Abstract
β-Turns are sites at which proteins change their overall chain direction, and they occur with high frequency in globular proteins. The Protein Data Bank has many instances of conformations that resemble β-turns but lack the characteristic N–H(i) → O=C(i − 3) hydrogen bond of an authentic β-turn. Here, we identify potential hydrogen-bonded β-turns in the coil library, a Web-accessible database utility comprised of all residues not in repetitive secondary structure, neither α-helix nor β-sheet (http://www.roselab.jhu.edu/coil). In particular, candidate turns were identified as four-residue segments satisfying highly relaxed geometric criteria but lacking a strictly defined hydrogen bond. Such candidates were then subjected to a minimization protocol to determine whether slight changes in torsion angles are sufficient to shift the conformation into reference-quality geometry without deviating significantly from the original structure. This approach of applying constrained minimization to known structures reveals a substantial population of previously unidentified, stringently defined, hydrogen-bonded β-turns. In particular, 33% of coil library residues were classified as β-turns prior to minimization. After minimization, 45% of such residues could be classified as β-turns, with another 8% in 310 helixes (which closely resemble type III β-turns). Of the remaining coil library residues, 37% have backbone dihedral angles in left-handed polyproline II structure.
Keywords: protein structure/folding, structure, computational analysis of protein structure, hydrogen bonds, thermodynamics, hydrodynamics
β-Turns (Venkatachalam 1968) represent the largest category of nonrepetitive protein secondary structure (Rose et al. 1985). Several β-turn subcategories have been identified (Lewis et al. 1971; Kuntz 1972; Chou and Fasman 1977; Richardson 1981; Ramakrishnan and Soman 1982; Kabsch and Sander 1983; Wilmot and Thornton 1988, 1990; Efimov 1993); all are four residues in length with an N–H(i) → O=C(i − 3) hydrogen bond. Upon adopting this motif, the polypeptide chain reverses its overall direction, a frequent occurrence in globular proteins.
Typically, turns are identified from X-ray coordinates based on investigator-defined geometric criteria (Ceccarelli et al. 1981; Jeffrey and Maluszynska 1982; Taylor and Kennard 1983, 1984; Rose et al. 1985; Dasgupta et al. 2004), a highly parameter-sensitive approach near the defined threshold values used to discriminate turns from non-turns (see Table V and Fig. 26 in Rose et al. 1985). In one widely used definition, a four-residue segment with a Cα(i) → Cα(i − 3) distance of 7.0 Å might be classified as a turn while a virtually indistinguishable structure with a corresponding distance of 7.01 Å might not.
We seek to take an accurate census of the hydrogen-bonded turns in proteins by testing whether likely turn conformers can be successfully minimized into tighter geometric criteria without deviating significantly from the original structure. In pursuit of this goal, we take advantage of the recently released coil library (Fitzkee et al. 2005b), a Web-accessible database utility comprised of all residues that do not participate in repetitive secondary structure, neither α-helix nor β-sheet. In a Pisces list of proteins (Wang 2003), this coil library accounts for 44% of total protein structure.
After minimization, we find that 53% of all residues in the coil library participate in β-turns, including 310 helices, which closely resemble type III turns. The largest remaining category is comprised of residues in polyproline II (PII) conformation (Stapley and Creamer 1999). In all, ~80% of protein residues are involved in four hydrogen-bonded backbone structures: α-helices, 310 helices, β-sheet, and β-turns.
Results
Using canonical turn definitions and stringent criteria, 46,409 β-turns were identified, comprising 142,069 (33.4%) of the coil library residues (Table 1); an additional 31,459 residues (7.4%) in 310 helices were found as well. These numbers can be augmented by 20,411 additional β-turns that satisfy relaxed criteria and survive the minimization protocol (see Materials and Methods), bringing our total census to 192,241 (45.2%) coil library residues (Table 1) and 34,848 (8.2%) additional residues in 310 helices. In all, slightly more than half of the residues in the coil library participate in either β-turns or 310 helices.
Table 1.
Fraction of residues in the coil library by category
| Before minimization | After minimization | |||
| Fraction (%) | No. of residues | Fraction (%) | No. of residues | |
| 310 helices | 7.4 | 31,459 | 8.2 | 34,848 |
| Stringent criteria | 33.4 | 142,069 | 45.2 | 192,241 |
| Relaxed criteria | 53.5 | 227,291 | — | — |
| Coil library | 100 | 424,981 | — | 424,981 |
The observed distribution of turn types (Table 2) is similar but not identical to that reported in the literature. The percentage of type I and III turns is slightly larger, and the percentage of type II turns slightly smaller than Richardson’s earlier classification (Richardson 1981). Upon adopting Richardson’s convention of combining type I and III turns—which are often indistinguishable in X-ray structures (Richardson 1981)—slightly more than two-thirds (68.6%) are type I and III turns, and 19.4% are type II turns, with other turn types accounting for the remaining 12%.
Table 2.
Distribution of residues by turn type
| Stringent criteria (%) | Relaxed criteria (%) | After minimization (%) | |
| Type I | 42.6 | 43.2 | 45.9 |
| Type I′ | 2.5 | 1.9 | 2.4 |
| Type II | 17.0 | 15.3 | 19.4 |
| Type II′ | 4.5 | 4.2 | 4.1 |
| Type III | 27.6 | 28.7 | 22.7 |
| Type III′ | 5.3 | 4.0 | 4.5 |
| Type VI(a) | 0.5 | 0.5 | 0.7 |
| Type VI(b) | 0.0 | 0.2 | 0.3 |
| No match | — | 2.0 | — |
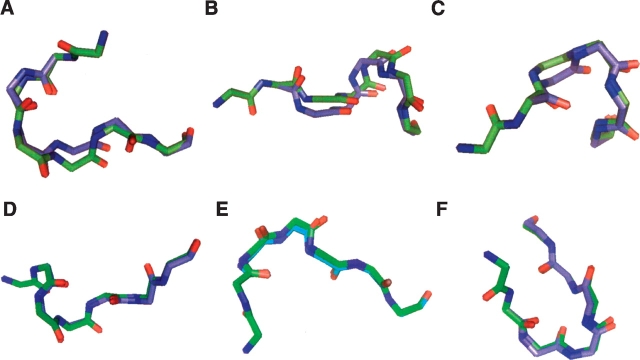
Within the set of 20,411 additional candidate β-turns surviving our protocol, differences in backbone conformation upon minimization were small (Table 3), with no essential change in overall chain direction. Figure 1 ▶ compares the peptide backbone of the experimental (green) and minimized (blue) structures for three typical cases and for three extreme cases. On average, the RMSD of backbone atoms between the starting and minimized turn segments was 0.27 ± 0.07 Å.
Table 3.
Average φ, ψ , variation upon minimization
| Average change in angle ± standard deviation | |
| φ1 | 0.67° ± 23.32° |
| ψ1 | −8.32° ± 21.6° |
| φ2 | 9.31° ± 12.90° |
| ψ2 | −8.05° ± 18.28° |
| φ3 | 5.57° ± 15.91° |
| ψ3 | 3.34° ± 14.67° |
| φ4 | 1.31° ± 26.14° |
| ψ4 | −2.53° ± 23.73° |
Figure 1.
Turns before and after minimization. Experimental (green) and minimized (blue) structures for three extreme examples (A–C) three typical examples (D–F). Each example is seven residues long, with the four-residue β-turn beginning at the second residue. (A) Residues 134–140 of 1bn8 (chain A). (B) Residues 136–142 of 1wos (chain A). (C) Residues 212–218 of 1q16 (chain B). (D) Residues 696–702 of 2nap (chain A). (E) Residues 386–392 of 1ht6 (chain A). (F) Residues 299–305 of 1vjp (chain A). For the entire set, the average RMSD of backbone atoms between the starting and the minimized structures was 0.268 ± 0.065 Å.
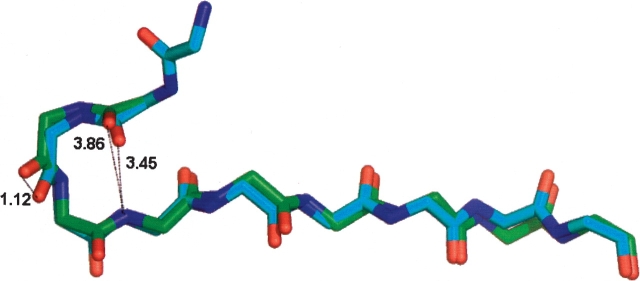
As a specific example, residues 210–213 of 6-phophonoglucolactonase from Thermotoga maritima (1vl1 chain A) fail to satisfy the stringent hydrogen-bonding criteria (donor to acceptor distance is 3.86 Å), but they were identified as a turn candidate using the relaxed angular criteria of Richardson (Fig. 2 ▶). The segment of the protein is clearly a turn by visual inspection. Minimization resulted in a conformation satisfying stringent criteria with only minor changes in φ,ψ angles [Δ (φ1, ψ1) = −14.55°, −9.29°; Δ(φ2, ψ2) = −7.74°, −6.48°; Δ(φ3 , ψ3) = 24.88°, 22.06°; Δ(φ4, ψ4) = 27.35°, 3.04°]. The minimized conformer is a type III turn with an N–H(213) → O=C(210) H-bond donor to acceptor distance of 3.45 Å. The RMSD of all backbone atoms between the initial and the minimized four-residue structure is 0.47 Å, with the largest difference resulting from a 1.12 Å shift in the carbonyl oxygen of residue 211.
Figure 2.
A worst-case example: residues 209 to 218 of 6-phophonoglucolactonase from Thermotoga maritima (1vl1 chain A). Residues 210–213 are a type III turn. The donor to acceptor distance of 3.86 Å in the experimental structure (green) decreases to 3.45 Å in the minimized structure (blue), with an RMSD of 0.47 Å for backbone atoms. The largest change is a 1.81 Å shiftin the carbonyl oxygen positionofresidue 211 in the turn. The positions of segment termini remain virtually superimposable.
In general, changes induced by minimization leave other residues almost completely unperturbed (Fig. 2 ▶), apart from atom positions in the turn segment proper. Such minor changes (Table 3) are usually within the range of uncertainty expected for a typical, X-ray-elucidated protein (Fabiola et al. 2002), although rigorous assessment would require re-refinement of the original structure, which has not been undertaken here.
Discussion
Current X-ray refinement procedures usually lack an explicit hydrogen-bond term (Brooks et al. 1983; Fabiola et al. 2002), and, when present in the force field, it is typically a distance-dependent function without any directional component (Fabiola et al. 2002). Thus, it is hardly surprising that structures refined in this way have fewer apparent hydrogen bonds than percentages found in high resolution X-ray studies of small molecules (Fabiola et al. 2002). Our approach of applying constrained minimization to post-refinement data reveals a substantial population of previously unidentified, stringently defined, hydrogen-bonded β-turns.
Prior to minimization, we find that 25% of geometrically identified β-turns do not participate in an i → (i − 3) hydrogen bond, as assessed by stringent hydrogen-bonding criteria. Following minimization, 87% of these can be reclassified as having a hydrogen bond. The increase in identified turns is accompanied by a corresponding reduction in the remaining fraction of residues that cannot be classified as participating in a hydrogen-bonded back-bone structure (α-helix, β-sheet, 310 helix, and β-turn).
Our study did not include type VIII turns (Wilmot and Thornton 1988, 1990), which are not hydrogen-bonded. When the hydrogen-bonded β-turns identified in our study are supplemented by type VIII turns, only 33% of the coil library (14% of total protein structure) remains unclassified.
There is an extensive literature on hydrogen-bond energy, which is better represented by a potential than a geometric threshold (see, for example, Fig. 2 ▶.1 in Jeffrey and Saenger 1991). Indeed, the energy of interaction between donor and acceptor does not drop to zero beyond some cutoff-value that was selected for computational convenience. Depending on how the figure of merit is defined, slight changes in backbone torsion angles that enhance hydrogen-bond energy may actually result in an improved experimental structure.
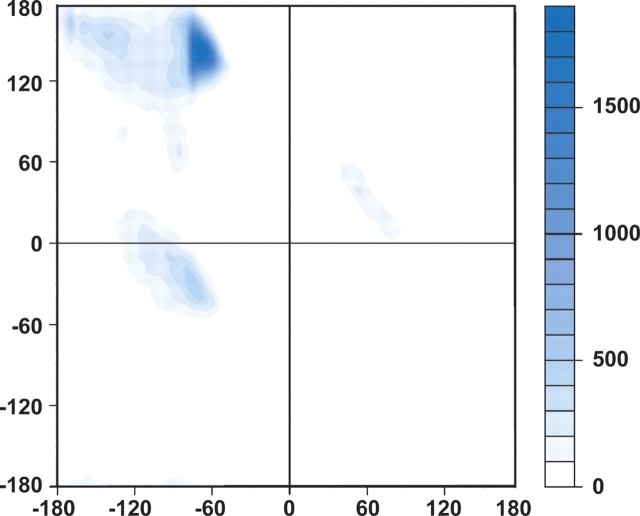
After subtracting all hydrogen-bonded β-turns from the coil library, 37% of the remaining residues are within the polyproline II (PII) region of the dipeptide map (Fig. 3 ▶), a conformation thought to optimize backbone exposure to hydrogen bonds with solvent water (Pappu and Rose 2002). This finding underscores the tendency of residues in folded proteins to participate either in hydrogen-bonded backbone structures or in hydrogen bonds with solvent. The results presented here lend further support to the hydrogen-bonding hypothesis, our proposal that essentially all potential hydrogen-bond donors and acceptors in proteins are satisfied a significant fraction of the time, either by intramolecular hydrogen bonds or by hydrogen bonds to solvent water (Fitzkee et al. 2005a; Fleming and Rose 2005).
Figure 3.
A population plot of φ, ψ-angles in the coil library that are not within β-turns or 310 helices. Populations are indicated by shading according to the scale on the right. The predominant population (37%) is localized to the PII region (Stapley and Creamer 1999).
Materials and methods
We analyzed structures from the Pisces list (Wang 2003) of November 22, 2004, comprising 4175 unique protein chains with <10% aligned sequence identity and with a resolution of at least 2.0 Å and an R factor of better than 25%. The list includes 975,348 residues in all; 424,981 (43.6%) are neither α-helix nor β-sheet and are included in the coil library (Fitzkee 2005b). In our analysis, the coil library was filtered initially to count and remove all 310 helices, after which β-turns were identified and tallied using stringent geometric criteria. Remaining residues were then screened using relaxed turn criteria and candidate structures were minimized; successfully minimized candidates were added to the turn census. These steps are now described in greater detail.
Identification of 310 helices
Residues participating in 310 helices were identified prior to assessing turn candidates. A 310 helix was defined as three or more consecutive residues with backbone dihedral angles of φ, ψ = −60 ± 30°, −30 ± 30°.
Identification of hydrogen-bonded turns using stringent criteria
Our turn definition is based on distance and angular constraints taken from well-resolved, small-molecule X-ray structures (Taylor and Kennard 1983, 1984). Specifically, a β-turn was identified as a four-residue segment in which backbone dihedral angles for the middle two residues (i + 1, i + 2) were within 30° of a canonical turn type (Rose et al. 1985), with an N–H (i + 3) → O=C(i) hydrogen bond (Stickle et al. 1992). Detailed hydrogen-bonding criteria are shown in Figure 4 ▶. In those rare cases where a candidate turn satisfied both type I and type III definitions, the turn was classified as type III. In the case of a type VI turn, the ω angle was required to lie within ±30° of planarity.
Figure 4.
Strict hydrogen-bonding criteria from Stickle et al. (1992). AA is an acceptor antecedent atom which is covalently bound to the acceptor oxygen, O, and AA′ is the antecedent to AA; DD and DD′ are donor antecedent atoms which are covalently bound to the donor nitrogen, N. Hydrogen-bonding criteria were evaluated using heavy atoms because coordinates for hydrogens are not available at the resolution of typical protein X-ray crystal structures. Hydrogen-bond evaluation was based on four criteria: (1) d, N…O distance ≤3.5 Å. This distance represents the sum of the hydrogen-bond radii for an sp2 hybridized nitrogen atom (1.90 Å) and an sp2 hybridized oxygen (1.60 Å). Note that these radii are 10% larger than the corresponding van der Waals radii to account for the presence of hydrogen atoms and to compensate for the fact that protein crystal structures often lack the resolution of small-molecule crystal structures. (2) The angle at the acceptor atom, i, (scalar angle N–O–AA) and the angle at the donor atom, ii, (scalar angle O–N–DD) were in the range 90°–180°. (3) The acceptor O was within the plane of the donor complex. Specifically, planes (N–DD–DD′) and (O–N–DD) were defined and the angle between their normals with within ±60°. (4) The donor was in the plane of the acceptor. Planes γ (O–AA–AA′) and δ (N–O–AA) were defined. The ideal angle between normals to γ and δ is 0°; deviations up to ±90° were accepted.
Identification of hydrogen-bonded turns using relaxed criteria
Some four-residue segments in the coil library have conformations that resemble turns, but with a geometry that fails to satisfy the stringent criteria listed above. Such segments were tested to determine whether they could be successfully minimized into our stringent turn definition without violating experimental constraints. Candidate segments were identified based on either the relaxed hydrogen-bond definition of Kortemme et al. (2003) or the relaxed backbone-angle definition of Hutchinson and Thornton (1994).
Counting turns
The two residues preceding and following a candidate structure were used in the identification of β-turns. For two- or three-residue coil segments, these flanking residues will adjoin an α-helix or β-strand external to the coil library. In such cases, only residues within the coil library proper were added to the count. When two adjacent turn segments had residues in common, the overlapping residues were counted only once.
Minimization protocol
Every potential turn candidate identified using relaxed criteria was subjected to a minimization protocol to determine whether it could be “tweaked” into a similar structure satisfying stringent criteria, including hydrogen-bond geometry derived from small molecule X-ray studies, canonical φ, ψ backbone angles, and satisfaction of hydrogen-bonding requirements for all polar atoms.
Up to 500 attempts of the minimization protocol were performed on an all-atom, nine-residue segment that included the four candidate turn residues. This nine-residue segment was excised from the original X-ray crystal structure, beginning with the residue preceding the potential turn (labeled residue i) and continuing through the four residues following the potential turn.
To determine whether a nearby backbone conformation satisfied strictly defined hydrogen-bonded turn criteria, a Monte Carlo search was performed (Srinivasan et al. 2004), with coordinates of residue i held constant and φ, ψ angles of residues (i + 1)–(i + 4) allowed to vary at random within ±30° of their experimentally determined X-ray values; ω was also allowed to vary by ±5°. Trial conformations were identified as those satisfying stringent hydrogen-bond criteria and within 30° of a canonical turn type (Rose et al. 1985). A total of 1000 trial conformations were generated for further testing.
To maintain downstream conformational integrity, trial conformations were eliminated if the RMSD between the last four α-carbons of the experimental structure and the trial structure exceeded 0.7 Å. Remaining trial conformations were then evaluated for steric clash using published hard-sphere radii (Srinivasan and Rose 1995). Clashes were relieved, if possible, by adjusting the positions of side chain atoms, using a combination of steepest descent and conjugate gradient minimization. Finally, surviving clash-free structures were evaluated using the CHASA procedure (Fleming et al. 2005) to assure that the hydrogen-bonding requirements of all polar atoms were satisfied (Fleming and Rose 2005), either by another H-bond partner within the protein or by a water molecule.
Acknowledgments
We thank Nicholas Fitzkee, Haipeng Gong, and Timothy Street for insightful suggestions and technical help and the Mathers Foundation for support.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051625305.
References
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., and Swaminathan, S. 1983. CHARMM: Chemistry at Harvard Macromolecular Mechanics. A program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem. 4 187–217. [Google Scholar]
- Ceccarelli, C., Jeffrey, G.A., and Taylor, R. 1981. A survey of O-H- - -O hydrogen-bond geometries determined by neutron diffraction. J. Mol. Struct. 70 255–271. [Google Scholar]
- Chou, P.Y. and Fasman, G.D. 1977. β etaturns in proteins. J. Mol. Biol. 115 135–175. [DOI] [PubMed] [Google Scholar]
- Dasgupta, B., Pal, L., Basu, G., and Chakrabarti, P. 2004. Expanded turn conformations: Characterization and sequence-structure correspondence in α-turns with implications in helix folding. Proteins 55 305–315. [DOI] [PubMed] [Google Scholar]
- Efimov, A.V. 1993. Standard structures in proteins. Prog. Biophys. Mol. Biol. 60 201–239. [DOI] [PubMed] [Google Scholar]
- Fabiola, F., Bertram, R., Korostelev, A., and Chapman, M.S. 2002. An improved hydrogen bond potential: Impact on medium resolution protein structures. Protein Sci. 11 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzkee, N.C., Fleming, P.J., Gong, H., Panasik Jr., N., Street, T.O., and Rose, G.D. 2005a. Are proteins made from a limited parts list? Trends Biochem. Sci. 30 73–80. [DOI] [PubMed] [Google Scholar]
- Fitzkee, N.C., Fleming, P.J., and Rose, G.D. 2005b. The Protein Coil Library: A structural database of nonhelix, nonstrand fragments derived from the PDB. Proteins 58 852–854. [DOI] [PubMed] [Google Scholar]
- Fleming, P.J. and Rose, G.D. 2005. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 14 1911–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, P.J., Fitzkee, N.C., Mezei, M., Srinivasan, R., and Rose, G.D. 2005. A novel method reveals that solvent water favors polyproline II over β-strand conformation in peptides and unfolded proteins: Conditional hydrophobic accessible surface area (CHASA). Protein Sci. 14 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1994. A revised set of potentials for β-turn formation in proteins. Protein Sci. 3 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey, G.A. and Maluszynska, H. 1982. A survey of hydrogen-bond geometries in the crystal structures of amino acids. Int. J. Biol. Macromol. 4 173–185. [Google Scholar]
- Jeffrey, G.A. and Saenger, W. 1991. Hydrogen bonding in biological structures. Springer-Verlag, Berlin.
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kortemme, T., Morozov, A.V., and Baker, D. 2003. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein–protein complexes. J. Mol. Biol. 326 1239–1259. [DOI] [PubMed] [Google Scholar]
- Kuntz, I.D. 1972. Protein folding. J. Am. Chem. Soc. 94 4009–4012. [DOI] [PubMed] [Google Scholar]
- Lewis, P.N., Momany, F.A., and Scheraga, H.A. 1971. Folding of polypeptide chains in proteins: A proposed mechanism for folding. Proc. Natl. Acad. Sci. 68 2293–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu, R.V. and Rose, G.D. 2002. A simple model for polyproline II structurein unfolded states of alanine-based peptides. Protein Sci. 11 2437–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, C. and Soman, K.V. 1982. Identification of secondary structures in globular proteins—A new algorithm. Int. J. Peptide Protein Res. 20 218–237. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. 1981. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34 168–340. [DOI] [PubMed] [Google Scholar]
- Rose, G.D., Gierasch, L.M., and Smith, J.A. 1985. Turns in peptides and proteins. Adv. Protein Chem. 37 1–109. [DOI] [PubMed] [Google Scholar]
- Srinivasan, R. and Rose, G.D. 1995. LINUS—A simple algorithm to predict the fold of a protein. Proteins 22 81–99. [DOI] [PubMed] [Google Scholar]
- Srinivasan, R., Fleming, P.J., and Rose, G.D. 2004. Ab initio protein folding using LINUS. Methods Enzymol. 383 48–66. [DOI] [PubMed] [Google Scholar]
- Stapley, B.J. and Creamer, T.P. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickle, D.F., Presta, L.G., Dill, K.A., and Rose, G.D. 1992. Hydrogen bonding in globular proteins. J. Mol. Biol. 226 1143–1159. [DOI] [PubMed] [Google Scholar]
- Taylor, R. and Kennard, O. 1983. Comparison of X-ray and neutron diffraction results for the N–H- - -O=C hydrogen-bond. Acta Crystallogr. 39 133–138. [Google Scholar]
- ———. 1984. Hydrogen-bond geometry in organic crystals. Acc. Chem. Res. 17 320–326. [Google Scholar]
- Venkatachalam, C.M. 1968. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers 6 1425–1436. [DOI] [PubMed] [Google Scholar]
- Wang, G. and Dunbrack Jr., R.L. 2003. PISCES: A protein sequence culling server. Bioinformatics 19 1589–1591. [DOI] [PubMed] [Google Scholar]
- Wilmot, C.M. and Thornton, J.M. 1988. Analysis and prediction of the different types of β-turns in proteins. J. Mol. Biol. 203 221–232. [DOI] [PubMed] [Google Scholar]
- ———. 1990. β-turns and their distortions: A proposed new nomenclature. Protein Eng. 3 479–493. [DOI] [PubMed] [Google Scholar]