Abstract
The designed peptide (denoted 20-mer, sequence VFITSDPGKTYTEVDPGOKILQ) has been shown to form a three-strand antiparallel β-sheet. It is generally believed that the DPro-Gly segment has the propensity to adopt a type II′ β-turn, thereby promoting the formation of this β-sheet. Here, we replaced DPro-Gly with Asp-Gly, which should favor a type I′ turn, to examine the influence of different type of turns on the stability of the β-sheet. Contrary to our expectation, the mutant peptide, denoted P6D, forms a five-residue type I turn plus a β-bulge between the first two strands due to a one amino-acid frameshift in the hydrogen bonding network and side-chain inversion of the first β-strand. In contrast, the same kind of substitution at DPro-14 in the double mutant, denoted P6DP14D, does not yield the same effect. These observations suggest that the SDGK sequence disfavors the type I′ conformation while the VDGO sequence favors a type I′ turn, and that the frameshift in the first strand provides a way for the peptide to accommodate a disfavored turn sequence by protruding a bulge in the formation of the β-hairpin. Thus, different types of turns can affect the stability of a β-structure.
Keywords: Protein folding, β-hairpin, β-sheet, turn, peptide, site-directed mutagenesis, NMR, structure, stability
De novo protein design has become an interesting and popular goal among biochemists. However, the knowledge we have obtained from protein structural biology has not been sufficient to accomplish this goal. Even for a simple peptide system, there are still many problems to be addressed and understood, including, for example, the phenomenon of peptide aggregation itself. β-structures are generally more difficult to design, since they involve two distinct structural motifs: extended strands and bent turns (or loops). In addition, long-range interstrand interactions can contribute to the stability of the β-structure. So far, there have only been a few successful designs of monomeric β-hairpins (Stanger and Gellman 1998; de Alba et al. 1999b), the more complicated three-stranded antiparallel β-sheet (Kortemme et al. 1998; Schenck and Gellman 1998; de Alba et al. 1999a), and a β-hairpin connected to an α-helix (Struthers et al. 1996). The unnatural amino acid D-form Pro has often been used to build a type II′ turn. It is generally believed that the NG or DG sequence offers the best choice for a stable type I′ turn.
A number of reports have discussed the β-forming propensity of amino acids in the strand region based on site-directed mutagenesis experiments (Kim and Berg 1993; Minor and Kim 1994a,b) or database surveys in the Protein Data Bank (PDB) (Swindells et al. 1995; Griffiths-Jones et al. 1998; Hutchinson et al. 1998). Typically, the β-branched amino acids such as Ile, Thr, and Val and aromatic amino acids such as Trp and Tyr show a higher propensity toward forming a β-structure due to steric interactions (Griffiths-Jones et al. 1998). However, sequences based on a combination of these residues do not always lead to β-structures. Several groups have examined the influence of the turn sequence on the formation of an anti-parallel β-hairpin (Ramirez-Alvarado et al. 1997; Blanco et al. 1998; de Alba et al. 1999b; Griffiths-Jones et al. 1999). Turns, with the ability to change the direction of the polypeptide chain, were suggested to be important in the formation of β-structures.
Toward improving our understanding of the role of the turn in the formation of β- structures, we chose a peptide that forms a triple-stranded sheet that was first described by the Gellman group in 1998 (Stanger and Gellman 1998) as the target of our study. This 20-residue peptide contains two DPro-Gly segments that adopt type II′ β-turns. We replace the unusual amino acid, DPro, with an aspartate to create an Asp-Gly sequence. This DG sequence, as well as the NG sequence, is a segment that favors the formation of a type I′ turn, according to the statistical data in the PDB. Using circular dichroism and NMR techniques, we show how the turn dominates the formation of the β-sheet and how the sequence of the turn affects the formation of the β-turn.
Results
Structure of wild-type peptide (WT 20-mer)
Part of the 2D-NOESY (Nuclear Overhauser Enhancement Spectroscopy) spectrum of the WT 20-mer is shown in Figure 1 ▶. The dαα crosspeaks between F2-T11, T4-T9, Y10-L19, and E12-K17 (Fig. 1A ▶), the dαN crosspeaks between T4-Y10, Y10-Q20, T11-I3, E12-I18, and K17-V13 (Fig. 1B ▶), and the dNN crosspeaks between I3-Y10, S5-K8, G7-K8, T11-I18, V13-O16, and G15-O16, clearly demonstrate that this short peptide forms a tight, three-stranded antiparallel β-sheet. The identification of the turn type is made on the basis of the observed NOEs. A distinction between type I and type II turn conformations can be made on the basis of the Nuclear Overhauser enhancements (NOEs) connectivity between the two central residues of the turn (usually referred to as residues 2 and 3 or residues L1 and L2): a dNN (i, i+1) connectivity between these two residues is expected for a type I turn, and a dαN (i, i+1) connectivity indicates a type II turn. The dNN (i, i+1) crosspeaks between G7-K8 and G15-O16, the dαN (i, i+2) crosspeaks between DP6-K8 and DP14-O16, and the stronger dαN (i, i+1) NOEs compared to the dδN (i, i+1) NOEs between DP6-G7 and DP14-G15 (CδH in proline occupies a position similar to that of NH of other amino acids), suggest that the two DPro-Gly segments form tight 2:2 type II′ turns. The dNδ (i, i+1) and dαδ (i, i+1) crosspeaks observed between S5-DP6 and V13– DP14 indicate that the DPro-6 and 14 are in the trans conformations. The interstrand NOEs and the NOEs in the turn regions of the WT 20-mer are summarized in Figure 2A ▶.
Fig. 1.
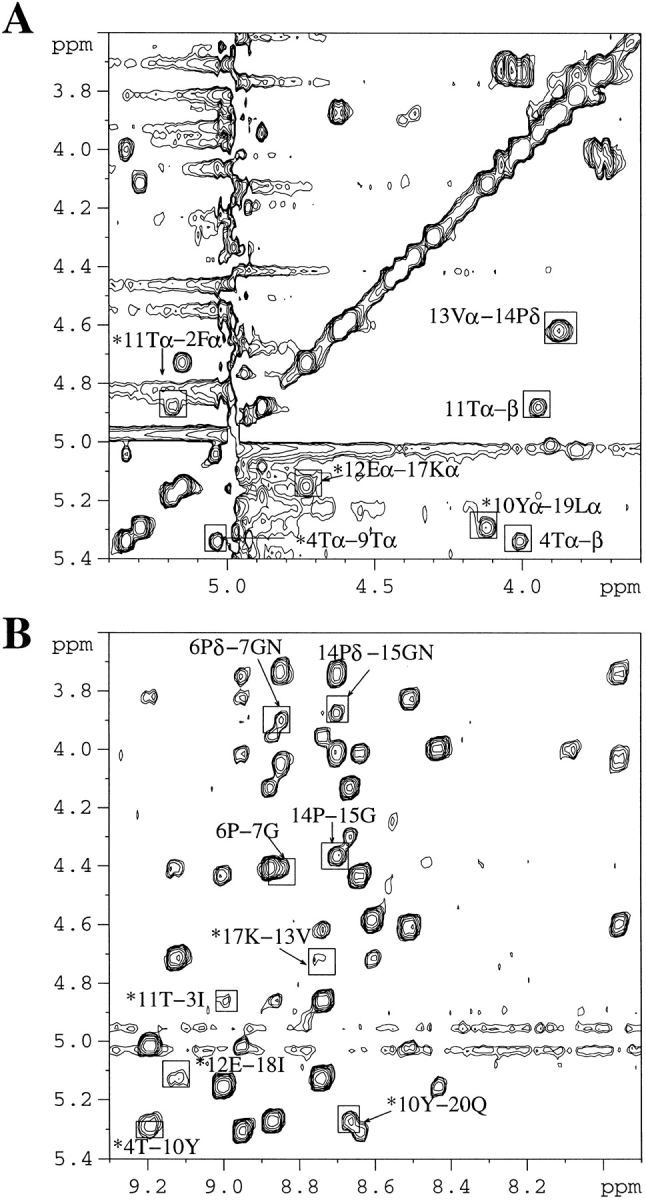
2D-NOESY spectrum of the WT 20-mer (0.4 mM) at pH 3.6 and 280K. (A) Hα–Hα region in D2O. (B) Hα–HN region in H2O:D2O (9:1). Important NOEs are highlighted. Asterisk-labeled peaks denote interstrand main chain — main chain NOEs. Unless described otherwise, the NOEs noted in B are Hα–HN NOEs. NOE for 5Sα–6Pδ were not observed since the α-proton of Ser-5 is too close to the water signal.
Fig. 2.
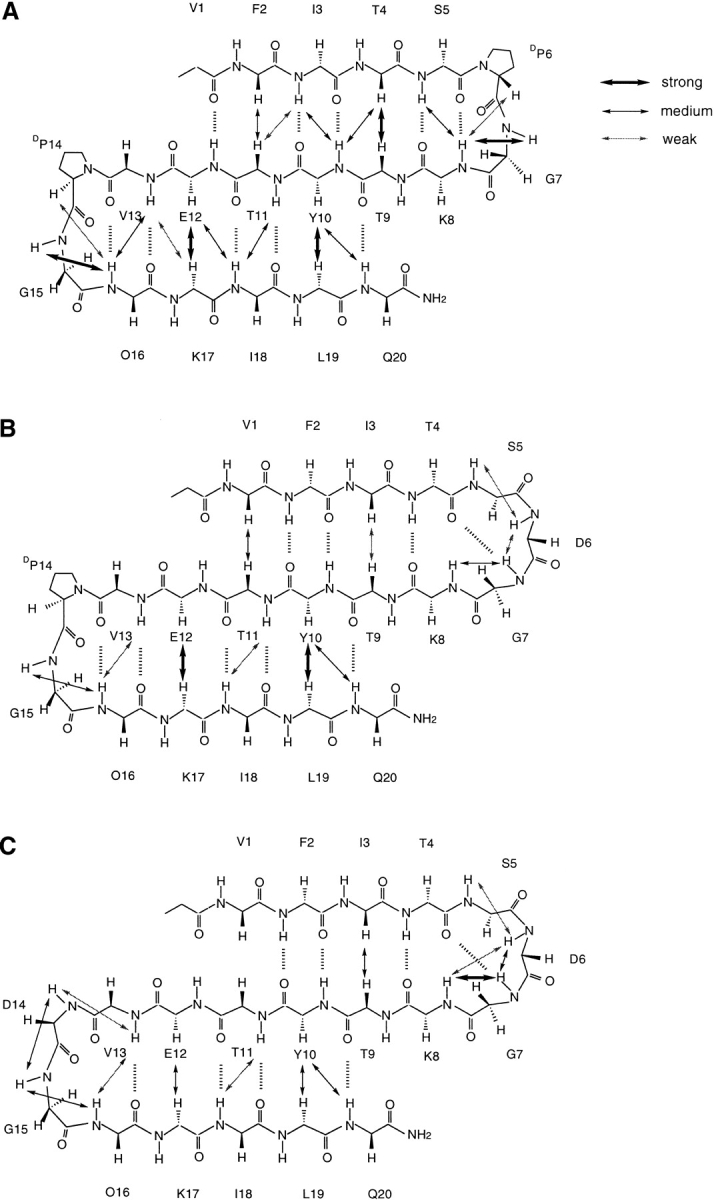
Interstrand NOEs and the NOEs in the turn regions observed in NOESY spectrum of the WT 20-mer and its mutants at 280K. (A) 20-mer; (B) P6D; (C) P6PD14D. Hα–Hα NOEs were measured in D2O, while the remaining NOEs were recorded in 9:1 H2O:D2O (pH 3.6). O = ornithine. Strong, medium, and weak intensity of NOEs are expressed by different thicknesses of the line.
Changes in the structure and stability of the β-sheet with P6D mutation
Part of the 2D-NOESY spectrum of P6D is shown in Figure 3 ▶. The dαα crosspeaks between V1-T11, I3-T9, Y10-L19, and E12-K17 (Fig. 3A ▶), the dNN crosspeaks between S5-D6, D6-D7, G7-K8, T11-I18, V13-O16, and G15-O16 (Fig. 3B ▶), and the dαN crosspeak between Y10-Q20 suggest that there has been a conformational change in the first hairpin of P6D relative to the wild-type peptide. Specifically, the data indicate that the first strand has one-residue shifted toward the turn region to form a five-residue turn. In the new turn formed by a TSDGK sequence, the dNN (i, i+1) crosspeaks between S5-D6, D6-G7, and G7-K8 suggest that these five residues form a type I plus G1 bulge turn. The interstrand NOEs and the NOEs in the turn regions of the P6D are highlighted in Figure 2B ▶.
Fig. 3.
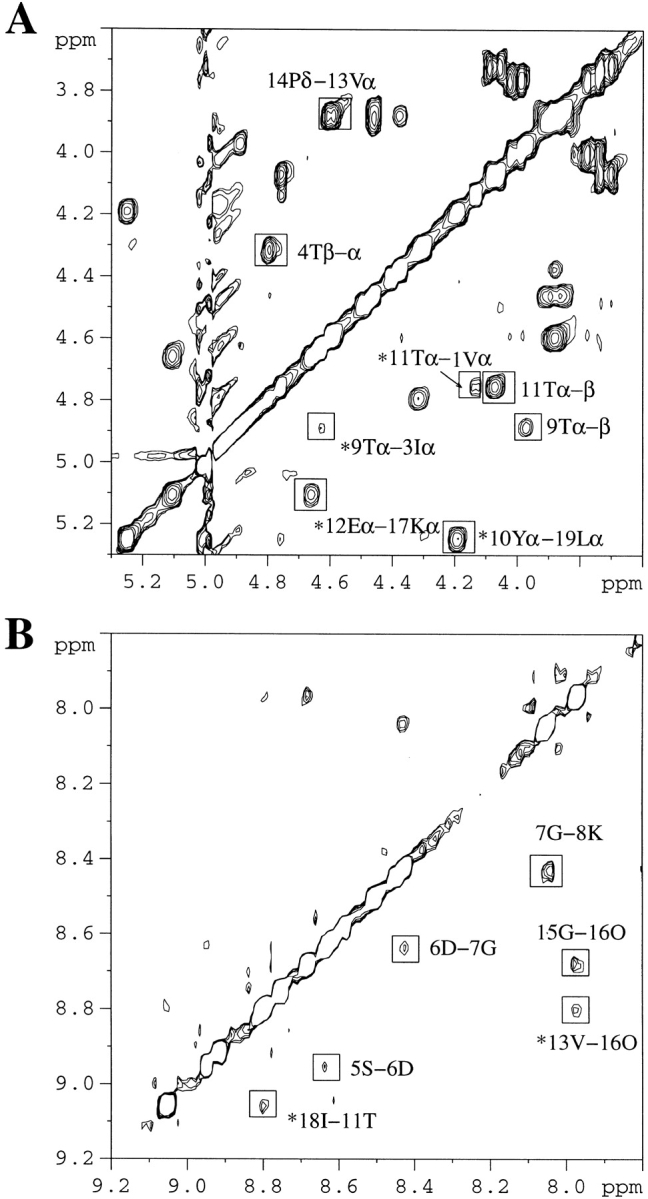
2D-NOESY spectrum of P6D (0.4 mM) at pH 3.6 and 280K. (A) Hα–Hα region in D2O. (B) HN–HN region in H2O:D2O (9:1). Important NOEs are highlighted. Asterisk-labeled peaks denote interstrand main chain — main chain NOEs. All of the NOEs noted in B are HN–HN NOEs.
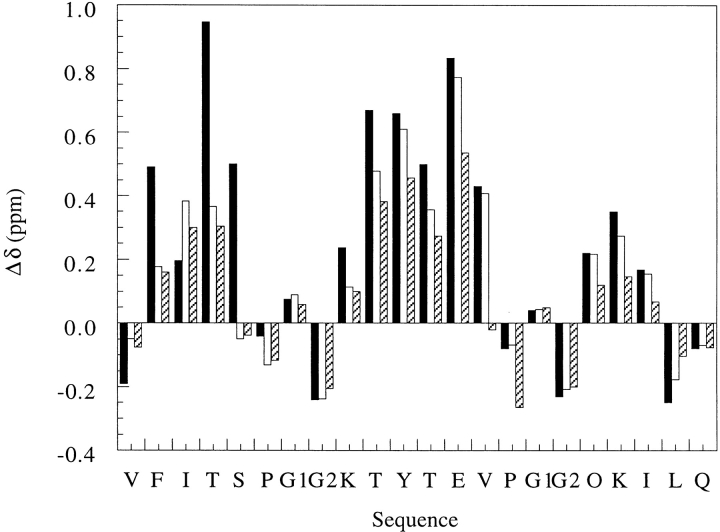
The CαH chemical shifts of the WT 20-mer show dramatic downfield shifts at residues 2–5, 8–13, and 16–18, corresponding to the three β-strand regions in the β-sheet (Fig. 4 ▶). The corresponding CαH shifts in the P6D mutant peptide are generally less downfield shifted than those in the WT 20-mer. These results suggest that the single residue mutation in the turn destabilizes the β-sheet. Moreover, the destabilization appears cooperative between the two hairpins. Evidence in support of this assertion comes from downfield CαH shifts of the third strand, though smaller in magnitude, and the weaker interstrand NOEs in the second hairpin of the P6D mutant peptide compared with those observed for the WT 20-mer. Finally, the CαH chemical shift of Ser-5 is near the value in the random-coil state, providing further evidence that Ser-5 is no longer in the strand region.
Fig. 4.
A plot of the deviations of CαH chemical shifts of the WT 20-mer and its P6D and P14D mutants from random-coil values. The NMR spectra were recorded in 9:1 H2O:D2O (pH 3.6) at 280K. The amino acid sequence of the WT 20-mer is shown on the x-axis. The two α-protons of glycine are denoted by G1 and G2. DProline is represented by a "P." Δδ = (observed δαH − random-coil δαH). The reported random-coil value (Wüthrich 1986) for lysine was used for ornithine because it has been shown that the δαH or ornithine is very close to the δαH of lysine (Stanger and Gellman 1998). Black bar, WT 20-mer; clear bar, P6D; hatched bar, P6DP14D.
The same substitution at position 14 does not lead to the same effect. In the spectra of P6DP14D, the dαα crosspeaks between I3-T9, Y10-L19, and E12-K17 (Fig. 5A ▶), the dNN crosspeaks between S5-D6, D6-D7, D6-K8, G7-K8, T11-I18, V13-D14, D14-G15, V13-O16, and G15-O16 (Fig. 5B ▶), and the dαN crosspeak between Y10-Q20 suggest that the second hairpin register is unchanged between the P6DP14D mutant and the WT peptide, whereas the P6D mutation alone causes a frameshift in the first hairpin (Fig. 5 ▶). The interstrand NOEs and the NOEs in the turn regions of P6DP14D are summarized in Figure 2C ▶. However, the CαH chemical shift of Val-13 is near the value in the random-coil state, suggesting that the hydrogen bonding network is weaker in the apex of the second hairpin (Fig. 4 ▶). The strong dNN (i, i+1) crosspeaks between D14-G15 and G15-O16 indicate that the VDGO sequence has formed a type I′ turn, as expected. Overall, the CαH chemical shifts of the P6DP14D mutant are generally even less downfield shifted than those of P6D, which suggests that the type I′ turn is less favored than the type II′ turn in the β-sheet. However, it is interesting that the ΔδHαs are comparable between the P6D and P6DP14D mutants for the first and second strands. Once again, this observation indicates cooperative interactions among the three strands of the β-sheet. Thus, turns are very important determinants in the stability of β-structures.
Fig. 5.
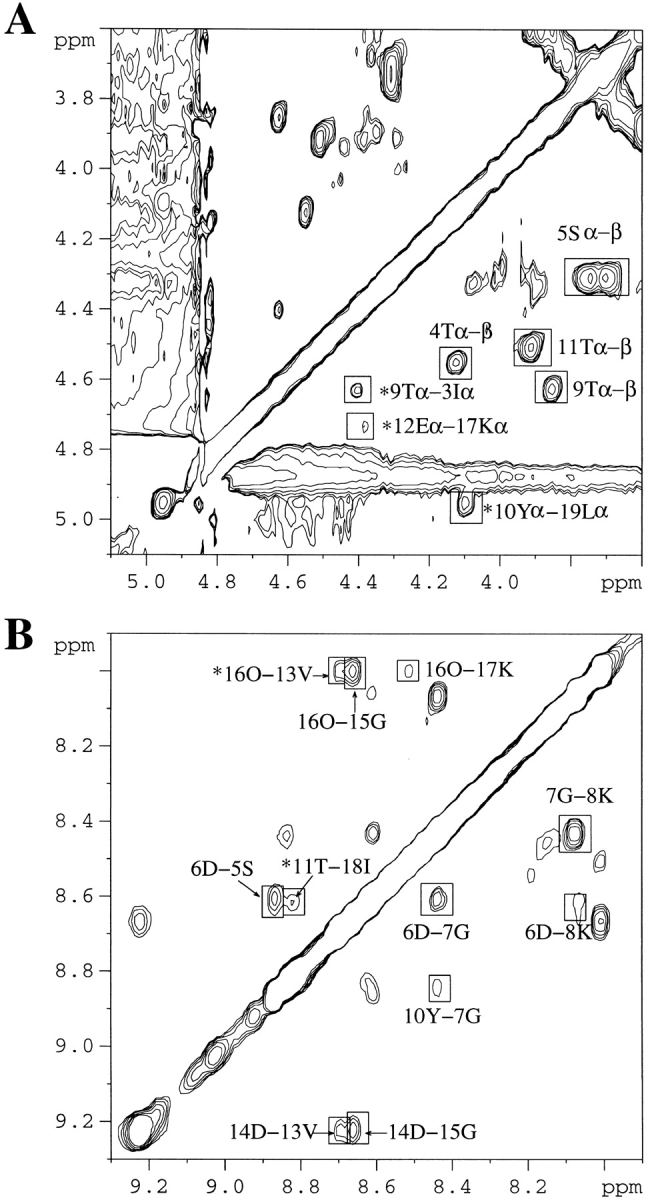
2D-NOESY spectrum of P6DP14D (1.6 mM) at pH 3.6 and 280K. (A) Hα–Hα region in D2O. (B) HN–HN region in H2O:D2O (9:1). Important NOEs are highlighted. Asterisk-labeled peaks denote interstrand main chain — main chain NOEs. All of the NOEs noted in B are HN–HN NOEs.
Circular dichroism of the peptides
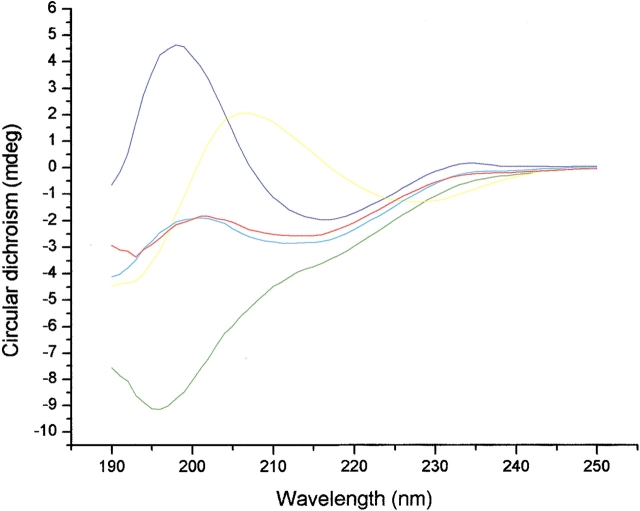
Circular dichroism (CD) spectra of the synthetic peptides are shown in Figure 6 ▶. The CD spectrum of the WT 20-mer shows a negative ellipticity at 216 nm and a positive one at 198 nm that are characteristic features for a β-structure. Surprisingly, the CD spectrum of the P6DP14D mutant, which has a negative ellipticity at 196 nm, looks more like the spectrum of a random-coil peptide, although the NMR data indicate that it has a β-sheet structure. We surmise that this unusual strong CD signal comes from the unnatural DPro-Gly turn. To verify this point, we have synthesized the short peptide TSDPGKT, denoted 6-mer, corresponding to the first DPro-Gly turn plus two flanking residues. The CD spectrum of 6-mer has a negative ellipticity around 228 nm and a positive one around 206 nm, which is red shifted compared with that of the 20-mer. The CD spectrum of P6D, which has only one DPro-Gly turn, appears to be a superposition of that for the 20-mer and P6DP14D. These results support our assertion that the CD signal arises from the turns rather than from the loose sheet of the peptide, and explain why most peptides found to populate β-structures by NMR often look like random coils by CD. Therefore, the prediction of the β-structure populations or the comparison of structural stability of short peptides with different turn sequences by CD could be unreliable. The relationship between the ellipticity in CD spectra and the content of secondary structure in the case of the β-structure-forming peptides remains to be clarified.
Fig. 6.
CD spectra of the peptides in 3 mM NaOAc (pH 3.8). Blue, WT 20-mer; red, P6D; green, P6DP14D; yellow, 6-mer; cyan, the average of the WT 20-mer and P6DP14D.
Discussion
The type I turn is the most favored turn type in proteins but not in β-hairpins, where type I′and II′are more common (Sibanda and Thornton 1985,1991). The reason is due to the incompatibility of the geometry of a type I turn with the right-handed twist of β-sheets. Therefore, one extra "bulge" residue normally follows a type I turn to release the strain and form a 3:5 turn. On the contrary, type I′and II′ turns, generally associated with 2:2 hairpins, have a geometry matching the right-handed twist. It has been shown that the replacement of the turn sequence by the sequence NPDG with the highest tendency to form a type I β-turn results in the formation of a 3:5 nonnative hairpin and causes a one-residue shift in the antiparallel alignment of the strand region (Blanco et al. 1993; Searle et al. 1995; de Alba et al. 1997). In the present study, we found that only a one-residue change in the P6D mutant produced the same effect.
The present mutagenesis studies on the WT 20-mer allow us to discuss the relative importance of the sequence effect on determining the turn type, as well as the importance of the turn on the stability of the β-sheet. Our results are consistent with the statistical preferences, which are based on all of the β-turns in the data bank (Hutchinson and Thornton 1994). According to the positional potentials of different turns given in this paper, Ser in the SDGK sequence has a lower score (1.10) than Val in the VDGO sequence (1.50) for a type I′ turn, while the TSDGK sequence has a higher score (Thr, 1.11; Ser, 1.50) than the EVDGO sequence (Glu, 0.74; Val, 0.72) for a type I + bulge turn.
Table 1 lists the turn sequences that have been published to date that form the turn of a stable β-hairpin in aqueous solution. From the sequence comparison, we were able to discern some preferred sequences for stable turn-formation in a short peptide. For example, Lys favors the +B1 position; DG and NG prefer the L1 + L2 positions in the type I′ turn of the 2:2 hairpin or the L2 + bulge positions in the type I + bulge turn of the 3:5 hairpin. The β-branched residues such as Val and Ile and aromatic residues such as Tyr, which prefer the β-conformation, favor the −B1 position of the type I′ turn instead of the L1 position of the type I+bulge turn, where an αR conformation is required. Moreover, the hydrophobic group is not welcomed for the −B1 position of the type I + bulge turn, even if it favors the β-conformation. Evans and Chen replaced the TLTGK sequence with the VLTGK sequence and found that the hydrophobic side chain destabilizes the hairpin structure in the peptide corresponding to the N-terminal segment of ubiquitin (P.A. Evans and P.Y. Chen, unpubl.). This explains why the hydrophobic residues such as Val and Ile have low statistical scores for the −B1 position of the type I turn and emphasizes the steric effect of residues in the turn formation.
Table 1.
The turn sequences of known peptides populating monomeric β-hairpin in aqueous solution
| Type I′turn −B1 L1 L2 + B1 β αL γLβ | type I turn + G1 β-bulge −B1 L1 L2 b + B1 β αR γR γLβ |
| I N G Ka | T L T G Kd |
| V N G Kb | T L D G Ke |
| V D G Kb | N P D G Tf |
| V G G Kb | N P D G Sg |
| V S G Kb | T S D G K |
| Y N G Kc | A P D G Th |
| V D G O |
a Maynard et al. 1998; Griffiths-Jones et al. 1999.
b Ramirez-Alvarado et al. 1997.
c de Alba et al. 1999b.
d Zerella et al. 1999.
e Zerella et al. 2000.
f Searle et al. 1995; de Alba et al. 1999b.
g Blanco et al. 1993.
h de Alba et al. 1997.
Turn sequence is important in determining the turn type and the turn length. Although we have not noted any side-chain interactions among the turn residues by NMR, we believe that the combination of side-chain groups could induce effects on the torsional preference of the turn backbone. Mutations in the turn sequence generally have less effects on protein compared to peptide conformation, where tertiary interactions are absent. In any case, this kind of conformational proclivity might play an important role in the early stages of protein folding.
Materials and methods
Peptide synthesis
Peptides, denoted 20-mer, 6-mer, P6D, and P6DP14D, were synthesized by the batchwise fmoc-polyamide method on a PS3 peptide synthesizer (Rainin). Rink Amide AM resin (substitution 0.69 mmol/g) from Novabiochem was used in the synthesis. Fmoc-amino-acid derivatives (four equivalents) were coupled on the resin with equivalent Benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP) and 4.45% N-methyl morpholine (NMM, by vol.) in dimethylformamide (DMF). The N-terminal ends were acetylated, and the C-terminal ends were amidated to avoid the electrostatic interactions at both ends. The acetylation was performed using four equivalents of acetic anhydride instead of an amino-acid derivative in the synthetic procedure. The fmoc-cleavage was performed with 20% (v/v) piperidine in DMF. The peptides were cleaved from the resin by stirring with a mixture of 10 mL trifluoroacetic acid, 0.75 g solid phenol, 0.5 mL thioanisol, 0.5 mL water, and 0.25 mL ethanedithiol (for less than 200 mg resin) at room temperature for 1–2 hours and precipitated with methyl t-butyl ether at 2000 g for 10 minutes three times and dried in a vacuum. The resulting white powder was purified by reverse-phase HPLC using a Vydac C18 column (10 mm × 250 mm) and acetonitrile-water mixtures containing 0.1% trifluoroacetic acid (v/v). The final products were analyzed by positive ion-electrospray mass spectroscopy. The fractions containing the desired products were lyophilized and stored at −20°C.
2D-NMR Spectroscopy
All NMR spectra were recorded on a Bruker AM 500 NMR spectrometer. Samples were dissolved in 0.5 mL of H2O/D2O (9/1) or in 0.5 mL D2O depending on the experimental requirement. The concentrations of the peptide samples were in the range of 0.4 ∼ 2.1 mM. NMR chemical shifts and line widths were shown to be independent of peptide concentration in the range of 2.1 mM to 40 μM, suggesting that the peptide remains monomeric in solution at the concentration used in 2D-NMR analysis. A 1/100 volume of sodium 3–(trimethylsilyl)-propionic-2, 2, 3, 3-d4 acid (TSP) solution (0.75% in D2O) was added as an internal reference. The pH values were adjusted to 3.6. Quoted pH values were not corrected for the D/H isotope effect. 2D-TOCSY and NOESY were recorded using standard phase-cycling sequences at 280K. Usually, spectra were acquired with 2K data points in direct dimension and 512 increments in indirect dimension. Typically, 64 or more scans were collected per increment depending on the peptide concentration. An 80 msec mixing time in TOCSY and 300 msec mixing time in NOESY were used. Data were processed by XWINNMR software (Bruker). The shifted square sine bell window functions in both dimensions were applied for all spectra. The Ansig program (version 3.3) was used to assign the spectra (Kraulis 1989).
Circular dichroism spectroscopy
CD spectra were recorded in a π* CD spectrometer (Applied Photophysics, UK). Peptides were dissolved in 3 mM NaOAc (pH 3.8) to give a concentration of about 30 μM. The concentration was determined from the UV absorbance. CD calibration was carried out by using 1.5 mg/mL D(−) Pantoyllactone at 219 nm. A CD scan of the sample was recorded in a 1 mm cell, between 190 and 250 nm, at room temperature. A scan interval of 1 nm with an integration of 200,000 points was employed. The spectrum of 3 mM NaOAc (pH 3.8) was collected as a baseline and subtracted automatically. The CD spectra of peptides were smoothed by averaging adjacent five points.
Electronic supplemental material
Complete 1H NMR assignments of the WT 20-mer and its P6D, P6DP14D mutant peptides are available as supplemental material.
Acknowledgments
We thank Ms. Su-Ching Lin for her kind assistance in recording the NMR spectra. This work is supported by grant NSC 89–2113-M-001–092 from the National Science Council, Taiwan, ROC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.49001.
References
- Blanco, F.J., Jiménez, M.A., Herranz, J., Rico, M., Santoro, J., and Nieto, J.L. 1993. NMR evidence of a short linear peptide that folds into a β-hairpin in aqueous-solution. J. Am. Chem. Soc. 115 5887–5888. [Google Scholar]
- Blanco, F., Ramirez-Alvarado, M., and Serrano, L. 1998. Formation and stability of β-hairpin structures in polypeptides. Curr. Opin. Struct. Biol. 8 107–111. [DOI] [PubMed] [Google Scholar]
- de Alba, E., Jiménez, M.A., and Rico, M. 1997. Turn residue sequence determines β-hairpin conformation in designed peptides. J. Am. Chem. Soc. 119 175–183. [Google Scholar]
- de Alba, E., Santoro, J., Rico, M., and Jiménez, M.A. 1999a. De novo design of a monomeric three-stranded antiparallel β-sheet. Protein Sci. 8 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alba, E., Rico, M., and Jiménez, M.A. 1999b. The turn sequence directs β-strand alignment in designed β-hairpins. Protein Sci. 8 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S.R., Maynard, A.J., and Searle, M.S. 1999. Dissecting the stability of a β-hairpin peptide that folds in water: NMR and molecular dynamics analysis of the β-turn and β-strand contributions to folding. J. Mol. Biol. 292 1051–1069. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S.R., Sharman, G.J., Maynard, A.J., and Searle, M.S. 1998. Modulation of intrinsic phi, psi propensities of amino acid by neighboring residues in the coil regions of protein structures: NMR analysis and dissection of a β-hairpin peptide. J. Mol. Biol. 284 1597–1609. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G., Sessions, R.B., Thornton, J.M., and Woolfson, D.N. 1998. Determinants of strand register in antiparallel β-sheets of proteins. Protein Sci. 7 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1994. A revised set of potentials for β-turn formation in proteins. Protein Sci. 3 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.W.A. and Berg, J.M. 1993. Thermodynamic β-sheet propensities measured using a zinc-finger host peptide. Nature 362 267–270. [DOI] [PubMed] [Google Scholar]
- Kortemme, T., Ramirez-Alvarado, M., and Serrano, L. 1998. Design of a 20-amino acid, three stranded β-sheet protein. Science 281 253–256. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1989. Ansig — a program for the assignment of protein H-1 2D-NMR spectra by interactive computer-graphics. J. Magn. Reson. 84 627–633. [Google Scholar]
- Maynard, A.J., Sharman, G.J., and Searle, M.S. 1998. Origin of β-hairpin stability in solution: Structural and thermodynamic analysis of the folding of a model peptide supports hydrophobic stabilization in water. J. Am. Chem. Soc. 120 1996–2007. [Google Scholar]
- Minor, D.L. and Kim, P.S. 1994a. Measurement of the β-sheet-forming propensities of amino-acids. Nature 367 660–663. [DOI] [PubMed] [Google Scholar]
- Minor, D.L. and Kim, P.S. 1994b. Context is a major determinant of β-sheet propensity. Nature 371 264–267. [DOI] [PubMed] [Google Scholar]
- Ramirez-Alvarado, M., Serrano, L., and Blanco, F.J. 1997. Conformational analysis of peptides corresponding to all the secondary structure elements of protein L B1 domain: Secondary structure propensities are not conserved in proteins with the same fold. Protein Sci. 6 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, H.L. and Gellman, S.H. 1998. Use of a designed triple-stranded antiparallel β-sheet to probe β-sheet cooperativity in aqueous solution. J. Am. Chem. Soc. 120 4869–4870. [Google Scholar]
- Searle, M.S., Williams, D.H., and Packman, L.C. 1995. A short linear peptide derived from the N-terminal sequence of ubiquitin folds into a water-stable nonnative β-hairpin. Nat. Struct. Biol. 2 999–1006. [DOI] [PubMed] [Google Scholar]
- Sibanda, B.L. and Thornton, J.M. 1985. β-Hairpin families in globular proteins. Nature 316 170–174. [DOI] [PubMed] [Google Scholar]
- Sibanda, B.L. and Thornton, J.M. 1991. Conformation of β-hairpins in protein structures — Classification and diversity in homologous structures. Methods Enzymol 202 59–82. [DOI] [PubMed] [Google Scholar]
- Stanger, H.E. and Gellman, S.H. 1998. Rules for antiparallel β-sheet design: D-Pro-Gly is superior to L-Asn-Gly for β-hairpin nucleation. J. Am. Chem. Soc. 120 4236–4237. [Google Scholar]
- Struthers, M.D., Cheng, R.P. and Imperiali, B. 1996. Design of a monomeric 23-residue polypeptide with defined tertiary structure. Science 271 342–345. [DOI] [PubMed] [Google Scholar]
- Swindells, M.B., MacArthur, M.W., and Thornton, J.M. 1995. Intrinsic phi, psi propensities of amino acids, derived from the coil regions of known structures. Nat. Struct. Biol. 2 596–603. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. John Wiley & Sons, NY, p. 17.
- Zerella, R., Chen, P.Y., Evans, P.A., Raine, A., and Williams, D.H. 2000. Structural characterization of mutant peptides derived from ubiquitin and implications for protein folding. Protein Sci. 9 2142–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerella, R., Evans, P.A., Ionides, J.M.C., Williams, D.H., Trotter, B.W., and Mackay, J.P. 1999. Autonomous folding of a peptide corresponding to the N-terminal β-hairpin from ubiquitin. Protein Sci. 8 1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]