of Vβ-specific Qa-1-restricted CD8+ T cells are induced during TCV by activated CD4+ T cells.
MATERIALS AND METHODS
Animals.
AKR (H-2k, Qa-1b) mice, B10.PL (H-2u, Qa-1a) mice (female, 6–12 weeks old), were purchased from The Jackson Laboratory and were maintained in our animal facilities.
Antibodies and Antisera.
Fluorescein (Fl)-, allophycocyanin (APC)-, or biotin (Bio)-coupled antibodies 53-6.72 (anti-mouse CD8), APC-GK1.5 (anti-mouse CD4), and Bio-F23.1 (anti-mouse TCR Vβ8.1-3) were purified from the ascites fluids of correspondent hybridomas and conjugated in our laboratory. Bio-RR4.7 (anti-mouse TCR Vβ6) was purchased from PharMingen (San Diego, CA). M1/42, rat IgG anti-mouse MHC class I-a; 16-1-2N, mouse IgG2a anti-H-2KkDk; 3-83P, mouse IgG2a anti-H-2k, crossreactive with H-2u; and Y3P, control mouse IgG2a anti-I-Ab were purified from hybridoma culture supernatants. Anti-Qa-1a and anti-Qa-1b antisera were prepared as described previously (14–16).
Transfectants.
4G4Vβ6, 4G4Vβ8, and 4G4Vβ10 transfectants were generated by electroporation of 4G4, a T cell hybridoma selected for the loss of TCRβ chain expression (17), with full-length TCRβ cDNAs. Transfectants were screened for the gain of surface CD3 expression and subcloned as needed to generate uniformly positive lines. Transfection with the appropriate cDNAs was verified by staining with the appropriate anti-Vβ antibody. Vβ6 and Vβ8 TCRβ transfectants of C1R and J1 were similarly prepared except that gene expression by the transfectants and subclones was confirmed by Northern analysis followed by reverse transcription (RT)-PCR using a Vβ8.2 (5′-CATGGAGGCTGCAGTCACCC-3′) or Vβ6 (5′-CAAAGAAAGTCCCTCCAAACTAT-3′) specific primer and a Cβ1-specific primer (5′-TCATGAATTCTTTCTTTTGACCAT-3′).
T Cell Clones and Lines.
CD4+, Vβ8+ or Vβ6+, MBP-reactive B10.PL clones (H-2u, Qa-1a) were derived by limiting dilution cloning of CD4+-purified T cells obtained from the spleen and lymph nodes of B10.PL mice immunized with the peptide 1–9NAc-MBP emulsified in complete Freund’s adjuvant. The SEB-reactive Vβ8+ clone in Table 3 was derived by limiting dilution cloning and SEB stimulation of CD4+-purified spleen cells from naive B10.PL animals. In assays of CD8+ T cell function, CD4+ T cells were used on days 5–7 after activation. SEB-activated purified CD4+Vβ8+ T cells (>95% CD4+Vβ8+) or MBP-activated CD4+Vβ8+ T cell clones were used to vaccinate mice. CD4+ T cells were activated as described (13) and used 5–7 days after activation.
Table 3.
Responses of CD8+ T hybridomas generated by vaccination with the MBP-specific CD4+Vβ8+ syngeneic T cell clone 1AE10
| Response
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stimulators
|
T cell hybridoma 21-5A9
|
T cell hybridoma 176-5F7
|
||||||||||||
| Name | Specificity | Vβ | H-2 | Qa-1 | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 |
| 1AE10 | MBP | 8.3 | u | a | 2+ | 0.461 | 0.846 | 0.324 | 0.309 | 2+ | 0.415 | 0.513 | 0.178 | 0.248 |
| 1AC3 | MBP | 8.2 | u | a | 2+ | 0.377 | 0.580 | 0.120 | 0.264 | ± | 0.060 | 0.054 | 0.000 | 0.037 |
| 3AC3 | MBP | 8.3 | u | a | 2+ | NT | 0.242 | 0.040 | 0.293 | 2+ | NT | 0.209 | 0.043 | 0.223 |
| SEB-Vβ8 | SEB | 8.2 | u | a | 1+ | 0.508 | 0.731 | 0.416 | 0.208 | 2+ | 0.514 | 0.637 | 0.386 | 0.222 |
| 3AB3 | MBP | 6 | u | a | ± | 0.164 | 0.127 | 0.055 | NT | 0 | 0.062 | 0.041 | 0.027 | NT |
| 3AF9 | MBP | 6 | u | a | 0 | 0.080 | 0.071 | 0.039 | NT | 0 | 0.183 | 0.117 | 0.068 | NT |
| 3AG2 | MBP | 6 | u | a | 0 | 0.131 | 0.054 | 0.043 | NT | 2+ | 0.478 | 0.534 | 0.314 | NT |
| 3BB8 | MBP | 6 | u | a | 0 | 0.172 | 0.101 | 0.082 | NT | 0 | 0.212 | 0.178 | 0.098 | NT |
| LBRM | — | 8.2 | k | a | 2+ | 0.844 | 1.434 | 0.926 | 0.611 | 2+ | 0.923 | 1.285 | 0.846 | 0.611 |
| J1-9f | — | 8.2 | HLA | b | 0 | NT | 0.114 | 0.096 | 0.028 | 0 | NT | 0.019 | 0.018 | 0.007 |
| 4G4-6B5 | — | 8.2 | k | b | 0 | NT | 0.106 | 0.079 | 0.017 | 0 | NT | 0.086 | 0.082 | 0.017 |
| PI | — | — | — | — | 3+ | 1.019 | 1.497 | 0.972 | 0.739 | 3+ | 0.782 | 1.268 | 0.484 | 0.327 |
The results for experiment 1 represent the visual estimates of the responses derived by counting the numbers of blue cells in each well containing 5 × 104 cells after fixation and incubation of the hybridomas with X-Gal (0 = <20; ± = <100; 1+ = 100–1,000; 2+ >1,000; and 3+ = virtually all). The results for experiments 2–6 represent the means minus the background (CD8 cells alone) of a colorimetric assay of β-galactosidase activity. NT, not tested. PI indicates the culture of the hybridomas for 4 hr with 20 ng/ml phorbol 12,13-dibutyrate and 0.4 μM ionomycin.
Generation of TCR Vβ8-Specific CD8+ T Cells.
In each experiment, four AKR (H-2k, Qa-1b) mice were injected intravenously with either SEB (50 μg per mouse) or 2–2.5 × 106 irradiated (3,000 R), SEB-activated CD4+Vβ8+ T cells (5–7 days after stimulation). CD8+ T cells were isolated from the mice 7–10 days after priming and retriggered with SEB-activated CD4+Vβ8+ T cells in vitro as described (13). Cytotoxicity of the CD8+ T cells was assayed after 10–14 days of culture.
Fluorescence-Activated Cell Sorting (FACS) Analysis for Measuring Specific CTL Activity.
FACS analysis was used for measuring specific CTL activity as described (13). 4G4Vβ6 transfectant cells that could not be killed by TCR Vβ8-specific CTL served as control cells for each target. In experiments using murine cells as targets (Table 1), 0.1 × 106 4G4Vβ6 cells were added to 0.1 × 106 of each of the different target cells in duplicate or triplicate tubes. Graded numbers of putative CD8+ CTL effector populations (effector-to-target ratio 1:1 or 2:1) were added to each mixed target population for 24 hr, and the ratio of Vβx+/Vβ6+ cells was measured by FACS. The control for each target was the ratio of Vβx+/Vβ6+ cultured for 24 hr in the absence of effector cells. Targets and effectors were distinguished by expression of CD8 as measured with the fluorescein-conjugated anti-CD8 antibody 53-6.72, and Vβx+ and Vβ6+ targets were distinguished by expression of Vβ6 as measured with biotin-conjugated anti-mouse Vβ6 antibody RR4.7 followed by phycoerythrin-streptavidin. Dead cells were excluded by using propidium iodide. For data analysis, the CD8+ T cells were gated out and the data were expressed as the ratio of Vβ6-negative cells (experimental targets) to Vβ6-positive cells (control targets). The percentage of specific cytolysis of each target was calculated as {[(Vβ6-negative cells/Vβ6-positive cells of control group) − (Vβ6-negative cells/Vβ6-positive cells of experimental group)]/(Vβ6-negative cells/Vβ6-positive cells of control group)} × 100%, in which the control group is samples without effector cells and the experimental group is samples with effector cells.
Table 1.
TCR Vβ8-specific CD8+ CTL are generated after vaccination with CD4+Vβ8+ T cells
| Exp. | CTL | Cytotoxicity for target cells, %
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 4G4 parent (Vβ−, H-2k, Qa-1b) | 4G4-6B5a (Vβ8+, H-2k, Qa-1b) | 4G4-6B5b (Vβ−, H-2k, Qa-1b) | 4G4-Vβ10 (Vβ10+, H-2k, Qa-1b) | D10 (Vβ8+, H-2k, Qa-1b) | RMA (Vβ12+, H-2b, Qa-1b) | BW-Vβ14 (Vβ14+, H-2k, Qa-1b) | BW-Vβ15 (Vβ15+, H-2k, Qa-1b) | ||
| 1 | TCV | <1 | 15.6 | <1 | <1 | 65.8 | <1 | <1 | <1 |
| 2 | TCV | <1 | 39.1 | <1 | <1 | 73.1 | <1 | <1 | <1 |
| 3 | SEB-primed | <1 | 16.5 | <1 | <1 | 34.4 | <1 | <1 | <1 |
| 4 | SEB-primed | 7.5 | 23.5 | <1 | 5.9 | 35.9 | <1 | <1 | <1 |
In each experiment, four AKR (H-2k, Qa-1b) mice were injected intravenously with syngeneic irradiated SEB-activated CD4+Vβ8+ T cells (experiments 1 and 2) or SEB (experiments 3 and 4). CD8+ T cells were isolated 7–10 days after priming, retriggered with SEB-activated CD4+Vβ8+ T cells in vitro as described in ref. 13, and assayed for cytotoxicity after 10–14 days of culture.
In the FACS cytotoxicity assays using human B cell C1R transfectants (Table 2), CD8+ T cells were added to a 1:1 mixture of mouse 4G4Vβ6 cells and each human target cell. FACS analysis and data reduction were performed as above except that Bio-W6/32 (anti-HLA class I mAb) and not the anti-Vβ6 antibody was used to distinguish human and mouse target cells.
Table 2.
TCR Vβ8-specific CD8+ T cells isolated from TCV mice kill human B cells cotransfected with murine Qa-1b and Vβ8 cDNAs
| Target | Expression | Cytotoxicity, %
|
|||
|---|---|---|---|---|---|
| of
| |||||
| Qa-1b | Vβ | CTL line 48 (3 exps.) | CTL line 49 (2 exps.) | CTL line 51 (5 exps.) | |
| C1R | − | − | <1 | <1 | <1 |
| C1R/Vβ8.2-6d | − | 8 | <1 | <1 | <1 |
| J1 | + | − | <1 | <1 | <1 |
| J1/Vβ6-S3 | + | 6 | <1 | <1 | <1 |
| J1/Vβ6-S7 | + | 6 | <1 | <1 | <1 |
| J1/Vβ8.2-b | + | 8 | 36.7 ± 2.8 | 49.5 ± 6.7 | 25.4 ± 3.8 |
| J1/Vβ8.2-2e | + | 8 | 30.9 ± 3.5 | 38.3 ± 1.9 | 30.3 ± 7.4 |
| J1/Vβ8.2-9f | + | 8 | 43.2 ± 6.5 | 49.9 ± 13.5 | 21.7 ± 5.0 |
T Cell Hybridomas.
BW-Vβ14 and BW-Vβ15 hybridomas were generated from alloreactive T cell clones described previously (18). To generate CD8 anti-Vβ hybridomas, B10.PL mice were injected intravenously with 3 × 106 antigen-activated, irradiated (3,000 R), 1AE10 cells (CD4+Vβ8+). Ten days later, a CD8-enriched population was prepared from the spleen by antibody and magnetic bead depletion of B cells and CD4+ cells. The resultant population was incubated with irradiated antigen-activated 1AE10 cells and irradiated syngeneic spleen cells. Interleukin 2 (10 units/ml) was added at 48 hr, and CD4+ cells present in the culture were removed again by antibody and magnetic bead separation. Cells were fused on the fourth day of culture with the CD8+, β-galactosidase-inducible fusion partner BWZ.36 (19). Hybridomas were assayed by incubating 5 × 104 hybridoma cells with an equal number of stimulator cells for 18 hr, followed by fixation and incubation with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (19) and direct visualization of blue cells by microscopy. Alternatively, the stimulated cultures were incubated with chlorophenol red β-galactopyranoside (CPRG; Calbiochem) as described (19) and absorbance at 570 nm minus absorbance at 650 nm was determined by using a 96-well plate reader (Molecular Devices, Menlo Park, CA).
RESULTS AND DISCUSSION
TCV Can Generate TCR Vβ-Specific CD8+ T Cells.
To determine whether TCV induces Vβ-specific CD8+ T cells, AKR mice were vaccinated with purified, activated syngeneic CD4+Vβ8+ T cells. Seven to 10 days later, CD8+ T cells were isolated from the animals, retriggered in vitro with activated CD4+Vβ8+ T cells, and tested for their TCR Vβ specificity. As a control, we also generated a TCR Vβ8-specific CD8+ line from SEB-treated animals as previously described. As shown in Table 1, the CD8+ lines from the TCV animals and from the SEB-treated animals have similar specificities. Thus, hybridoma cells and T cell transfectants (H-2k, Qa-1b) expressing Vβ8 TCRs are efficiently lysed by the AKR (H-2k, Qa-1b) CD8+ T cells, whereas targets expressing other Vβ TCRs, including Vβ6, Vβ10, Vβ14, and Vβ15 are not lysed. For example, the AKR-derived 4G4 T cell line, which does not express TCRβ (17), is not lysed by the anti-Vβ8 CD8+ T cells. In contrast, 4G4 cells transfected with cDNA encoding Vβ8 TCRs but not Vβ10 are lysed by the anti-Vβ8 CD8+ T cells. Moreover, one of the 4G4 Vβ8.2 transfectants, 4G4–6B5a, is efficiently lysed, but a subclone, 4G4–6B5b, that spontaneously lost expression of the Vβ8 transgene (as confirmed by RT-PCR) is not killed. Thus, CD8+ T cells isolated from both T cell-vaccinated and SEB-primed mice kill targets in a Vβ-specific manner. Taken together, these data are consistent with the model that TCR Vβ8-specific CD8+ T cells isolated from SEB-primed mice are induced to differentiate in vivo by SEB-activated CD4+ Vβ8+ T cells independent of SEB.
Co-expression of Murine Qa-1 and a Relevant TCRβ Chain Is Necessary and Sufficient for Recognition by Murine CD8+ Anti-Vβ8 CTL Induced During TCV.
We had previously shown that the TCR Vβ8-specific CD8+ CTL isolated from SEB-primed mice were inhibited by allele-specific anti-Qa-1 antisera and not by anti-MHC class I-a antibodies. To more precisely investigate the TCR Vβ and Qa-1 specificity of the CD8+ T cells induced during TCV, we asked whether target cell co-expression of murine Qa-1 and a relevant TCRβ chain is sufficient for recognition or whether other murine T cell surface molecules are required. A human B cell line (C1R) that had been stably transfected with a murine Qa-1b gene and that expresses high levels of Qa-1b on its cell surface (J1) was used for further study. The J1 cells previously had been shown to serve as targets of Qa-1-restricted CTL specific for a peptide derived from the leader of murine MHC class I-a molecules (20). For our studies the J1 cells were further transfected with either the allospecific Vβ8.2 (J1/Vβ8) or Vβ6 TCR cDNA (J1/Vβ6). The parental line, C1R, was also transfected with the same Vβ8 TCR cDNA (C1R/Vβ8) and used as control. Transfectants were screened by Northern blot analysis of TCRβ mRNA expression and cloned by limiting dilution to derive target cell populations expressing both TCRβ and Qa-1. The clones were further assayed by Vβ-specific RT-PCR to confirm their Vβ expression (Fig. 1) and used in the following set of experiments. Putative Vβ-specific CD8+ CTL were isolated from mice previously vaccinated with activated CD4+Vβ8+ T cells. These CD8+ T cells were then retriggered in vitro and assayed for CTL specificity on the C1R transfectants. The results of multiple experiments are depicted in Table 2. The CD8+ T effector cells did not kill C1R (Qa-1−, Vβ8−), C1R/Vβ8 (Qa-1−, Vβ8+, Vβ6−), J1 (Qa-1b+, Vβ8−, Vβ6−), or J1/Vβ6 (Qa-1b+, Vβ8−, Vβ6+) cells. In contrast, the same CD8+ effector T cells efficiently killed the target cells expressing both murine Qa-1b and Vβ8. Taken together, these results demonstrate that co-expression of murine Qa-1b and Vβ8 cDNAs is sufficient for human B cells to serve as targets for the murine CD8+ anti-Vβ8 CTL induced during TCV. Because these target B cells do not express cell surface TCRs, it is clear that the Vβ8 sequences do not need to be expressed on the cell surface as part of a mature TCRβ polypeptide. In this regard, it is well known that MHC class I molecules can efficiently present processed intracellular protein antigens. Moreover, it is also known that Qa-1 molecules can present foreign antigens (21) as well as self MHC class I-a peptides (20). In light of these considerations, our results are most consistent with the idea that CD8+ T cells generated by TCV recognize TCR Vβ peptide/Qa-1 complexes expressed on target T cells.
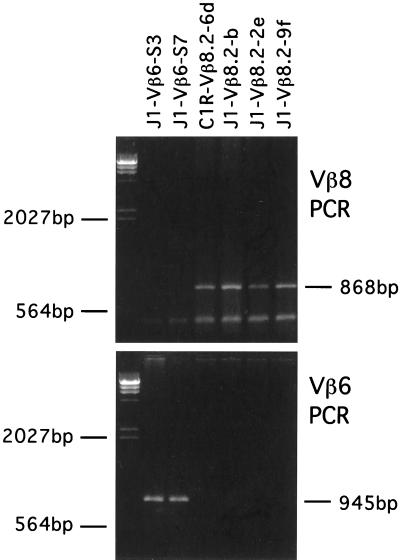
Figure 1.
RT-PCR analysis of Vβ8.2 and Vβ6 C1R and J1 transfectants. PCR products were analyzed on a 1% agarose gel and stained with ethidium bromide.
The Generation of CD8+ T Cell Hybridoma Clones from TCV Mice That Recognize Targets in a Vβ-Specific Qa-1-Restricted Fashion.
The specificity of the CD8+ CTL populations isolated from either T cell-vaccinated or SEB-primed mice could, in principle, reflect the effector functions of distinct CD8+ T clones. To address this issue and because immortalized CD8+ anti-Vβ8 clones would greatly facilitate the analysis of the fine specificity of these cells, we generated CD8+ T cell hybridoma clones from T cell-vaccinated mice. To this end, B10.PL mice (H-2u, Qa-1a) were vaccinated with the syngeneic, MBP-specific, CD4+ Vβ8+ T cell clone 1AE10. CD8+ T cells from these vaccinated animals were restimulated in vitro with the same CD4+ T cell and fused with the αβ− BW5147 fusion partner that had been transfected with both the CD8α gene and a variant of the lacZ gene under control of a nuclear factor of activated T cells (NFAT)-inducible promoter (19). Hybridomas were assayed by incubating hybridoma cells with stimulator cells for 18 hr, followed either by fixation and incubation with X-Gal and direct visualization of blue cells by microscopy or by colorimetry (see Materials and Methods). This assay system relies solely on TCR triggering and is less dependent on the variety of nonspecific surface interactions involved in CTL assays. Thus, hybridomas were screened colorimetrically for the ability to respond to a Qa-1a+ CD4+Vβ8+ clone and not to a Qa-1a+ CD4+Vβ6+ clone, and two putative Vβ-specific hybridomas were derived. Their specificities are shown in Table 3. The first hybridoma, 21–5A9, is stimulated by the inducing clone, 1AE10, as well as other Vβ8+ antigen or superantigen-specific, syngeneic T cells but is not stimulated or is only weakly stimulated by four different syngeneic Vβ6+ T cells. The second hybridoma, 176–5F7, is similarly stimulated by three of the Vβ8+ clones. However, it does not respond to a fourth Vβ8+ clone, 1AC3, and it strongly crossreacts with one of the four Vβ6 clones, 3AG2. Despite these differences, both hybridomas can be triggered by the LBRM T cell lymphoma (Vβ8.2, H-2k, Qa-1a) but not by 4G4–6B5a (Vβ8.2, H-2k, Qa-1b) or J1/Vβ8 (Vβ8.2, Qa-1b). Furthermore, as shown in Table 4, the reactivity of both hybridomas is inhibited by allele-specific anti-Qa-1a sera but not by anti-Qa-1b sera or by mAbs to framework regions of MHC class I-a (M1/42). We emphasize that the anti-Qa-1b antisera used was capable of blocking TCR Vβ8-specific Qa-1b-restricted cytotoxicity and that M1/42 efficiently blocked MHC class 1-a allospecific CTL (13). Thus, the first T hybridoma clone (21–5A9) reacts with target T cells expressing TCR Vβ8 in a Qa-1-restricted manner and precisely reflects the specificity of the splenic CTL described in detail in this report. This hybridoma demonstrates at a clonal level that Qa-1-restricted, Vβ-specific CD8 T cells are generated by TCV. The second T cell hybridoma (176–5F7) has a more complex specificity that is not completely understood at present. Nonetheless, like the first hybridoma, it also distinguishes between syngeneic activated CD4+ T cell clones in a Qa-1-restricted fashion.
Table 4.
Antibody blocking of the CD8 anti-Vβ8 T hybridoma responses
| Response to stimulators
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LBRM positive control | 4G4Vβ8 negative control | LBRM with blocking antibody
|
1AE10 positive control | 3AF9 negative control | 1AE10 with blocking antibody
|
||||||||||||
| Exp. | T cell hybridoma | Anti-Qa-1a | Anti-Qa-1b | M1/42 | Rat Ig | 16-1-2N | Y3P | AntiQa-1a | Anti-Qa-1b | M1/42 | Rat Ig | 3-83P | Y3P | ||||
| 1 | 21-5A9 | 0.618 | 0.051 | 0.023 | 0.637 | 0.637 | 0.660 | 0.502 | 0.629 | 0.389 | 0.004 | 0.160 | 0.406 | 0.314 | 0.411 | 0.294 | 0.382 |
| (—) | (—) | (99.6)* | (0) | (0) | (0) | (18.7) | (0) | (—) | (—) | (58.9) | (0) | (19.3) | (0) | (24.2) | (0) | ||
| 2 | 21-5A9 | 0.751 | 0.037 | 0.014 | 0.758 | 0.774 | 0.790 | 0.636 | 0.759 | 0.435 | NT | 0.206 | 0.552 | 0.495 | 0.558 | 0.444 | 0.547 |
| (—) | (—) | (98.1) | (0) | (0) | (0) | (15.3) | (0) | (—) | (52.6) | (0) | (0) | (0) | (2.1) | (0) | |||
| 3 | 176-5F7 | 0.781 | 0.058 | 0.055 | 0.812 | 0.830 | 0.857 | 0.656 | 0.774 | 0.375 | NT | 0.032 | 0.267 | 0.339 | 0.344 | 0.279 | 0.547 |
| (—) | (—) | (92.9) | (0) | (0) | (0) | (16.0) | (4.7) | (—) | (91.5) | (28.8) | (9.6) | (8.3) | (25.6) | (0) | |||
Anti-Qa-1a and Qa-1b antisera (13) were used at a 1:100 final dilution. The mAbs (M1/42, rat IgG anti-mouse MHC class I-a; 16-1-2N, mouse IgG2a anti-H-2KkDk; 3-83P, mouse IgG2a, anti-H-2k, crossreactive with H-2u; Y3P, control mouse IgG2a anti-I-Ab) and purified rat Ig were used at a final concentration of 20 μg/ml except for the 16-1-2N antibody, which was used as supernatant at 1:4 final dilution. Results are expressed as means of triplicates as in Table 3. NT, not tested.
Anti-Qa-1a antiserum did not inhibit phorbol 12,13-dibutyrate/ionomycin stimulation of hybridoma cells. Percent inhibition is indicated in parentheses where applicable and was calculated as [(positive control − experimental)/positive control] × 100%.
It is of interest that a population of CD8+ T cells can be generated that specifically recognize TCR Vβ peptide/Qa-1 structures expressed on the surface of autologous CD4+ T cells. The concept that CD8+ T cells regulate immune responses and that they are specifically induced by and in turn suppress activated CD4+ T cells was proposed more than a decade ago (22–25). Both antigen- specific and nonspecific mechanisms have been proposed whereby CD8+ T cells regulate CD4+ T cells (15, 23, 26–29). Antigen-specific regulation effected by CD4–CD8 T cell interactions could be accounted for, in principle, by postulating that the regulatory CD8+ T cells are specifically triggered by the same protein antigen that activates the CD4+ T cells. The activated CD8+ T cells would then suppress or inactivate CD4+ cells in the same microenvironment by either cell contact or cytokine-mediated processes. Alternatively, TCR-related structures such as TCR-derived peptide/MHC complexes expressed on antigen-activated CD4+ T cells may represent one set of target structures that induce and are recognized by the regulatory CD8+ T cells. Evidence in support of this idea has been generated by a number of investigators (3, 6–9, 29–32). The general idea that antigen receptors of immunocompetent lymphocytes serve as regulatory elements in the control of immune responses was originally proposed by Jerne (33). Our studies show that at least one type of interaction between CD8+ T cells and CD4+ T cells is mediated by CD8+ T cell recognition of TCR Vβ structures associated with Qa-1 molecules on the surface of activated CD4+ T cells. This is the major CTL specificity identified in polyclonal CD8+ T cell lines developed after TCV and is confirmed by the hybridoma clone 21–5A9. It is of interest that Qa-1 molecules, unlike conventional MHC class I-a molecules, are not expressed on resting T cells but are induced following antigen activation (34). We propose that after T cell activation by antigen, TCR Vβ peptide(s) bind to Qa-1 molecules intracellularly and are then expressed on the surface of activated CD4+ T cells to function both as inducers of TCR Vβ-specific CD8+ T cells and as targets for down-regulation. Resting T cells that do not express Qa-1/Vβ structures are spared the suppressive and/or potentially injurious effects of the regulatory CD8+ T cells. In this way TCV may function to control autoimmunity by selectively down-regulating activated autoimmune CD4+ T cell clones.
Acknowledgments
We thank N. Shastri for kindly sending us the β-galactosidase CD8+ αβ− BW5147 T cell hybridoma fusion partner and Jun-Tao Liu and Bin Yu for their excellent technical assistance. This work was supported by National Institutes of Health Grants AI39630 to H.J., AI39635 and AI39790 to L.C., and AI34930 and AI37942 to J.F.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TCV, T cell vaccination; EAE, experimental allergic encephalomyelitis; TCR, T cell receptor; MBP, myelin basic protein; SEB, staphyloccal enterotoxin B; CTL, cytotoxic T lymphocytes; β2m, β2-microglobulin; MHC, major histocompatibility complex; RT-PCR, reverse transcription–PCR; FACS, fluorescence-activated cell sorting; X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside.
References
- 1.Ben-Nun A, Wekerle H, Cohen I R. Nature (London) 1981;292:60–61. doi: 10.1038/292060a0. [DOI] [PubMed] [Google Scholar]
- 2.Cohen I R, Weiner H L. Immunol Today. 1988;9:332–335. doi: 10.1016/0167-5699(88)91330-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Medaer R, Stinissen P, Hafler D, Raus J. Science. 1993;261:1451–1454. doi: 10.1126/science.7690157. [DOI] [PubMed] [Google Scholar]
- 4.van Laar J M, de Vries R R, Breedveld F C. Clin Exp Rheumatol. 1993;11:S59–S62. [PubMed] [Google Scholar]
- 5.Mor F, Cohen I R. Int Arch Allergy Immunol. 1995;108:345–349. doi: 10.1159/000237180. [DOI] [PubMed] [Google Scholar]
- 6.Sun D, Qin Y, Chluba J, Epplen J T, Wekerle H. Nature (London) 1988;332:843–845. doi: 10.1038/332843a0. [DOI] [PubMed] [Google Scholar]
- 7.Offner H, Hashim G A, Vandenbark A A. Science. 1991;251:430–432. doi: 10.1126/science.1989076. [DOI] [PubMed] [Google Scholar]
- 8.Howell M D, Winters S T, Olee T, Powell H C, Carlo D J, Brostoff S W. Science. 1989;246:668–670. doi: 10.1126/science.2814489. [DOI] [PubMed] [Google Scholar]
- 9.Vandenbark A A, Hashim G, Offner H. Nature (London) 1989;341:541–544. doi: 10.1038/341541a0. [DOI] [PubMed] [Google Scholar]
- 10.Zamvil S, Nelson P, Trotter J, Mitchell D, Knobler R, Fritz R, Steinman L. Nature (London) 1985;317:355–358. doi: 10.1038/317355a0. [DOI] [PubMed] [Google Scholar]
- 11.Acha-Orbea H, Mitchell D J, Timmermann L, Wraith D C, Tausch G S, Waldor M K, Zamvil S S, McDevitt H O, Steinman L. Cell. 1988;54:263–273. doi: 10.1016/0092-8674(88)90558-2. [DOI] [PubMed] [Google Scholar]
- 12.Urban J L, Kumar V, Kono D H, Gomez C, Horvath S J, Clayton J, Ando D G, Sercarz E E, Hood L. Cell. 1988;54:577–592. doi: 10.1016/0092-8674(88)90079-7. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Ware R, Stall A, Flaherty L, Chess L, Pernis B. Immunity. 1995;2:185–194. doi: 10.1016/s1074-7613(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 14.Boyse E A, Flaherty L, Stockert E, Old L J. Transplantation. 1972;13:431–432. doi: 10.1097/00007890-197204000-00013. [DOI] [PubMed] [Google Scholar]
- 15.Eardley D D, Hugenberger J, McVay-Boudreau L, Shen F W, Gershon R K, Cantor H. J Exp Med. 1978;147:1106–1115. doi: 10.1084/jem.147.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanton T H, Boyse E A. Immunogenetics. 1979;3:525–531. [Google Scholar]
- 17.Digiusto D L, Palmer E. Mol Immunol. 1994;31:695–699. doi: 10.1016/0161-5890(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 18.Berkowitz N, Braunsten N S. J Immunol. 1992;148:309–317. [PubMed] [Google Scholar]
- 19.Sanderson S, Shastri N. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 20.Aldrich C J, DeClousc A, Woods A S, Cotter R J, Woloski M J, Forman J. Cell. 1994;79:649–659. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 21.Bouwer H G A, Seaman M S, Forman J, Hinrichs D J. J Immunol. 1997;159:2795–2801. [PubMed] [Google Scholar]
- 22.Cantor H, Hugenberger J, McVay-Boudreau L, Eardley D D, Kemp J, Shen F W, Gershon R K. J Exp Med. 1978;148:871–877. doi: 10.1084/jem.148.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorf M E, Benacerraf B. Ann Rev Immunol. 1984;2:127–158. doi: 10.1146/annurev.iy.02.040184.001015. [DOI] [PubMed] [Google Scholar]
- 24.Thomas Y, Sosman J, Irigoyen O, Friedman S M, Kung P C, Goldstein G, Chess L. J Immunol. 1980;125:2402–2408. [PubMed] [Google Scholar]
- 25.Green D R, Flood P M, Gershon R K. Annu Rev Immunol. 1983;1:439–463. doi: 10.1146/annurev.iy.01.040183.002255. [DOI] [PubMed] [Google Scholar]
- 26.Bloom B R, Modlin R L, Salgame P. Annu Rev Immunol. 1992;10:453–488. doi: 10.1146/annurev.iy.10.040192.002321. [DOI] [PubMed] [Google Scholar]
- 27.Jandinski J, Cantor H, Tadakuma T, Peavy D L, Pierce C W. J Exp Med. 1976;143:1382–1390. doi: 10.1084/jem.143.6.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas Y, Sosman J, Rogozinski L, Irigoyen O, Kung P C, Goldstein G, Chess L. J Immunol. 1981;126:1948–1951. [PubMed] [Google Scholar]
- 29.Gaur A, Ruberti G, Haspel R, Mayer J P, Fathman C G. Science. 1993;259:91–94. doi: 10.1126/science.8418501. [DOI] [PubMed] [Google Scholar]
- 30.Kumar V, Sercarz E E. J Exp Med. 1993;178:909–916. doi: 10.1084/jem.178.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanda N K, Sercarz E. J Exp Med. 1996;184:1037–1043. doi: 10.1084/jem.184.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang H, Zhang S I, Pernis B. Science. 1992;256:1213–1215. doi: 10.1126/science.256.5060.1213. [DOI] [PubMed] [Google Scholar]
- 33.Jerne N K. Harvey Lect. 1975;70:93–110. [PubMed] [Google Scholar]
- 34.Stanton T H, Carbon S. Immunogenetics. 1982;16:435–444. doi: 10.1007/BF00372102. [DOI] [PubMed] [Google Scholar]