Abstract
ALL1, the human homologue of Drosophila trithorax, is directly involved in human acute leukemias associated with abnormalities at 11q23. Using the differential display method, we isolated a gene that is down-regulated in All1 double-knockout mouse embryonic stem (ES) cells. The gene, designated ARP1 (also termed RIEG, Ptx2, or Otlx2), is a member of a family of homeotic genes containing a short motif shared with several homeobox genes. Using a bacterially synthesized All1 polypeptide encompassing the AT-hook motifs, we identified a 0.5-kb ARP1 DNA fragment that preferentially bound to the polypeptide. Within this DNA, a region of ≈100 bp was protected by the polypeptide from digestion with ExoIII and DNase I. Whole-mount in situ hybridization to early mouse embryos of 9.5–10.5 days indicated a complex pattern of Arp1 expression spatially overlapping with the expression of All1. Although the ARP1 gene is expressed strongly in bone marrow cells, no transcripts were detected in six leukemia cell lines with 11q23 translocations. These results suggest that ARP1 is up-regulated by the All1 protein, possibly through direct interaction with an upstream DNA sequence of the former. The results are also consistent with the suggestion that ALL1 chimeric proteins resulting from 11q23 abnormalities act in a dominant negative fashion.
Abnormalities at 11q23 are common in acute lymphocytic, acute myelomonocytic, acute monocytic, and acute myeloid leukemia, drug-induced secondary leukemias, and myelodisplastic syndrome. We and others have isolated a gene at 11q23 that is involved in these chromosome aberrations (1, 2). The gene was designated ALL1 (also HRX, HTRX, and MLL). It exhibits strong homology to the Drosophila trithorax (trx) gene (3), particularly in the region encoding multiple zinc fingers (PHD fingers), as well as in the C-terminal (SET) domain. In addition, the All1 protein contains a cluster of “AT-hook” motifs involved in DNA binding (2), a region with homology to DNA-methyltransferase (4), and a transcriptional activation domain (5, 6). In Drosophila, trx maintains the expression of homeotic genes of the bithorax and Antennapedia complexes (BX-C and ANT-C), which direct body segmentation during Drosophila embryonic development (7, 8). Recent analysis of All1 knockout mice suggests that ALL1 plays a role in mammals that is similar to the role of trx (9).
Antibodies directed against the trx protein detected multiple, specific binding sites on polytene chromosomes of larvae salivary glands (10, 11). The sites included the regulatory region of a known trx target gene, fork head, and mutations of trx caused reduced expression of this target gene (10). These findings suggested that trx exerts its effects by binding directly or indirectly to specific DNA sequences in target genes. Some of the targets for ALL1 are likely to be the mammalian Hox genes. This notion is supported by the demonstration (9) of spatially altered expression of Hoxa 7 and Hoxc 9 in All1 +/− knockout mice.
In acute leukemias with aberrations at 11q23, the ALL1 gene recombines with many partner genes from a variety of chromosomes (12, 13). In addition, in a significant number of AML patients, ALL1 undergoes partial tandem duplication (14, 15). Finally, several patients with acute lymphocyte leukemia show an identical deletion spanning ALL1 exon 8 and flanking intronic sequences (16). The partner genes are a diverse lot. Some are transcription factors (17, 18), and others are involved in signal transduction (19). AF9 and ENL (20), as well as AF10 and AF17 (17), are highly homologous. Because of these diverse features, the underlying mechanism for ALL1 leukemogenicity is still unknown. It is conceivable that ALL1 triggers leukemia through its effects on target gene(s) involved in hematopoiesis. We have proposed previously (14, 21) that ALL1 rearrangements result in its loss of function and that the chimeric proteins exert a dominant negative effect on the normal All1 protein encoded by the intact allele present in the leukemic cells.
We recently have generated All1 knockout embryonic stem (ES) cell lines and investigated their ability to differentiate along hematopoietic lineages after withdrawal of leukemia inhibitory factor (LIF) (22). Whereas wild-type ES cells formed lineage-restricted mature colonies, +/− and −/− knockout ES cells developed, at high frequency, immature and/or “multiphenotypic” colonies. This phenotype is similar to that of acute leukemia cells with 11q23 abnormalities. These results suggested that ALL1 gene rearrangements in humans and All1 knockout in mice result in similar abnormalities in hematopoietic cells. We reasoned that the All1 −/− knockout ES cells might fail to express transcripts of genes induced by All1. Applying the differential display methodology (23), we isolated and characterized a gene that is underexpressed in All1 knockout ES cells. The properties of this gene, ARP1, support the notion that it is a target for ALL1.
MATERIALS AND METHODS
Cell Lines and Differential Display.
ES AB2.1 (wild type), 48all1+/−, 48Sall1−/−, and 48Nall1−/− cells were maintained in gelatinized tissue culture dishes in DMEM supplemented with 15% heat-inactivated fetal bovine serum, 2 mM glutamine, 0.1 mM 2-mercaptoethanol, and 1,000 units/ml of recombinant leukemia inhibitory factor (GIBCO/BRL).
Using Genhunter RNA image kit (Genhunter, Nashville, TN), we performed the differential display comparing total RNAs of AB2.1, 48Sall1−/−, and 48Nall1−/− cells.
Binding-Site Selection, Exonuclease III Protection, and DNase I Footprint Analysis.
Two regions of ALL1 cDNA, encoding the AT-hooks domain (ATH) spanning nucleotides 331–948 and DNA methyltransferase (MTase) homologous region encompassing nucleotide 3346–3753, were cloned into the pET-23d vector (Novagen), which provides a stretch of six histidines. The BL21 (DE3) bacteria strain (Novagen) was used for expression. The 25-kD ATH and 18-kD MTase peptides were affinity-purified by Ni-NTA resin (Qiagen) and dialysed against buffer D (20 mM Hepes, pH7.9/0.1 M KCl/20% glycerol/0.2 mM EDTA/1 mM DTT). Aliquots of 1 μg each of the ATH or MTase peptides were mixed either with 1 μg of BSA or 1 μg of HeLa cell nuclear extract in 10 μl of buffer D, spotted on 1 cm2 of nitrocellulose filter (BA85, Schleicher & Schuell) and air-dried for 10 min. The filter was subsequently blocked for 30 min at 4°C with 100 μl of binding buffer (25 mM Hepes, pH 7.9/40 mM KCl/5 mM MgCl2/1 mM DTT) containing 0.5% nonfat milk and 10 μg of poly (dI-dC) and washed with binding buffer containing 0.25% nonfat milk. Ten micrograms of ARP1 genomic cosmid clone 2 was completely digested with Sau3AI and ligated to a Sau3AI adapter formed by annealing the 24-mer 5′-GATCAGAAGCTTGAATTCGAGCAG-3′ and 20-mer 5′-CTGCTCGAATTCAAGCTTCT-3′ oligonucleotides. After incubation for 2 hr at 4°C in 100 μl of binding buffer, containing 500 ng of the segmented DNA preparation, the filter was washed three times with binding buffer including 0.25% milk and once with binding buffer and transferred into an Eppendorf tube. DNA was eluted by incubation with a buffer containing 20 mM Tris, pH 8.0, and 1 M KCl for 10 min on ice with occasional vortexing. The elute was diluted with water and precipitated with isopropanol in the presence of glycogen, and the DNA pellet was resuspended in 20 μl water. One milliliter aliquot of the DNA was amplified by 25 cycles of PCR, using the 20-mer adapter oligonucleotide, and the PCR product was resolved on a 2% MetaPhor XR agarose (FMC) and cloned into pBluescript SK(−) (Stratagene).
Exonuclease III (ExoIII) protection assay was carried out by the method of Wu (24). DNase I footprint analysis was carried out by using the Sure track DNase footprint kit (Pharmacia).
Whole-Mount in Situ Hybridization.
Embryos were obtained from natural matings of FVB/N or C57B/6J mice. The time point of vaginal plug observation was designated at 0.5 day postcoitum, and embryonic stages are listed as days postcoitum (E followed by the number of the day). Hybridization experiments were performed as described (25). Digoxigenin-labeled RNAs were prepared by using an RNA labeling kit (Boehringer Mannheim) with ARP1 cDNA. Anti-digoxigenin antibodies (Fab fragment) conjugated with alkaline phosphatase were purchased from Boehringer Mannheim. Some E10.5 embryos were consequently incubated in isopropanol, isopropanol-Paraplast (1:1), and Paraplast (58°C) for several hours and finally embedded in Paraplast Plus (Oxford Scientific, St. Louis, MO). Sections (20–25 μm) were cut in sagittal and transverse planes, mounted in slides, and photographed under Nomarski optics using an Olympus Vanox microscope.
RESULTS AND DISCUSSION
Identification of a Gene Down-Regulated in All1 Knockout ES Cells.
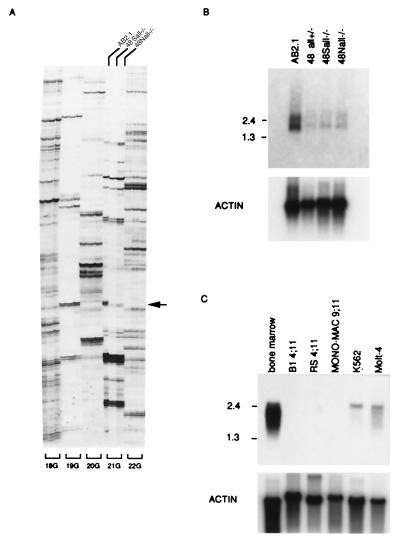
Recently we established All1 +/− and −/− knockout ES cell lines, 48all1+/−, 48Sall1−/−, and 48Nall1−/−, from parental AB2.1 ES cells and showed that these ES cells are deficient in hematopoietic differentiation in an in vitro colony-formation assay (22). To identify genes down-regulated in the All1 knockout ES cells, we examined the expression of approximately 15,000 genes in these, as well as in parental ES cells, by using a differential display kit. We carried out 240 primer-combination arbitrary reverse transcription–PCRs (RT-PCRs) and compared the bands derived from AB2.1 RNA to those derived from RNAs of 48Sall1−/− and 48Nall1−/− cells. As expected, the great majority of bands displayed similar intensity in the patterns of the three cell lines. The intensity of several bands was reduced in All1 −/− knockout ES cells. However, these differences were proved to be false or minor by subsequent RT-PCR or Northern blot analysis. Nevertheless, one band (set 21G composed of the primer 5′-anchor dN6-TCTCTGG-3′ and dTG-5′-TTTTTTTTTTTG-3′) was reproducibly reduced in All1 −/− knockout ES cells (indicated by arrow in Fig. 1A, 21G; lanes 48Sall1−/− and 48Nall1−/−). DNA corresponding to this band was excised from the gel, reamplified by PCR using the same primers, cloned, and sequenced. Using this DNA of 161 bp as a probe in Northern blot analysis, two mRNA species (1.4 and 2.5 kb) were observed in parental AB2.1 cells. These transcripts were considerably less abundant in 48Sall1−/−, 48Nall1−/− cells, as well as in 48all+/− cells (Fig. 1B). Sequence of this DNA indicate no relationships to known genes present in the database, but exhibited homology with the human anonymous sequences—expressed sequence tags (ESTs) gb/T64905 and gb/R77812. By PCR amplification, we obtained a 300-bp fragment of the human homologue. Using the 161- and 300-bp DNA fragments as probes, the expression of the gene was examined in mouse and human adult tissues. Transcripts were detected in mouse skeletal muscle, mouse kidney, human skeletal muscle, human placenta (not shown), and human bone marrow (Fig. 1C). In contrast, other tissues showed little expression. In analysis of polyadenylated RNAs from leukemic cell lines in which ALL1 is rearranged, ARP1 transcripts were not detected in RS4;11 [with t(4:11)], B1B [with t(4:11)], Mono-Mac6 [with t(9:11)] (Fig. 1C), or MV4;11[with t(4:11)], THP-1 [with t(9;11)], and ML-2 [with t(6;11)] (data not shown). In contrast, ARP1 transcripts are detectable in human leukemia cell lines and bone marrow cells without 11q23 gene rearrangements (Fig. 1C).
Figure 1.
Identification of a transcript down-regulated in All1 knockout ES cells. (A) Differential display. 18G, 19G, 20G, 21G, and 22G indicate specific primer sets. The three lanes for each set correspond to the ES cell line from which the RNA was amplified: AB2.1 wild type (left lane), 48Sall−/− (center lane), and 48Nall−/− (right lane). Arrow points to the band apparent only in the wild-type cells. (B) Northern analysis of Arp1 expression in ES cell lines. Two micrograms of polyadenylated RNA were electrophoretically resolved and examined for hybridization to the 161-bp probe (see text). (C) Northern analysis of ARP1 expression in human leukemia cell lines. Analysis as in B. The 300-bp human cDNA (see text) was utilized as a probe. Numbers on the left correspond to size markers (kb).
Isolation and Characterization of Mouse and Human Full Length ARP1 cDNAs.
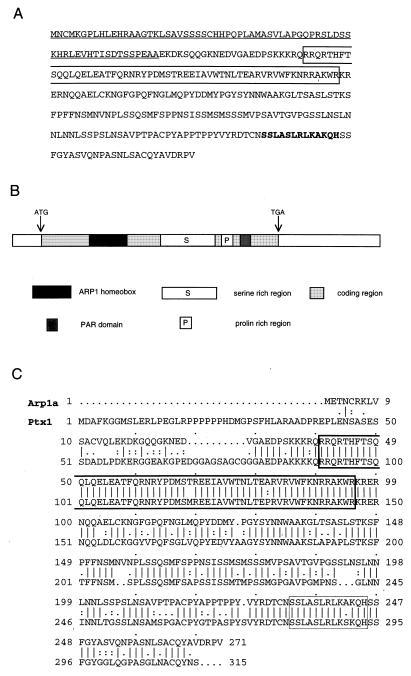
To isolate the full-length mouse cDNA, we prepared a cDNA library from the mouse AB2.1 wild-type ES cell line. One million independent colonies were screened, and 17 positive clones were isolated. All clones were sequenced, and 10 appeared to contain full-length cDNA. We designated the corresponding gene ARP1 (ALL1 responsive gene 1). Physical mapping and sequence analysis of the cDNAs indicated two species (ARP1a and ARP1c) that varied at 5′ segments including the initiation codon ATG (see later in text). The predicted ORFs for the murine Arp1a and Arp1c encoded 271 and 324 aa, respectively. The human 300-bp DNA fragment was used as a probe to clone the human homologue. Two of the cDNA species (ARP1a and ARP1c) corresponded to the mouse species, and a third (ARP1b) contained additional sequences derived from an internal, alternatively spliced exon (see later in text). Comparison of the sequences of human and murine ARP1a and ARP1c cDNAs showed striking conservation, 90 and 89% nucleotide identity and 99 and 97% amino acid identity, respectively. The database search indicated that ARP1a/Arp1a and ARP1b are likely to be identical to Ptx2a (26)/Otlx2 (27)/RIEG (28) and Ptx2b (26), respectively. However, the N-terminal domain of ARP1c did not correspond to any known gene. Amino acid sequence and domains of ARP1c are shown in Fig. 2A and B, respectively. ARP1 also has striking homology with Ptx1/Potx, which is thought to be a pituitary cell fate determinant (29, 30). The amino acid sequences of the two genes are 82% similar along the whole coding sequence, increasing to 86% without the N-terminal regions and increasing further to 98% when considering only homeodomains (Fig. 2C). The predicted Arp1 protein contains a previously unknown motif (Fig. 2), which we also found at the C terminus of some homeobox genes that belong to the “Phox-Aristaless” group (31). We named this motif PAR (Phox-ARP1-Aristaless) motif. This motif was also noticed by Semina et al. (28).
Figure 2.
Amino acids sequence, domains, and homology of ARP1. (A) Amino acids sequence of ARP1c. Underline indicates unique domain of ARP1c; box indicates homeodomain. Bold letters are short motifs mentioned by Semina et al. (28). (B) Schema of domain of ARP1c. (C) Similarity between the Arp1a and Ptx1 proteins. The sequences were aligned by using the algorithm bestfit.
RIEG and Ptx1/Potx were described as bicoid-related homeobox transcription factors (28–30) because of lysine residue at position 9 of the third helix according to the classification of Burglin (32). We will further specify the ARP1/RIEG/Ptx2/Otlx2 and Ptx1/Potx proteins as members of a distinctive family within the “Ortoid-Ortodentical” class of homeobox genes based on the definition of “group-specific patterns” and “intergroup-specific motifs” (31).
Chromosome Location and Genomic Structure of ARP1.
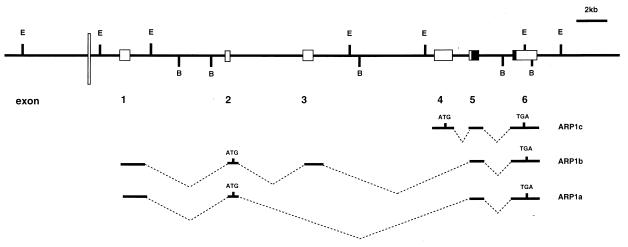
Using a fragment of the ARP1 gene as a probe in Southern analysis of DNAs from somatic cell hybrids and applying in situ hybridization, we mapped human ARP1 to chromosome region 4q24–25. For analysis of the genomic structure, cosmid clones 2, 4, and 5 were isolated by screening 5 × 105 colonies of human genomic cosmid library (CLONTECH). Physical mapping and sequencing of these cosmid clones enabled us to determine the genomic structure of the human ARP1 gene (Fig. 3). The gene spans 30 kb. Alternative splicing gives rise to the three transcripts (APR1a, ARP1b, and ARP1c). All transcripts include sequences of exon 5, which encodes the most 5′ region of the homeodomain, and of exon 6, encoding the 3′ 11 aa of the homeodomain.
Figure 3.
Genomic structure of human ARP1. Exons are depicted by lightly shaded boxes. Solid box indicates ARP1 homeodomain. Box sizes do not correspond to precise length of exons. The All1 AT-hook binding site is noted by thin, white bar. E and B correspond to EcoRI and BamHI sites, respectively. Exons included within ARP1 transcripts are shown in the middle.
The AT-Hooks Domain of the All1 Protein Binds to a Sequence Positioned Upstream of the ARP1 Transcription Start Site.
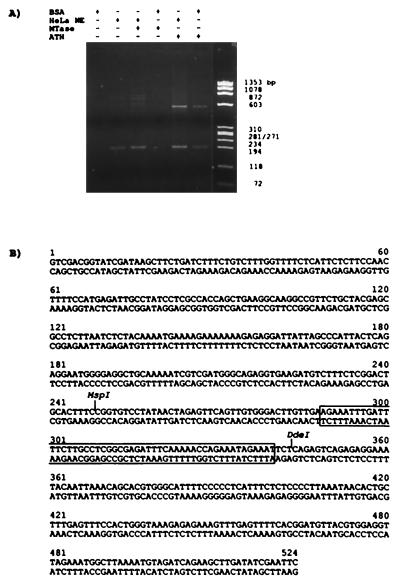
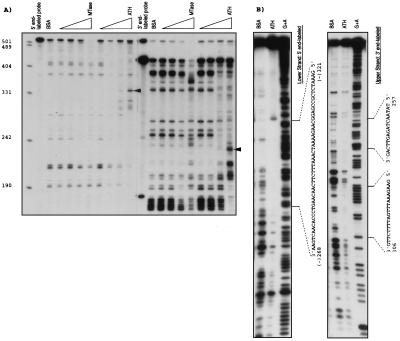
To investigate the possibility that ALL1 regulates ARP1 expression through direct protein–DNA association, we searched for All1-binding sites within the human ARP1 locus. “AT-hook” domains of several proteins were previously shown to be involved in direct binding to DNA (33, 34). Therefore, we expressed in E. coli All1 ATH, tagged at the C terminus with six histidines, and inserted it into the PET bacterial expression vector (Novagen). As a control, we expressed a similar-sized All1 polypeptide spanning the MTase region. Cosmid 2, containing the entire ARP1 gene, was digested with Sau3AI enzyme, and the fragments were ligated to PCR adapters. These fragments were subjected to binding-site selection (BSS) on nitrocellulose filters spotted with each of the two bacteria-synthesized All1 polypeptides in the presence or absence of either BSA or HeLa cell nuclear extracts. The bound DNAs were eluted, PCR-amplified, and analyzed electrophoretically (Fig. 4A). A unique 0.5-kb DNA fragment was amplified after selection with the ATH polypeptide in the presence of BSA or HeLa cell nuclear extract. This fragment was not selected with the MTase polypeptide, but could be selected with ATH alone in a second cycle of binding and PCR amplification. A fragment of 0.19 kb that was selected by HeLa cell nuclear extract in the presence or absence of ATH or MTase polypeptides was not studied further. The selected 0.5-kb fragment containing the ATH peptide-binding site was subcloned and its sequence was determined (Fig. 4B). This DNA was found to reside within a 2-kb EcoRI fragment located at the 5′ end of cosmid clone 2, 1.7 kb upstream of the 5′ terminus of ARP1a and ARP1b cDNAs.
Figure 4.
Binding-site selection of ARP1 genomic fragments. (A) Bacterially synthesized All1 polypeptides spanning the “AT-hooks” domain (ATH) or the region homologous to methyltransferases (MTase), together or without BSA or HeLa nuclear extract (NE), were reacted with the fragmented ARP1 genomic DNA. Bound fragments were PCR-amplified and identified by agarose gel electrophoresis. (B) The sequence of the ATH-selected DNA fragment. Sequence protected by ATH from ExoIII digestive is boxed.
To narrow the binding site to ATH within the 524-bp DNA, an ExoIII protection experiment was carried out. As shown in Fig. 5A, the ATH peptide protected a 340-bp fragment of the 5′ end-labeled DNA (Fig. 5A Left) and 235-bp fragment of the 3′ end-labeled DNA (Fig. 5A Right) in a dose-dependent manner; no protection was found by using the MTase peptide. This indicated that the ATH-binding site spanned nucleotides 289–340 (Fig. 4B, open box). Next, a 220-bp DNA composed of nucleotides 202–421 (includes the ExoIII-protected region) was subcloned and used in a DNase I footprinting study. Nucleotides 268–321 of the lower strand (numbering starts at nucleotide 1 in Fig. 5B) and nucleotides 242–257 and 306–325 of the upper strand were consistently protected (Fig. 5B). Thus, the results of two types of assays indicated that All1 “AT hooks” polypeptide binds ARP1 DNA in a region of around 100 bp located ≈1.7 kb upstream of the transcription start site. Utilizing this region (nucleotides 250–344) in electrophoretic mobility shift assay we observed specific binding to a protein complex in nuclear extracts from cultured cells (not shown).
Figure 5.
Fine mapping of ARP1 genomic sequence that binds the ATH polypeptide. (A Left and Right) Protection from ExoIII digestion of ARP1 DNA segment end-labeled at the 5′ and 3′ ends, respectively. Bands protected specifically by increasing amounts of the ATH polypeptide are indicated by arrows. (B) Protection from DNaseI digestion of ARP1 DNA segment labeled at the 5′ or 3′ end.
Expression of Arp1 During Mouse Embryogenesis.
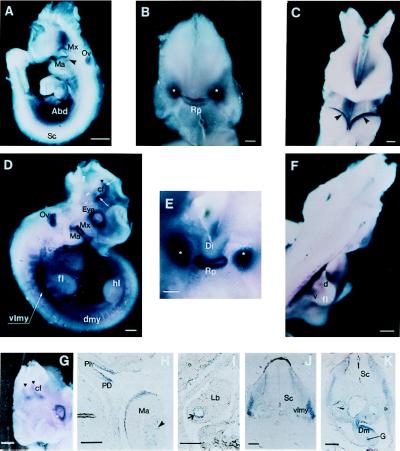
We examined the expression of the Arp1 gene at different stages of mouse embryo development by Northern analysis. No expression was detected at E7.0, whereas we found strong expression at E11, E13, and E15 embryos (not shown). Expression of the Arp1 gene was also analyzed by whole-mount in situ hybridization in day E8.0–E15.5. As shown and mentioned in Fig. 6, we detect Arp1 mRNA in a number of tissues in day E9.5–E10.5. The All1 gene has also been shown to be widely expressed throughout embryonic development in the neural and somitic tissues from E7.0 into adulthood (9). Interestingly, Arp1 mRNA is detected in ectodermal cells in first branchial arch (Fig. 6 A and C), ventrolateral dermomyotome of early somites (Fig. 6 D and F), and neural tissues (Fig. 6 D and G), overlapping the expression of All1.
Figure 6.
Spatial distribution of Arp1 transcripts during mouse development. (A) Lateral view of E9.5 embryo. Hybridizable RNA is observed in the abdominal area (Abd), in surface ectoderm of the maxillary (Mx), and mandibular (Ma) regions of the first branchial arch. Arrowhead points out the mesenchymal component of the mandibular arch. Staining of the optic vesicle (Ov) is because of the nonspecific trapping of colored conjugates as evidenced by reaction with a sense probe. No signal was observed in spinal cord (Sc). (B) Anterior view of the head of E9.5 embryo. Facial part has been removed for better visualization of the signal. Staining is observed in the developing Rathke’s pouch (Rp) and in two bilateral structures (marked by white asterisks), still to be identified. (C) Anterior view of the first branchial arches of E9.5 embryo. Arrowheads point to the sharp bands of Arp1 expression along the surface ectoderm of the mandibular component of the first branchial arch. (D) Lateral view of E10.5 embryo. Expression is detected in a number of new structures: in the dermomyotomal compartments of the somites (dmy), in the presumable migratory myogenic precursors (of the limb muscle) localized in the ventrolateral myotome (vlmy) at the forelimb level. The signal is also seen at the periphery of the eye. Strong staining is observed in the restricted domains of the forebrain (white arrows) at the area of the cranial flexure (cf). Black triangle corresponds to the midbrain/forebrain boundary; fl, forelimb; hl, hindlimb. (E) High magnification of the anterior view of head of E10.5 embryo (facial part has been removed); expression of Arp1 is sharply localized to the Rathke’s pouch. Bilateral staining previously detected becomes stronger and more compact. (F) Dorsolateral view of E10.5 embryo. Arp1 specific signal is seen in presumable ventral (v) and dorsal (d) myoblast cells within the forelimb (fl). (G) Anteriolateral view of E10.5 head showing bilateral distribution of Arp1 signal in the forebrain. (H) Sagittal section of the prestained E10.5 embryo. Expression is stronger in the pars dislis (PD) than in pars intermedia (PI) within the Rathke’s pouch. Expression is seen in the ectoderm and mesenchyme (arrowhead) of the mandibular branch (Ma) of the first branchial arch. (I) Staining was detected on sagittal section in epithelial cells of the bronchi (arrow) within the lung bud (Lb). (J and K) Transverse sections of the E10.5 embryo at the level of the forelimbs. Open triangle points to the myoblast precursor cells migrating into the proximal portion of the forelimb. Staining is also revealed in the mesenchyme of the dorsal mesentery (Dm); G, gut. [Bars = 200 mm (A, D, and F–I), 100 mm (J and K), and 50 mm (B and C).]
ARP1 Is a Target for ALL1.
The ARP1 gene was isolated by virtue of its down-regulation in ES cells, in which both alleles of ALL1 were disrupted by homologous recombination (22). The disruption eliminated ALL1 third exon, which encodes 885 aa, and contains the “AT-hook” motifs. It is conceivable that the deletion of this large and highly conserved segment of the protein disrupted its function. The elimination of the “AT-hooks” domain presumably interferes with the capacity of the protein to bind its DNA targets. In three ES cell lines in which one or two alleles of ALL1 were disrupted, transcription of ARP1 was reduced by more than 10-fold. In addition, in six leukemic cell lines in which ALL1 is rearranged ARP1 is not expressed. This is consistent with the idea (12, 14, 21) that ALL1 rearrangements result in loss of function of the gene and, therefore, in down-regulation of its targets, such as ARP1 (shown in this work) or Hox genes (22). The suggestion that ARP1 is a primary target of ALL1 is based on our findings that the All1 “AT hooks” domain binds in vitro specifically to an ARP1 sequence located upstream to the transcription initiation point and that the expression of All1 coincides with Arp1 expression pattern in several tissues including the dermomyotome. However, this suggestion would have to be supported by the demonstration in vivo, and possibly in vitro, of All1-dependent transcription of a reporter gene linked to the ARP1 sequence. Our preliminary gel-shift experiments show that the sequence binds specifically to a protein complex present in mammalian nuclear extracts (data not shown). It still remains to be shown that the All1 protein is a constituent of this complex.
We have identified a genomic ARP1 sequence to which the All1 AT-hook polypeptide binds in vitro. It was previously shown that AT hooks binding domains may bind to AT-rich sequences in a structure-specific rather than sequence-specific manner (5, 35). However, in selecting a 0.5-kb fragment out of 40 kb, the binding of All1 polypeptide appears to be more specific in our assay. All chimeric proteins produced as a result of ALL1 translocations, as well as All1 proteins resulting from partial tandem duplications, retain both the All1 AT-hook domain and the MTase domain (12, 15), suggesting that these domains are critical for the leukemogenic activity of the proteins. The DNA-binding capacity of the All1 AT-hook polypeptide suggests that the chimeric All1 proteins can bind to DNA targets of the normal All1 protein. However, this binding might be nonproductive.
Potential Role of ARP1 in Oncogenesis.
ALL1 is frequently rearranged in different kinds of acute leukemias, such as acute lymphocytic (ALL), acute myelomonocytic, acute monocytic, and acute myeloid leukemias. Furthermore, leukemic cells of the majority of patients with the t(4;11) chromosomal translocation show both precursor B cell and myeloid markers (36) and are considered to be “biphenotypic.” These features suggest that alteration in expression of the target gene(s) due to ALL1 abnormalities may affect differentiation of multipotential hematopoietic stem cell along the lymphoid and myeloid lineages. Thus, ALL1′s target genes involved in leukemogenesis could play an important role in the differentiation of hematopoietic stem cells to specific cell lineage. Interestingly, ARP1 has striking homology with Ptx1/Potx, a pituitary cell fate determinant that plays a role in the differentiation of pituitary cells. The strong expression of ARP1 in bone marrow cells and in skeletal muscle cells raises the possibility that it is involved in the differentiation of hematopoietic cells (and skeletal muscle cells) and therefore could be a mediator of leukemogenesis resulting from ALL1 alterations. The lack of ARP1 expression in all six leukemia cell lines with 11q23 translocations examined is consistent with this notion and the hypothesis (12, 14, 21) that ALL1 rearrangements result in a loss of function of the gene and therefore in down-regulation of its targets.
While this work was in progress, ARP1a and ARP1b were isolated by others in three different contexts (26–28). The major point in those studies was that ARP1, termed RIEG, or Otlx2, or Ptx2 is expressed in the pituitary gland, in the eye, and in restricted brain tissues. Moreover, the gene was found to be directly involved in Rieger syndrome (28).
Based on binding of the trx protein to many sites on polytene chromosomes (10, 11), we expect ALL1 to regulate a variety of mammalian genes in addition to the Hox loci. ARP1 appears to be the first example of these target genes.
Acknowledgments
We thank Dr. Victor M. Gindilis for helpful discussions and valuable suggestions. We are grateful to Dr. Kay Heubner and Dr. Jim Jaynes for critical reading of the manuscript. We particularly acknowledge the contribution of Drs. Richard R. Schmidt and Kenneth P. Chepenik to the analysis of whole-mount hybridized embryos. These studies were supported by National Cancer Institute Grants CA 39860 and CA 50507, and by grants from the Tobacco Research Council, U.S.–Israel Binational Fund, and the Minerva and Forschheimer Foundations.
ABBREVIATIONS
- ATH
AT-hooks domain
- ES
embryonic stem
- MTase
DNA methyltransferase
Footnotes
References
- 1.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 2.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 3.Mazo A M, Huang D H, Mozer B A, David I B. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Q, Alder H, Nelson K K, Chatterjel D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeleznick-Le N J, Harden A M, Rowley J D. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Lehskowitz S, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingham P W. Nature (London) 1983;306:591–593. doi: 10.1038/306591a0. [DOI] [PubMed] [Google Scholar]
- 8.Kennison J A. Annu Rev Genetics. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 9.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 10.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 11.Chinwalla V, Jane E P, Harte P J. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canaani E, Nowell P C, Croce C M. Adv Cancer Res. 1995;66:213–234. doi: 10.1016/s0065-230x(08)60255-9. [DOI] [PubMed] [Google Scholar]
- 13.Bernard O A, Berger R. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 14.Schichman S A, Caligiuri M A, Gu Y, Strout M P, Canaani E, Bloomfield C D, Croce C M. Proc Natl Acad Sci USA. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schichman S A, Canaani E, Croce C M. J Am Med Assoc. 1995;273:571–576. [PubMed] [Google Scholar]
- 16.Lochner K, Siegler G, Fuhrer M, Gereil J, Berk J D, Fey G H, Marschalek R. Cancer Res. 1996;56:2171–2177. [PubMed] [Google Scholar]
- 17.Chaplin T, Ayton P, Bernard O A, Saha V, Della V, Hillion J, Gregorini A, Lillington D, Berger R, Young B D. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 18.Cairns B R, Henry N L, Kornberg R D. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuriyama M, Harada N, Karoda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C M, Canaani E, Kaibuchi K. J Biol Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W N, Nowell P C, et al. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad R, Lehkowitz D, Gu Y, Alder H, Nakamura T, Saito H, Hubner K, Berger R, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1994;9:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidanza V, Melotti P, Yano T, Nakamura T, Bradley A, Canaani E, Calabretta B, Croce C M. Cancer Res. 1996;56:1179–1183. [PubMed] [Google Scholar]
- 23.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 24.Wu C. Nature (London) 1985;317:84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- 25.Conlon R A, Rossant J. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 26.Gage P J, Camper S A. Hum Mol Genet. 1997;6:457–464. doi: 10.1093/hmg/6.3.457. [DOI] [PubMed] [Google Scholar]
- 27.Muccielli M L, Martinez S, Pattyn A, Goridis C, Brunet J F. Mol Cell Neurosci. 1996;8:258–271. doi: 10.1006/mcne.1996.0062. [DOI] [PubMed] [Google Scholar]
- 28.Semina E V, Reiter R, Leysens N J, Alward W L M, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, et al. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 29.Lamonerie T, Tremblay J J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 30.Szeto D P, Ryan A K, O’Connell S M, Rosenfeld M G. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gindilis V, Banikazemi M, Vyasankin A, Verlinsky O, Matveyev I, Verlinsky Y. J Assist Reprod Genet. 1994;11:244–269. doi: 10.1007/BF02214344. [DOI] [PubMed] [Google Scholar]
- 32.Bürglin T R. In: A Guidebook for Homeobox Genes. Duboule D, editor. Oxford: Oxford Univ. Press; 1993. pp. 25–71. [Google Scholar]
- 33.Thanos D, Maniatis T. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 34.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 35.Solomon M J, Strauss F, Varshavsky A. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stong R C, Korsmeyer S J, Parkin J L, Arthur D C, Kersey J H. Blood. 1986;67:391–397. [Google Scholar]