Abstract
Many of the protein fusion systems used to enhance the yield of recombinant proteins result in the addition of a small number of amino acid residues onto the desired protein. Here, we investigate the effect of short (three amino acid) N-terminal extensions on the equilibrium denaturation and kinetic folding and unfolding reactions of the FK506-binding protein (FKBP) and compare the results obtained with data collected on an FKBP variant lacking this extension. Isothermal equilibrium denaturation experiments demonstrated that the N-terminal extension had a slight destabilizing effect. NMR investigations showed that the N-terminal extension slightly perturbed the protein structure near the site of the extension, with lesser effects being propagated into the single α-helix of FKBP. These structural perturbations probably account for the differential stability. In contrast to the relatively minor equilibrium effects, the N-terminal extension generated a kinetic-folding intermediate that is not observed in the shorter construct. Kinetic experiments performed on a construct with a different amino acid sequence in the extension showed that the length and the sequence of the extension both contribute to the observed equilibrium and kinetic effects. These results point to an important role for the N terminus in the folding of FKBP and suggest that a biological consequence of N-terminal methionine removal observed in many eukaryotic and prokaryotic proteins is to increase the folding efficiency of the polypeptide chain.
Keywords: Protein folding, kinetic intermediate, expression system, N terminus, stopped-flow
FK506-binding protein (FKBP) is a small (107 amino acid residues) globular protein composed of five antiparallel β-strands, an amphiphilic α-helix, and three loops. There are no disulfides and all seven proline residues are in the trans conformation in the native state. The structure of the protein has been determined to high resolution in several forms by X-ray crystallography and NMR (Michnik et al. 1991; van Duyne et al. 1991, 1993; Meadows et al. 1993). FKBP catalyzes peptidyl-prolyl cis-trans isomerization and there is significant interest in the stability and folding of FKBP due to the possibility of autocatalyzed folding (Egan et al. 1993; Scholz et al. 1996; Veeraraghavan et al. 1996; Main et al. 1998, 1999; Fulton et al. 1999).
We are using FKBP as a model system to investigate how nonrandom structure present in the urea-unfolded state affects subsequent folding events. Specifically, we are using an FKBP variant in which the single Cys at position 22 has been replaced by Ala (C22A). This mutation has little effect on the thermodynamic stability, structure, or chemical mechanism of FKBP (Park et al. 1992), but allows us to perform reversible differential scanning calorimetry experiments with minimal loss of protein (data not shown). To facilitate the purification of mutant proteins, we have generated a fusion construct between a modified form of the pMALc2 plasmid vector and the FKBP C22A structural gene. The modification adds eight histidine residues (H8) in a 17-residue linker after the maltose-binding protein (MBP) gene and immediately preceding a thrombin proteolytic site (Pryor and Leitung 1997), which replaces the less efficient Factor Xa cleavage site. Removal of the MBP-H8 affinity tag with thrombin leaves three additional amino acid residues on the N terminus of the C22A gene.
Many of the current plasmids used for expressing recombinant proteins in Escherichia coli result in N-terminal extensions of one or more amino acids because of the particular restriction endonuclease sites used for cloning and from proteolytically cleavable or noncleavable affinity tags used to aid purification. These tags are generally innocuous to the recombinant protein, and may even provide additional stability and solubility (Davis et al. 1999). Here, we report isothermal equilibrium denaturation and kinetic-folding studies of two extended FKBP constructs. These data show that N-terminal extension has a small, but significant impact on the thermodynamic stability. This result may be anticipated from the three dimensional structure of FKBP. However, the extension has an unexpected and substantial effect on the folding kinetics and apparent folding pathway of this small, single-domain protein. We show that this effect has contributions both from the size and amino acid sequence of the N-terminal extension.
Results and Discussion
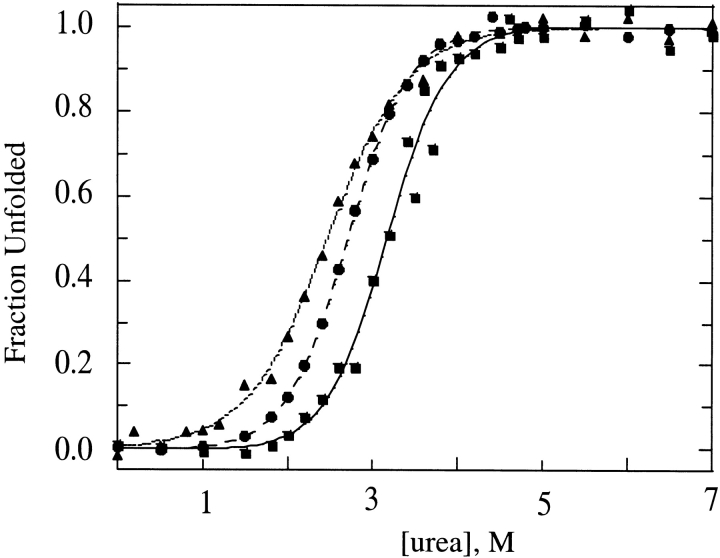
The addition of three amino acids at the N terminus destabilized the C22A variant of FKBP. Figure 1 ▶ compares the isothermal equilibrium denaturation profiles for gsmFKBP and gsgFKBP with that of C22A, as measured by changes in the intrinsic fluorescence of Trp59 as a function of urea concentration. All three proteins exhibit highly cooperative unfolding transitions with an apparent lack of equilibrium unfolding intermediates, consistent with the behavior of true wild-type FKBP (Egan et al. 1993; Main et al. 1998). The normalized integrated fluorescence intensity data were fitted to a two-state model (equation 1) in which the native and completely unfolded states are the only significantly populated conformations throughout the transition. These data (Table 1) provide an estimate of the D50 value, which is the concentration at which the free energy for unfolding, ΔG°unf, is zero, and of the equilibrium m-value, which is related to the free energy of transfer of the protein between two solvents and is interpreted structurally as being proportional to the change in nonpolar surface area of the protein exposed to the solvent upon (un)folding (Schellman 1987a,b, 1994). As shown in Table1, the equilibrium m-values for gsmFKBP and C22A are nearly equivalent (1.65 kcal/mole•M−1 and 1.62 kcal/mole•M−1, respectively), indicating that denaturation occurs between similar thermodynamic states in the two proteins. On the other hand, the equilibrium m-value for gsgFKBP (1.26 kcal/mole•M−1) is considerably lower than the other two variants, indicating a less cooperative unfolding transition and/or a slight change in the structure of the unfolded state.
Fig. 1.
Isothermal equilibrium denaturation profiles for C22A (▪; solid line), gsmFKBP (•; broken line), and gsgFKBP (▴; dotted line) plotting fraction unfolded as a function of urea concentration at 30°C. Curves show the best fit of the data to equation 1. Each data point represents the average of at least two independent determinations. Equilibrium denaturation parameters resolved from the fitting are collected in Table 1.
Table 1.
Equilibrium and kinetic parameters for C22A, gsmFKBP and gsgFKBP
| Parameter | C22A | gsmFKBP | Intermediate | gsgFKBP |
| D50 (M) | 3.17 (0.02) | 2.69 (0.01) | — | 2.48 (0.08) |
| meq (kcal/moleM) | 1.65 (0.17) | 1.63 (0.04) | — | 1.26 (0.13) |
| ΔG°unf (kcal/mole) | 4.87 (0.54) | 4.38 (0.11) | — | 3.12 (0.34) |
| k°u (sec−1) | 0.014 (0.004) | 0.054 (0.006) | 0.162 (0.036) | 0.007 (0.004) |
| mu (M−1) | 0.64 (0.05) | 0.61 (0.02) | 0.76 (0.08) | 0.86 (0.09) |
| k° (sec−1) | 4.46 (0.50) | 2.85 (0.18) | 3.64 (0.25) | 2.73 (0.28) |
| mf (M−1) | −1.29 (0.08) | −0.97 (0.06) | −1.74 (0.15) | −0.74 (0.08) |
Numbers in parentheses represent the error associated with a simultaneous fit of Equations 1 or 2 to three independent data sets for the equilibrium and kinetic data, respectively. Kinetic data for Intermediate was obtained from an independent fit to Equation 2 of the kinetic folding data for gsmFKBP from 0.35 to 3.09 M urea.
We observed a D50 of 3.17 M urea for C22A, giving a ΔG°unf of 4.87 kcal/mole, which is slightly lower than values published previously for true wild-type FKBP (Egan et al. 1993; Main et al. 1998). In contrast, the D50 for gsmFKBP and gsgFKBP were reduced to 2.69 M and 2.48 M urea, yielding ΔG°unf values of 4.38 and 3.12 kcal/mole at 30°C, respectively. In fact, the gsgFKBP construct was so destabilized that the purification and cleavage procedure had to be modified to increase the yield following thrombin digestion. Interestingly, the Schmid group also observed a destabilization (D50 = 2.9 M urea) coupled with an increased tendency for precipitation when FKBP was expressed with a sixteen residue N-terminal extension containing a histidine tag (Scholz et al. 1996).
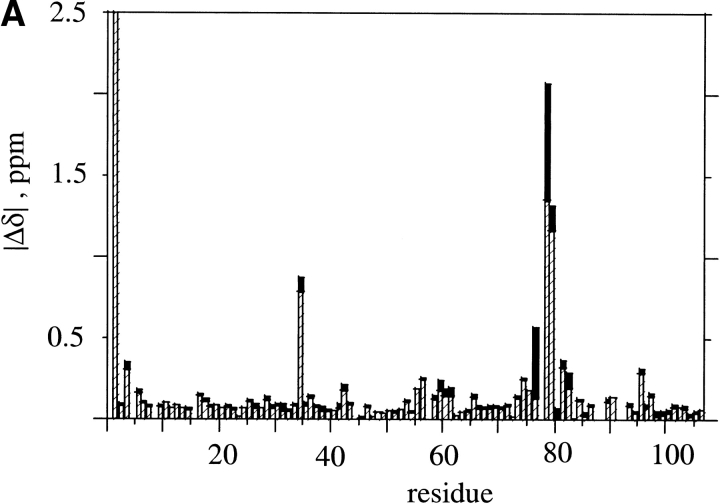
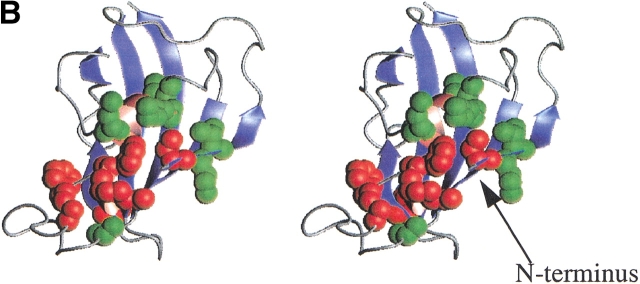
A structural basis for the decreased stability in gsmFKBP was apparent from a comparison of 2D 1H,15N correlation experiments (Kay et al. 1992). Figure 2A ▶ shows the differences in chemical shifts of the backbone amide 1H and 15N resonances between the C22A and gsmFKBP constructs, and Figure 2B ▶ shows the location of the perturbed residues in the native structure of FKBP. The largest differences in chemical shift are observed for residues V2, K35, S77, D79, Y82, and G83. Except for V2, these residues are located in the single-turn 310 helix and at the base of the Ω-shaped loop connecting β-strands II and III of native FKBP (red balls, Fig. 2B ▶). Smaller chemical shift differences are observed for residues Q3, V4, I56, R57, W59-G62, and A95. Residues I56–G62 are in the single α-helix, which is located above the plane of the five-stranded β-sheet, whereas Q3, V4, and A95 are in βI and βII, adjacent to the N terminus. The NMR results suggested that steric contacts between the gsm extension and residues adjacent to the N terminus were the likely cause for the decreased equilibrium stability of gsmFKBP relative to C22A. Because the gsgFKBP construct is further destabilized relative to gsmFKBP and C22A, the steric interactions are not likely to arise solely from the methionine side chain.
Fig. 2.
(A) Magnitude of the chemical shift difference, Δδ, between C22A and gsmFKBP. Hashed and solid bars represent the difference in 15N and 1H shifts, respectively. The precision in measuring the observed chemical shift value is limited by the digital resolution in the 1H and 15N dimensions of the HSQC spectra (0.008 and 0.1 ppm, respectively, in HSQC spectra of the two variants). The shift difference for residue V2 is off scale. (B) Stereoview of FKBP generated by use of the 1fkj pdb file (Wilson et al. 1995). The ligand and solvent molecules are not shown for clarity. β-strands are in blue, whereas helical regions are in pink. The red balls indicate resonances that exhibit large chemical shift changes; green balls indicate residues that experience smaller, but still significant chemical shift changes in gsmFKBP relative to C22A. The N terminus is indicated as N; β-strands are numbered consecutively starting from the right-most strand in this diagram; the gsm extension is not shown. Figure generated by use of Molmol (Koradi et al. 1996).
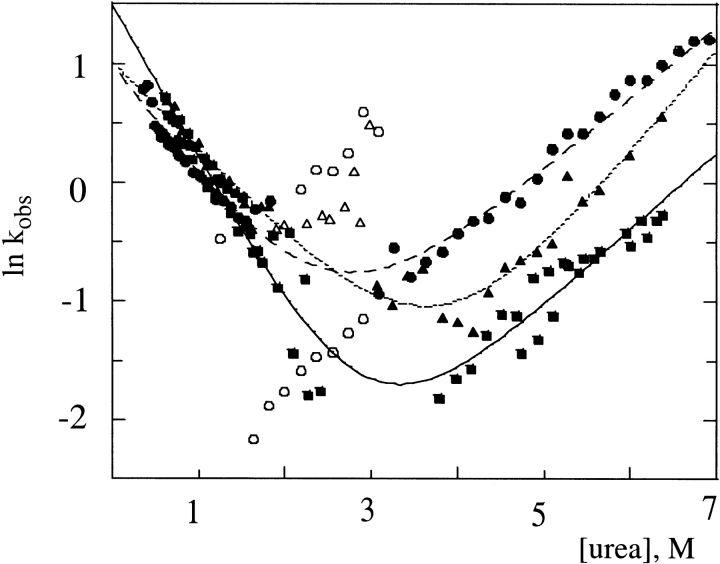
The three additional N-terminal residues strongly impact the folding and unfolding kinetics of FKBP. As shown in Figure 3 ▶, the folding and unfolding kinetics of C22A (solid boxes) fit well to a two-state model over an extended range of urea concentrations, in agreement with results obtained on the true wild-type FKBP (Main et al. 1999). The gsmFKBP and gsgFKBP constructs also appear to fold and unfold via a two-state process at the extremes of urea concentrations (solid circles and solid triangles, respectively). Table 1 shows that the folding rate of gsmFKBP in the absence of denaturant, k°f, is ∼40% slower than C22A (2.85 s−1 vs. 4.46 s−1, respectively), whereas the unfolding rate, k°u, is more than three times faster (0.054 s−1 vs. 0.014 s−1, respectively). The gsgFKBP variant folds with a rate similar to gsmFKBP (2.73 s−1), but unfolds more slowly than C22A FKBP (0.007 s−1). The ratio of the unfolding and folding rates does not equal the equilibrium constant for denaturation obtained from ΔG°unf in each protein construct due, at least in part, to the fact that the kinetic values reported here do not account for subsequent kinetic phases occurring on significantly longer time frames that are due to prolyl isomerization (Veeraraghavan et al. 1996; Main et al. 1999).
Fig. 3.
Chevron plot summarizing the kinetic data for C22A (▪; solid line), gsmFKBP (•; broken line), and gsgFKBP (▴; dotted line). Curves represent best fit of data at high and low urea concentrations to equation. 2. Each data point represents the average of three independent determinations each comprising the average of 15 individual kinetic traces. Kinetic parameters were determined by using only the solid symbols. The open symbols represent kinetic data for gsmFKBP (○) and gsgFKBP (▵) near the equilibrium denaturation midpoint.
We observed a more interesting effect on the folding and unfolding reactions due to the N-terminal extension when the kinetics were measured near the midpoint of the equilibrium denaturation. Specifically, the observed kinetic rates of gsmFKBP in 1.0–3.0 M urea strongly depended on whether the reaction was initiated from the native or unfolded states, as evidenced from the divergence in the chevron plot in this region (Fig. 3 ▶, open circles). The additional kinetic phase observed in gsmFKBP refolding is characterized by a faster rate of unfolding (top set of open circles) than was observed for unfolding from the native state (bottom set of open circles) at the same urea concentrations (listed as Intermediate in Table 1). The gsgFKBP construct also deviates from the classical V-shaped chevron plot at urea concentrations near the equilibrium denaturation midpoint, but the effect is less pronounced than in gsmFKBP. The chevron plot for gsgFKBP resembles the superposition of two chevron plots that intersect near the equilibrium denaturation midpoint for this protein (open and closed triangles in Fig. 3 ▶). In general, nonlinearity on the unfolding arm of the chevron plot, that is, at denaturant concentrations greater than the equilibrium denaturation midpoint, indicates a broad transition state the changes slightly in response to increasing denaturant concentration (Otzen et al. 1999). On the other hand, nonlinearity on the low concentration side of the denaturation midpoint indicates the formation of kinetic intermediate(s) on the folding pathway (Parker et al. 1995; Raschke and Marqusee 1997). The discontinuity observed at urea concentrations near the D50 of gsmFKBP and gsgFKBP, but not C22A FKBP, presents an extreme example of nonlinearity in a chevron plot. Evidently, the C22A molecule folds and unfolds via a simple two-state mechanism, whereas the extended FKBP variants do not.
The folding pathway of FKBP is not documented as thoroughly as that of other proteins. Therefore, in contrast to the thermodynamic results, a structural explanation for the observed kinetic changes is not immediately apparent, and mechanisms proposed for intermediate formation must be regarded as speculative at this stage. One potential source of folding intermediates in FKBP is the loop crossing (Michnik et al. 1991). Loop crossings are not commonly observed in antiparallel β-sheet proteins, but are thought to slow folding and possibly generate kinetic folding intermediates when present (Richardson 1977; Ptitsyn and Finkelstein 1980). However, the loop crossing seems an unlikely source for the folding intermediates observed here because wild-type FKBP and C22A, which contain this loop crossing, do not exhibit kinetic folding intermediates on this time scale.
The observed folding intermediate must reflect a change in how the N terminus packs into the rest of the protein in the extended variants. Φ-value analysis and molecular dynamics simulations indicate that the β-sheet structure in FKBP is loosely formed in the folding transition state, in particular with respect to βI (Fulton et al. 1999). Mutations to residue I7, which is at the C-terminal end of βI and near the loop crossing, have a low φ-value (0.1) compared with mutations to V2, which is at the N-terminal end of βI (φ-value of 0.5) (Fulton et al. 1999). Fractional φ-values are difficult to interpret structurally, but it is possible that the N-terminal part of β1 is more native like at the transition state than residues at the C-terminal end of β1. In other words, the N-terminal end of β1, at the free end of the polypeptide chain, may form the initial contacts that lead to β-strand formation, rather than strand formation starting from the C-terminal end of β1. Perhaps the three additional residues at the N terminus of FKBP alter interaction of βI with residues in the single-turn helix during folding. These interactions may force this β-strand to pack into the rest of the protein via a different route, resulting in the formation of a kinetic-folding intermediate.
Our results point to an important role for the N terminus in the folding of FKBP, which exhibits considerable sequence variability among FKBP homologs. Homologs such as the E. coli SlyD (Hottenrott et al. 1997) and the MbFK from Methanobacterium thermoautotrophicum (Ideno et al. 2000), lack the N-terminal β-strand, β1 (using the nomenclature for the FKBP12). In these proteins, the N terminus begins in the loop connecting β1 and β4 in FKBP12, which presumably eliminates the loop crossing found in longer FKBPs. On the other hand, the FKBP domain from hsp56 (FKBP59) contains an additional 30 residues preceding the N terminus of FKBP12 (Craescu et al. 1996). These residues form a small loop and a short β-strand that pairs with the strand that is structurally homologous to β1 in FKBP12. Investigating the folding and unfolding reactions of these variants may provide additional insights into the role of the N terminus in FKBP folding.
In conclusion, a three-residue extension on the N terminus of FKBP slightly alters the thermodynamic stability, but more dramatically, alters the kinetic-folding pathway of the protein. Both the extension and its the amino acid sequence contribute to this effect. Like many proteins, the N-terminal Met residue is not present in mature FKBP12 isolated from eukaryotic sources (Harding et al. 1988), and is also removed when expressed recombinantly in prokaryotes (Standaert et al. 1990). In other words, translation of the FKBP cDNA results in a protein that is apparently too long by one residue at the N terminus. Methionine residues are frequently removed proteolytically from polypeptide chains either co- or post-translationally (Bradshaw et al. 1998), and it is interesting to speculate about why the N-terminal Met residue of FKBP is removed. The N-terminal residue is linked to the proteins in vivo half-life, as developed in the N-end rule (Varshavsky 1996). Gly, the first residue in mature FKBP, is a stabilizing residue against proteolytic degradation via the ubiquitin pathway, as is Met, so this cannot explain the Met removal in FKBP. Instead, our data suggests that the N-terminal Met residue in FKBP, and perhaps that of other proteins, is removed to stabilize the protein against denaturation and to improve the folding efficiency by removing a kinetic intermediate on the folding pathway.
Materials and methods
Protein expression and purification
The DNA fragment encoding the FKBP structural gene including the initiator methionine was cloned into a modified pMALc2 plasmid by PCR. In the modified plasmid, the proteolytic cleavage site for Factor Xa was replaced with that for thrombin. In addition, a spacer containing eight consecutive histidine residues was introduced between MBP and the thrombin protease cleavage site to improve the separation of FKBP from the MBP carrier protein after cleavage (Pryor and Leitung 1997). Protein was expressed in BL21(DE3) cells in LB medium, and induced by addition of 0.3 mM isopropylthiogalactoside (IPTG). After a 4-hour-expression period, cells were collected by centrifugation and lysed by passage through a French pressure cell. The clarified lysate was loaded onto an amylose column. The fusion construct was eluted by use of 10 mM maltose and cleaved with thrombin, using 1 unit/mg fusion construct, overnight at 30°C, leaving an additional three-amino-acid residue at the N terminus, Gly–Ser–Met (gsmFKBP). After exchange into loading buffer (20 mM TRIS at pH 7.9, containing 500 mM NaCl and 5 mM imidazole), the cleaved sample was applied to a Ni2+-column, and gsmFKBP was collected in the flow through. Trace MBP contaminants remaining after the Ni2+-column were removed by purification over Q-Sepharose (pH 8.0) in 20 mM TRIS containing 25 mM NaCl. A derivative of gsmFKBP was created in which the methionine residue was modified to glycine (gsgFKBP) by use of PCR. Purification of gsgFKBP was performed as described above except that cleavage was performed on a smaller scale to maximize the reaction yield. C22A FKBP was expressed by use of the pFKSAL di-cistronic plasmid (Pilot-Mathias et al. 1993), and purified as described previously (Zhang et al. 1997).
Isothermal equilibrium denaturation
Purified protein was concentrated and exchanged into 50 mM sodium phosphate buffer containing 25 mM NaCl (pH 6.4) for storage. Samples for isothermal equilibrium denaturation were generated by adding the appropriate amount of protein to solutions containing the desired final urea concentration, and allowed to come to equilibrium at 30°C prior to monitoring the extent of unfolding by fluorescence. Urea concentrations were determined by measuring the refractive index of the solution (Pace 1986). Isothermal equilibrium denaturation was monitored by the change in the intrinsic fluorescence of W59 (emission spectra) following established protocols (Egan et al. 1993). Fluorescence intensity was integrated from 310–500 nm and normalized to protein concentration. The data were fit to a model that assumes a two-state transition (Equation 1) by use of a Marquardt-Levenson algorithm implemented in Kaleidagraph.
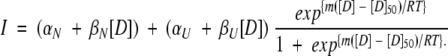 |
1 |
In this equation, derived from the linear extrapolation model (Santoro and Bolen 1988), αN (αU) and βN (βU) are the intercepts and slopes of the pre-(post)-transition baselines, respectively, D is the denaturant concentration, m is the molar cosolvation free energy, and D50 is the denaturant concentration at which the protein is 50% in the native state. The free energy of unfolding in the absence of denaturant, ΔG°unf, was determined by
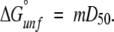 |
2 |
Stopped-flow kinetic folding and unfolding
Stopped-flow experiments were performed by use of a Kintec SF-100 at 30°C. Folding and unfolding reactions were followed by fluorescence emission selected by use of a 340-nm interference filter after excitation at 280 nm. Unfolding or folding were initiated by diluting protein solution into unfolding or folding buffer starting from native or unfolded protein, respectively. The kinetic traces, averaged from 13–15 single shots, were fitted to a single exponential term for the time frame between 50 ms and 1 sec. The rate for each individual concentration below 2.0 M and above 3.0 M urea was fitted to a kinetic scheme that assumes a two-state reaction in which the dependence of the unfolding and refolding rates are assumed to have a linear dependence on urea concentration,
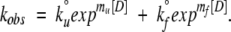 |
3 |
In Equation 3, k°f and k°u represent the folding and unfolding rates in the absence of denaturant. The folding (mf) and unfolding (mu) kinetic m-values, which are analogous to the equilibrium m-values, report on structural changes as the protein approaches the transition state from the native and unfolded states, respectively.
Acknowledgments
This work was supported by National Institutes of Health grant no. GM-54035 (awarded to T.M.L.) and by the National High Magnetic Field Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/
References
- Bradshaw, R.A., Brickely, W.W., and Walker, K.W. 1998. N-Terminal processing: The methionine aminopeptidase an Nα-acetyl transferase families. Trends Biochem. Sci. 23 263–276. [DOI] [PubMed] [Google Scholar]
- Craescu, C.T., Rouviére, N., Popescu, A., Cerpolinin, E., Lebeau, M.-C., Baulieu, E.-E., and Mispelter, J. 1996. Three dimensional structure of the immunophilin-like domain of FKBP59 in solution. Biochemistry 35 11045–11052. [DOI] [PubMed] [Google Scholar]
- Davis, G.D., Elisee, C., Newham, D.M., and Harrison, R.G. 1999. New fusion protein systems designed to give soluble expression in Eschericia coli. Biotechnol. Bioengin. 65 382–388. [PubMed] [Google Scholar]
- Egan, D.A., Logan, T.M., Liang, H., Matayoshi, E., Fesik, S.W., and Holzman, T.F. 1993. Equilibrium denaturation of recombinant human FK binding protein in urea. Biochemistry 32 1920–1927. [DOI] [PubMed] [Google Scholar]
- Fulton, K.F., Main, E.R.G., Daggett, V., and Jackson, S.E. 1999. Mapping the interactions present in the transition state for unfolding/folding of FKBP12. J. Mol. Biol. 291 445–461. [DOI] [PubMed] [Google Scholar]
- Harding, M.W., Galai, A., Uehling, D.E., and Schreiber, S.L. 1988. A receptor for the immuno-supressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341 758–760. [DOI] [PubMed] [Google Scholar]
- Hottenrott, S., Schumann, T., Plückthun, A., Fischer, G., and Rahfield, J.U. 1997. The Eschericia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans isomerase. J. Biol. Chem. 272 15697– 15701. [DOI] [PubMed] [Google Scholar]
- Ideno, A., Yoshida, T., Furutani, M., and Maruyama, T. 2000. The 28.3 kDa FK506 binding protein from a thermophilic archaeum, Methanobacterium thermautotrophicum, protects the denaturation of proteins in vitro. Eur. J. Biochem. 267 3139–3148. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Keifer, P., and Saarinen, T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114 10663–10665. [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Main, E.R.G., Fulton, K.F., and Jackson, S.E. 1998. Context-dependent nature of destabilizing mutations on the stability of FKBP12. Biochemistry 37 6145–6153. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Folding pathway of FKBP12 and characterisation of the transition state. J. Mol. Biol. 291 429–444. [DOI] [PubMed] [Google Scholar]
- Meadows, R.P., Nettesheim, D.G., Xu, R.X., Olejniczak, E.T., Petros, A.M., Holzman, T.F., Severin, J., Gubbins, E., Smith, H., and Fesik, S.W. 1993. Three-dimensional structure of the FK506 binding protein/ascomycin complex in solution by heteronuclear three- and four-dimensional NMR. Biochemistry 32 754– 765. [DOI] [PubMed] [Google Scholar]
- Michnik, S., Rosen, M.K., Wandless, T.J., Karplus, M., and Schreiber, S. 1991. Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science 252 836. [DOI] [PubMed] [Google Scholar]
- Otzen, D.E., Kristensen, O., Proctor, M., and Oliveberg, M. 1999. Structural changes in the transition state of protein folding: Alternative interpretations of curved chevron plots. Biochemistry 38 6499–6511. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods. Enzymol. 131 266–280. [DOI] [PubMed] [Google Scholar]
- Park, S.T., Aldape, R.A., Futer, O., DeCenzo, M.T., and Livingston, D.J. 1992. PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J. Biol. Chem. 267 3316–3324. [PubMed] [Google Scholar]
- Parker, M.J., Spencer, J., and Clarke, A.R. 1995. An integrated kinetic analysis of intermediates and transition states in protein folding rections. J. Mol. Biol. 253 771–786. [DOI] [PubMed] [Google Scholar]
- Pilot-Mathias, T., Pratt, S.D., and Lane, B.C. 1993. High-level synthesis of the 12-kDa human FK506-binding protein in Eschericia coli using translational coupling. Gene 128 219–225. [DOI] [PubMed] [Google Scholar]
- Pryor, K.D. and Leitung, B. 1997. High-level expression of soluble protein in Eschericia coli using a His6-tag and maltose binding protein double affinity fusion system. Protein Exp. Purific. 10 309–319. [DOI] [PubMed] [Google Scholar]
- Ptitsyn, O.B. and Finkelstein, A.V. 1980. Similarities of protein topologies: Evolutionary divergence, functional convergence, or principles of folding? Q. Rev. Biophys. 13 339–386. [DOI] [PubMed] [Google Scholar]
- Raschke, T.M. and Marqusee, S. 1997. The kinetic folding intermediate of ribonuclease H resembles the acid molten globule and partially unfolded molecules detected under native conditions. Nature Struct. Biol. 4 298–304. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. 1977. β-Sheet topology and the relatedness of proteins. Nature 268 495–500. [DOI] [PubMed] [Google Scholar]
- Santoro, M.W. and Bolen, D.W. 1988. Unfolding free energy changes determined by the linear extrapoloation method. I. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27 8063–8068. [DOI] [PubMed] [Google Scholar]
- Schellman, J.A. 1987a. Selective binding and solvent denaturation. Biopolymers 26 549–559. [DOI] [PubMed] [Google Scholar]
- ———. 1987b. The thermodynamic stability of proteins. Annu. Rev. Biophys. Chem. 16 115–137. [DOI] [PubMed] [Google Scholar]
- ———. 1994. The thermodynamics of solvent exchange. Biopolymers 34 1015–1026. [DOI] [PubMed] [Google Scholar]
- Scholz, C., Zarnt, T., Kern, G., Lang, K., Burtscher, H., Fischer, G., and Schmid, F.X. 1996. Autocatalytic folding of the folding catalyst FKBP12. J. Biol. Chem. 271 12703–12707. [DOI] [PubMed] [Google Scholar]
- Standaert, R.F., Galat, A., Verdine, G., and Schreiber, S.L. 1990. Molecular cloning and overexpression of the human FK506-binding protein FKBP. Nature 346 671–674. [DOI] [PubMed] [Google Scholar]
- van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L., and Clardy, J. 1991. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252 839–842. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229 105–124. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A. 1996. The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. 93 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraraghavan, S., Holzman, T.F., and Nall, B.T. 1996. Autocatalyzed protein folding. Biochemistry 35 10601–10607. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Yamashita, M.M., Sintchak, M.D., Rotstein, S.H., Murcko, M.A., Boger, J., Thompson, J.A., Fitzgibbon, M.J., and Navia, M.A. 1995. Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unligated for and in complex with FK506 and rapamycin. Acta Cryst. D. 51 511–517. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Li, W., Logan, T.M., Li, M., and Marshall, A.G. 1997. Human recombinant [C22A] FK506 binding protein amide hydrogen exchange rates from mass spectrometry match and extend those from NMR. Protein Sci. 6 2203–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]