Abstract
Replacement of a cis-proline by glycine at position 114 in ribonuclease A leads to a large decrease in thermal stability and simplifies the refolding kinetics. A crystallographic approach was used to determine whether the decrease in thermal stability results from the presence of a cis glycine peptide bond, or from a localized structural rearrangement caused by the isomerization of the mutated cis 114 peptide bond. The structure was solved at 2.0 Å resolution and refined to an R-factor of 19.5% and an Rfree of 21.9%. The overall conformation of the protein was similar to that of wild-type ribonuclease A; however, there was a large localized rearrangement of the mutated loop (residues 110–117—a 9.3 Å shift of the Cα atom of residue 114). The peptide bond before Gly114 is in the trans configuration. Interestingly, a large anomalous difference density was found near residue 114, and was attributed to a bound cesium ion present in the crystallization experiment. The trans isomeric configuration of the peptide bond in the folded state of this mutant is consistent with the refolding kinetics previously reported, and the associated protein conformational change provides an explanation for the decreased thermal stability.
Keywords: RNase A, ribonuclease A, proline, cis, trans, structure, x-ray crystallography, P114G, cesium binding site, protein structure/folding, conformational changes, stability and mutagenesis
Kinetic studies of the folding of bovine pancreatic ribonuclease A (RNase A) have been carried out to identify the pathway that leads to the folded protein. Interpretation of the results from these studies has been complicated due to the cis ↔ trans isomerization of the peptide bonds preceding proline residues that are responsible for forming slowly refolding molecules, as outlined in the proline isomerization model of Brandts et al. (1975). In a number of systems, site-directed mutagenesis has been used to replace the cis prolines with other amino acids in order to simplify the refolding kinetics. In some cases (if a trans peptide bond is formed), this has simplified the refolding kinetics, as predicted by the proline isomerization model. Frequently, however, the refolding kinetics are still complex. One factor that may be partly responsible for the complexity is the persistence of a cis peptide bond preceding the substituted non-proline amino acid, due to conformational constraints imposed by the rest of the protein (Schultz and Baldwin 1992; Schultz et al. 1992; Dodge and Scheraga 1996). This bond would then still have to isomerize from the trans to the cis configuration during the folding process.
Replacement of cis-prolines can also have large effects on protein stability. A conformational change in the protein resulting from isomerization of the mutated cis peptide bond could cause destabilization. Alternatively, because the cis configuration of non-prolyl peptide bonds is unstable relative to that of cis-prolyl peptide bonds, the replacment of cis-proline by any other naturally occurring amino acid that remains cis in the folded state would be expected to destabilize a protein (Drakenberg et al. 1972; Ramachandran and Mitra 1976; Jorgensen and Gao 1988; Radzicka et al. 1988; Schultz et al. 1992).
The current study focuses on the structural consequences of cis-proline replacement in bovine pancreatic ribonuclease A (RNase A), and its relationship to protein folding, stability, flexibility, conformational change, and evolution. RNase A is 124 amino acids in length and contains two cis-prolines, Pro93 and Pro114 (Richards and Wyckoff 1971; Wlodawer and Sjolin 1983; Wlodawer et al. 1988; Robertson et al. 1989; Santoro et al. 1993). Pro114, the focus of this study, is located in a solvent-exposed type VIb-2 β-turn (Pal and Chakrabarti 1999), which is anchored by the Cys58–Cys110 disulfide bond, and at the other end by Pro117, whose mobility is restricted by tightly packed neighboring residues. Measurements of the isomeric ratio in unfolded RNase A (63%) favor the trans configuration of the Asn113–Pro114 peptide bond (Adler and Scheraga 1990). Thus, the cis isomer found for Pro114 in the folded protein must be held in place by constraints imposed by the rest of the protein. The loop containing the cis configuration may be preferentially stabilized relative to the trans configuration by hydrogen bonding, electrostatics, and van der Waals interactions. Alternatively, the formation of a low energy trans configuration of this loop may be prevented by steric clash between residues in the loop with the remainder of the folded protein. Replacement of a cis-proline by glycine at Pro114 in ribonuclease A leads to a large decrease in thermal stability and simplifies the refolding kinetics (Schultz and Baldwin 1992; Schultz et al. 1992; Dodge and Scheraga 1996).
A crystallographic structure determination of a cis Pro114 to Gly mutant (P114G) was undertaken to ascertain whether a trans configuration of the loop is adopted, and to determine whether the structural effects are localized to the loop or the rest of the protein imposes anchorage constraints that force the peptide linkage to remain cis. In addition, we examine the hypothesis (based on the results from kinetic experiments) that the Asn113–Gly114 peptide bond is trans in the folded P114G protein.
Results
Ribonuclease A P114G crystallization
Crystals of the ribonuclease A P114G variant were grown as described in Materials and Methods. The crystallization conditions (30% saturated ammonium sulfate, 3.0 M cesium chloride, 0.15 M sodium phosphate [pH 6.6], 25°C) are similar to those used for the crystallization of ribonuclease S and a high-salt crystal form of ribonuclease A (Kim et al. 1992; P. Harkins and H. Wyckoff, pers. comm.). However, the P114G crystals grew in a new space group, P43212. This crystal form was prone to radiation decay unless stabilized in nearly saturating amounts of ammonium sulfate at 15°C. The crystals diffract to 2.0 Å resolution.
Refinement of the P114G crystal structure
The results of the final refinement using all data from 30.0 to 2.0 Å resolution are summarized in Table 1. Electron density was seen for all residues except parts of the side chains of Lys1, Lys91, and Tyr115. Although the side chains of several residues in RNAse A have been modeled in alternative conformations in highly refined structures (Svensson et al. 1986; Kim et al. 1992), we modeled all side chains in a single conformation. According to the PROCHECK program (Laskowski et al. 1993), the variations from ideality in bond properties lie within the expected ranges, 89% of the residues lie in the most favored regions of a Ramachandran plot, and 100% lie in the allowed regions.
Table 1.
Crystallographic and geometrical parameters at the end of the refinement
| Space group | P43212 |
| Unit cell lengths | |
| a,b (Å ) | 40.87 |
| c (Å ) | 129.25 |
| Resolution range (Å ) | 30.0–2.0 |
| No. of reflections | 7592 |
| Rsymmetrya | 0.053 |
| Rfactor (work)b | 0.195 |
| Rfree (test)c | 0.219 |
| No. of total atoms | 1024 |
| No. of protein atoms | 956 |
| No. of water molecules | 62 |
| No. of cesium atoms | 1 |
| No. of sulfate ions | 1 |
| Deviations from ideal geometry | |
| RMSD on bond lengths (Å ) | 0.005 |
| RMSD on bond angles (°) | 1.20 |
| RMSD on dihedrals angles (°) | 24.2 |
| RMSD on improper angles (°) | 0.6 |
a Rmerge = ∑hkl | Ihkl –〈Ihkl〉| /∑Ihkl, where 〈Ihkl〉 is the weighted
average of all the symmetry-related reflections.
b R = ∑hkl | |Fo| – |Fc| | /∑|Fo|, where Fo and Fc are observed and
calculated structure factors.
c Rfree is calculated as in b but with the test set of reflections.
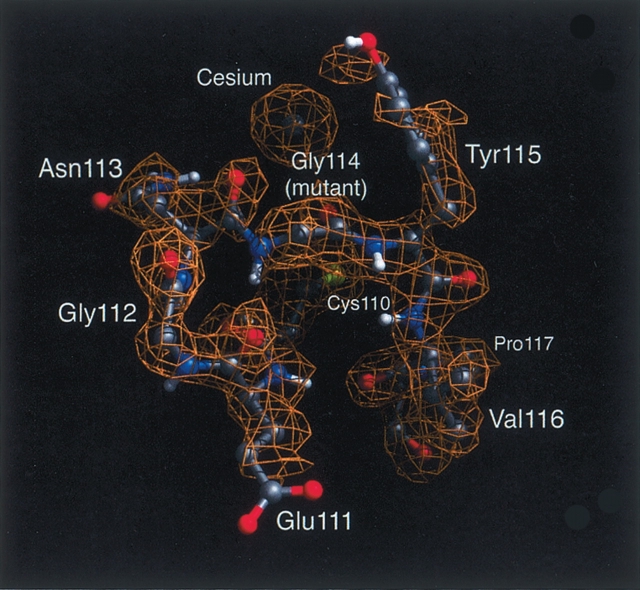
Early in the refinement, before residues 110–116 had been included in the model, examination of the 2Fo–Fc electron density revealed unambiguous density for this loop, which could be modeled only with a trans peptide bond between Asn113 and the mutant Gly114. Further refinement (including residues 110–116, water, and sulfate and cesium ions) (see below) led to a final R-factor of 19.5%. As confirmation of the trans configuration, residues 110–116 and the cesium ion were removed from the model, followed by simulated annealing and Powell minimization to remove model bias (Hodel et al. 1992). The resulting 2Fo–Fc simulated annealing omit map clearly defined the trans peptide bond, including strong density for the carbonyl oxygen of Asn113 (Fig. 1 ▶).
Figure 1.
Electron density surrounding the site of the P114G substitution, with coordinates from the final refined structure. The configuration of the peptide bond preceding the mutated residue 114 is now trans. The fit of the carbonyl groups for residues 113 and 114 to the density confirms the trans configuration. The density corresponding to the bound cesium ion is also easily observed. Nitrogen, carbon, oxygen, and amide hydrogen atoms are colored blue, gray, red, and white, respectively.
Comparison of the P114G and wild-type structures
While several native RNase A structures have been previously reported (Kim et al. 1992), for this study we compared the mutant structure with a native structure crystallized under similar conditions (P. Harkins and H. Wyckoff, pers. comm.). A superposition of the Cα back-bone of this crystal form and P114G is shown in Figure 2A ▶. The differences between the two structures were investigated by the difference distance matrix approach (Kundrot and Richards 1987). The overall mean distance difference between the two structures is 0.35 Å (ES = 0.64 Å) for the Cα’s of all residues, and 0.25 Å (ESD = 0.30 Å) for Cα’s excluding residues 110–116. The latter deviation is not highly significant when compared with the predicted mean atomic coordinate error of 0.20 Å as calculated from a Luzzati plot (Luzzati 1952). The average temperature factors for the backbone and side-chain atoms of P114G and the wild-type structure are similar, further suggesting that only limited changes have occurred in the global protein structure. The position of residues forming the active site (His12, Lys41, Val43, Asn44, Thr45, His119, Phe120, Asp121, and Ser123) are the same in the mutant and the wild type, consistent with the retention of nearly wild-type catalytic activity (Schultz and Baldwin 1992; Schultz et al. 1992).
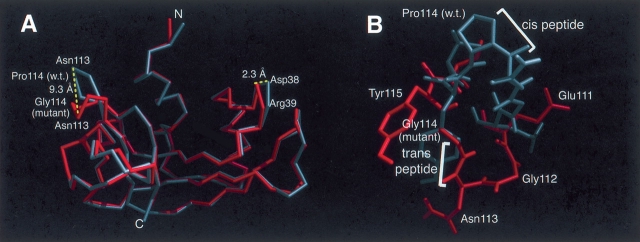
Figure 2.
(A) An alignment of the Cα atoms of the P114G and wild-type RNase A structures (Kim et al. 1992), colored gray and red, respectively. The overall structure is similar; however, there is a large structural rearrangement localized primarily to the loop containing the P114G substitution. The largest movement is the 9.3 Å shift of the Cα of residue 114. (B) A comparison of the P114G and wild-type structures (see Fig. 1 ▶), colored red and gray, respectively, showing the configuration of the mutated loop (residues 111–116) and the associated side chains. Wild-type Pro114 and all of the P114G side chains are labeled. In the wild-type structure, the peptide bond preceding Pro114 is cis, whereas in the P114G structure the peptide bond between Asn113 and Gly114 is trans.
Based on the difference distance plot (Fig. S1), the most significant structural changes are confined to the site of mutation (residues 110–116) (Fig. 2B ▶). Minor structural differences occurred in the 65–70, 35–40, and 91–96 turns. The region of residues 35–40 is known to be variable between different RNase structures (Kim et al. 1992), although the type of turn remains constant. In contrast, in the mutated loop, a new turn type was adopted, resulting in a 9.3 Å movement of the Cα atom of residue 114 (Fig. 2A ▶). The isomerization of the peptide bond preceding residue 114 from cis to trans altered the φ,ψ dihedral angles of the neighboring residues; Gly112 moved from the ~p region (wild type) to the I region (P114G), and the introduced Gly114 was found in the b region (Table S1). Nevertheless, all non-glycine/-proline residues in both structures had dihedral angles that fall in the allowed, low-energy areas of the Ramachandran plot. Along with the isomeric state of the 113–114 peptide bond, the most striking difference between the P114G and wild-type loop conformations is the rearrangement of the hydrogen-bond network. Three hydrogen bonds and two bifurcated hydrogen bonds present in the cis wild-type structure were replaced by four hydrogen bonds and two bifurcated hydrogen bonds in the trans P114G structure (Table S2). In the P114G protein, a new hydrogen bond was formed between Gly114 amide and Glu111 carbonyl oxygen, whereas in the wild-type protein, the proline side chain prevents hydrogen bonding to residue 114. An additional hydrogen bond between Cys110 and Asn113 was observed in the trans P114G structure. The observed dihedral angles and hydrogen-bonding pattern in P114G define the new β-turn as type IIB (Richardson 1981). Figure 2B ▶ shows the alignment of the wild-type and P114G structures, with cis and trans peptide bonds, respectively, preceding residue 114.
Identification of associated ligands using anomalous density
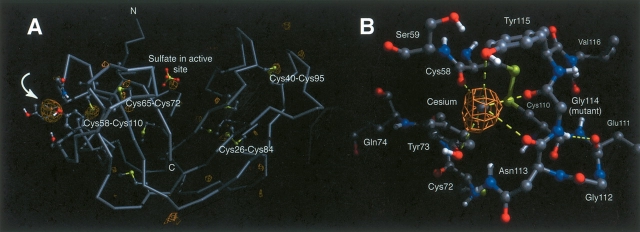
Anomalous difference maps were calculated to confirm the presence of a sulfate ion in the active site (Wlodawer et al. 1988) and to help identify the ligand giving rise to the strong electron density peak next to residue 114. Anomalous electron density (>3σ) was observed for the four disulfide bonds, and two of the five methionine residues. The density also showed a sulfate or phosphate present in the active site, and a very large (>15σ) peak observed in close proximity to residue 114 (Fig. 3A ▶). A comparison of the (relative) anomalous density peak heights suggested that the anomalous scattering was too large to be accounted for by a sulfate ion (f″ = 0.6 e−). Consideration of the ligand coordination geometry and strong anomalous density suggested that a cesium ion (f″ = 8.3 e−) was bound to the protein. While 3 M CsCl was present in the original crystallization conditions, most of it was removed when transferring the crystal into stabilizing solution. The anomalous density and associated ligands are shown in Figure 3B ▶. The predominant interactions and distances are listed in Table 2. The occupancy of the cesium ion (Q = 0.46) was calculated using CNS (Brünger et al. 1998). The B-factor of 24.8 Å2 shows that the cesium is not extraordinarily mobile. The conformation of the loop must be similar with and without bound cesium as no additional density is observed for an alternative loop conformation, despite the partial occupancy of the cesium ion (Q = 0.46). This is further supported by the similarity in B-factors for the cesium and the loop, suggesting that the observed loop conformation is at full occupancy, while the cesium ion is only partially occupied.
Figure 3.
(A) The calculated anomalous difference map (Fo − Fc) for the P114G structure. Electron density is observed at the expected sites of the disulfide bonds, and also in the active site, where phosphate or sulfate is known to bind. The largest patch of anomalous electron density (noted by arrow) is localized near the site of the P114G mutation, and is attributed to the coordination of a cesium ion to the mutated loop. The side chains of all cysteines, methionines, and residues in close spatial proximity to the associated cesium ion are also displayed. (B) Structure of P114G, displaying residues in close proximity to the bound Cs+ ion. The anomalous electron densities for the cesium ion and five protein ligands are highlighted. The remaining coordination ligands are solvent- or symmetry molecule–based.
Table 2.
Cesium ligand distances for P114G ribonuclease A
| Ligand | Atom | Ligand–Ion Distance (Å) |
| Cys58 | Backbone oxygen | 2.96 |
| Tyr73 | Oxygenη | 3.32 |
| Asn113 | Backbone oxygen | 3.18 |
| Gly114 | Backbone oxygen | 3.07 |
| Tyr115 | Oxygenη | 4.32 |
η, ring oxygen.
Discussion
Analysis of the P114G mutant structure
The crystal structure of the P114G RNase A mutant was solved in order to determine the structural consequence of replacing a cis proline, and, in particular, the role that Pro114 plays in influencing RNase A’s stability and refolding. We initially addressed the questions of whether the energetic penalty of placing the substituted amino acid into the cis configuration was larger than the energetic penalty caused by structural rearrangements that would result from the peptide bond’s isomerization, and whether the structural effects of the perturbation are localized or global.
Relationship between evolution and the occurrence of cis peptide bonds
The low frequency of direct replacement of cis-proline in proteins throughout evolution suggests that there is an energetic penalty. Ribonuclease sequences from different species show that although cis Pro114 is usually conserved, it is replaced by Leu in the capybara, and is deleted in the kangaroo, wallaby, and turtle (Beintema et al. 1986). This latter result coincides with the finding that, during evolution, cis-proline replacement is frequently accompanied by a neighboring insertion or deletion. The additional mutations may be necessary to provide increased flexibility to the substituted region so that it can rearrange to a stable structure with all trans peptide bonds. The frequency of cis non-proline peptide bonds is very low (0.03%–0.05%) based on an examination of known crystal structures (Stewart et al. 1990; Weiss et al. 1998; Jabs et al. 1999), whereas cis X-Pro peptide bonds occur commonly in folded proteins (5.2%–6.5%) (MacArthur and Thornton 1991). Therefore, itis not surprising that after cis-proline replacement, the peptide bond adopts the trans configuration (Stewart et al. 1990; Hynes et al. 1994). Further support for this expectation is provided by the unfolded state where the trans isomeric form is the dominant configuration for non-proline peptide bonds (99% trans), whereas for X-Pro peptide bonds (Grathwohl and Wüthrich 1976)the cis form is significantly populated. In the unfolded state of RNase A, the Asn113–Pro114bond is 37% cis (Adler and Scheraga 1990). So what are the structural consequences of simple cis-proline replacement? It necessarily creates either a disfavored cis non-proline peptide bond, or a change in backbone geometry with a trans peptide bond.
Structural effects of the mutation
The pivotal change in the structure is the isomerization of the cis peptide bond to trans. The other large movements in the loop probably are a consequence of the trans isomer. The differences between the structures of wild-type RNase A and P114G are largely confined to the loop containing the mutation, residues 110–117 (Figs. 2A ▶, S1). More subtle long-range effects of the mutation are minor, and we have made no attempt to interpret them because they may also result from crystal packing forces. Remarkably, a large adjustment in this loop (9.3 Å movement) can be accommodated without compromising the integrity of the rest of the protein. It should be noted that although there was no observed electron density (or other evidence) consistent with a cis peptide bond preceding Gly114, an extremely minor population of this isomeric form cannot be ruled out based solely on crystallographic analysis.
Why is the glycine variant trans, whereas the proline in wild-type RNase A is cis?
With the insight gained from the P114G structure we can now address a related question, why is the turn cis in wild-type RNase A when proline is present and what are the different factors that contribute to the isomeric preferences of the folded proteins? In the unfolded state of both P114G and the wild-type protein, the preferred isomeric configuration for the peptide bond preceding Pro114 is trans (wild type ~63% [Adler and Scheraga 1990] vs. 84%–94% in the mutant [Houry and Scheraga 1996]). Our finding that static substitution of the proline side chain of residue 114 in the P114G trans structure introduces steric clashes between the Cγ and Cδ of proline (1.5 Å and 1.4 Å respectively) and the carbonyl of Glu111 provides support for a model where the wild-type trans configuration is strained. Energy minimization of the proline trans structure relieves the steric clash but weakens the hydrogen bonding network, specifically, the hydrogen bond between E111N and Val116O (Table S2). However, when the 113–114 proline peptide bond is cis, less strained backbone conformations are available, which presumably compensate for the energy required for formation of a cis peptide bond. In contrast, in the P114G and P114A mutants, there is a larger driving force to remain trans due to the unfolded state equilibrium and the fact that both glycine and alanine are accommodated in the folded trans configuration without steric clash. A possible additional factor contributing to the formation of a trans peptide bond in the P114G variant is that in the wild-type protein the φψ values of Asn113 (−135.7°, 106.8°) and Pro114 (−58.9°, 151.8°) are not readily compatible with the average values of φψ for residues flanking non-proline cis peptide bonds (123°, 121° and 102°, 152°). Presumably, a more negative value of φ would be required to accommodate a non-proline cis peptide bond in the structure (Jabs et al. 1999; Pal and Chakrabarti 1999).
Influence of the cesium ligand on the cis/trans equilibrium configuration
Interpretation of the structural effects of the mutation must be examined carefully in light of the observation that a Cs+ ion is bound to the P114G variant through the side-chain oxygens of Tyr73 and Tyr115 and the backbone carbonyl oxygens of residues 58, 113, and 114 (Table 2). Several of these residues (113, 114, and 115) are present in the turn variant. The Cs+ ion appears to bind to a pre-existing loop conformation. If the Cs+ ion were driving a distinct conformation, we would expect an alternative conformation to have been visible in the density with an occupancy of up to 0.54. If the putative unbound form were disordered, the observed conformation would reflect this with a partial occupancy or higher B factors. The fact that the 0.46 occupancy Cs+ ion and the full occupancy loop have similar and moderate B factors indicates that the Cs+ ion is not perturbing the conformation of the loop, but is instead binding to a pre-existing conformation.
Comparison with other structural studies of cis-proline replacements in proteins
Interestingly, the generation of an Asn113–trans Gly114 peptide linkage is found here, in contrast with the X-ray structures of carbonic anhydrase II (P202A) (Tweedy et al. 1993), aspartate aminotransferase (P195A) (Birolo et al. 1999), Escherichia coli aspartate transcarbamoylase (P268A) (Jin et al. 2000), and the NMR structural analysis of ribonuclease T1 (P39A), where cis peptide bonds are retained after the substitution (Mayr et al. 1994). Conformational changes associated with either the isomerization of pre-proline peptide linkages, or cis-proline replacement have also been structurally examined in bovine pancreatic ribonuclease A (Pro93 variants) (Pearson et al. 1998; Schultz et al. 1998; Xiong et al. 2000), staphylococcal nuclease variants (Evans et al. 1987, 1989; Hinck et al. 1993; Hodel et al. 1993, 1994, 1995a, Hodel et al. b; Hynes et al. 1994; Maki et al. 1999), and calbindin (Svensson et al. 1992). These studies demonstrate that either isomer can be accommodated, sometimes both isomers are found, or the isomer choice is dependent on the characteristics of the introduced amino acid. The cis peptide bond adopts a trans configuration in a P137A variant of aspartate aminotransferase (Birolo et al. 1999) and in a P76A variant of a protein fragment complementation system of thioredoxin (Yu et al. 2000).
Proline isomerization and effects on protein folding
The effects of cis-proline replacement on protein-folding kinetics have been examined in thioredoxin (Kelley and Richards 1987), RNase T1 (Kiefhaber et al. 1990a,b; Mayr and Schmid 1993; Mayr et al. 1993), aspartate aminotransferase (Birolo et al. 1999), pectate lyase C (Kamen and Woody 2002), and ribonuclease A (Schultz and Baldwin 1992; Schultz et al. 1992; Dodge and Scheraga 1996; Wedemeyer et al. 2002). In the case of RNase A (Pro93) and RNase T1 (Pro39), TEM-1 β-lactamase (Pro167) (Vanhove et al. 1996), α subunit of trp synthase (Pro28) (Wu and Matthews 2002), the folding kinetics were found to be consistent with the presence of introduced non-proline cis peptide bonds. This was later confirmed by structural analysis (Odefey et al. 1995; Pearson et al. 1998; Xiong et al. 2000). In contrast to the folding kinetic complexity that might occur if a non-Pro residue can adopt the cis isomer (in the examples above), the folding kinetics of the P114G variant are simplified. In addition, the result reported here that the P114G mutant has a trans peptide bond preceding residue 114 is consistent with the finding that the unfolded wild-type species UF, which is postulated to contain a trans Asn113–Pro114 peptide bond, can fold into a stable trans conformation before isomerization of the peptide bond to the cis configuration (Dodge and Scheraga 1996; Houry and Scheraga 1996; Juminaga et al. 1997; Bhat et al. 2003). Our finding of a trans P114G peptide bond also agrees with the results of several theoretical and experimental studies showing that a nonnative Pro114 does not perturb the native conformation or interfere with the refolding of RNase A significantly (Stimson et al. 1982; Pincus et al. 1983; Oka et al. 1984; Ihara and Ooi 1985; Schultz and Baldwin 1992).
Materials and methods
Expression, purification, and crystal growth
The ribonuclease A P114G variant was expressed and purified as described previously (Schultz and Baldwin 1992; Schultz et al. 1992). The P114G protein (5 mg/mL) was crystallized from a solution containing sodium phosphate and cesium chloride (0.15 M Na2HPO4, 3 M CsCl2 [pH 6.6]) using the precipitant ammonium sulfate (30% of saturation) at 25°C. Diamond-shaped crystals of dimensions 0.3 × 0.2 × 0.2 mm were obtained after a few days. The crystals were transferred to >95% ammonium sulfate, 0.1 M sodium phosphate (pH 6.6) for stabilization. Crystals were mounted in 0.8-mm glass capillaries with a small amount of mother liquor on both sides. The crystals obtained belonged to the tetragonal space group P41212 or its enantiomorph (P43212), with unit cell dimensions a = b = 40.87 Å, c = 129.25 Å.
Data collection and processing
Diffraction data were collected from a single crystal of P114G at 15°C using an R-axis II imaging plate system. CuKα radiation generated from a Rigaku RU300 rotating anode operating at 50 kV and 90 mA and focused with double Franks’ focusing mirrors. Each 1° oscillation image was collected for 10 min. Images were reduced using DENZO, and scaled and post-refined with SCALEPACK (Otwinowski and Minor 1997). Only frames that did not show significant radiation damage were included. Data were included to a d spacing of 2.0 Å, the highest resolution shell with an average intensity >2σ. 97,542 observations of 8030 reflections covering 97% of the possible reflections to 2.0 Å were collected. The R-merge was 10.7% and the R-factor between Friedel mates was 5.3%.
Structure solution and refinement
The refined structure of wild-type RNase A crystallized under similar high-salt conditions (Kim et al. 1992; P. Harkins and H. Wyckoff, pers. comm.), with solvent molecules, sulfate ligand, and residues 110–116 removed, was used as the molecular replacement search model. All molecular replacement was done with data between 8.0 and 3.5 Å resolution and Fo ≥ 2σ. Rotation functions were performed in XPLOR (Brünger 1987) via a Patterson search method. The orientation of the top solution (7.1σ) was refined using the Patterson correlation coefficient of the squared normalized structure factors as the target. The refined orientation was used in translation searches with the same target using the enantiomorphic space groups P43212 and P41212. Comparison of the solutions indicated the space group as P43212 with a 15.6σ peak in the translation function. The R-factor was 38.1% after rigid body refinement.
Crystallographic least squares refinement was performed using the wild-type high-salt RNase A coordinates (Kim et al. 1992; P. Harkins and H. Wyckoff, pers. comm.) as the starting model. To minimize model bias, residues 110–116, which are the solvent-exposed loop and the site of mutation, were omitted from the starting model. Solvent molecules, the sulfate ligand, and the side chain of Lys1 and Lys91 were also excluded in the initial rounds of refinement. Additionally, the extra methionine at the N terminus, present as a consequence of heterogonous expression in E.coli, was not included in the model. Rigid body minimization, positional, and B-factor refinement were carried out with the program XPLOR (Brünger et al. 1990). The edited model, after application of the rotation and translation solutions, was rigid-body refined against the low-resolution diffraction data (8–3.5 Å, Fo ≥ 2σ) using the crystallographic residual as the target function. An overall temperature factor was then applied to the model. An active-site sulfate ion is typically found in high-salt RNase A and S crystals (Kim et al. 1992). Density was also observed in the active-site region of the P114G mutant and modeled as a sulfate. The model was then further refined against the high-resolution data (6–2 Å, Fo ≥ 2σ) by alternate cycles of positional and individual restrained B-factor refinement to an R-factor of 24.1%. The omitted loop residues (110–116) were manually fit to the electron density of an Fo–Fc map by visual inspection using FRODO (Jones 1985). The final refinement using CNS included all observations (30.0–2.0 Å) and utilized the PMB/CNS automated refinement interface (Scott et al. 2004). Sixty-two waters were placed in the map using CNS (Brünger et al. 1998) and edited by manual inspection using XtalView (McRee 1999). Occupancy of all waters was fixed at unity, waters with a B > 60 Å2 after refinement were removed from the model. The N-terminal Met, originally omitted from the model, was located in a σA-weighted 2mFo–nFc map.
Identification of a bound cesium ion through anomalous differences
A large difference density peak was observed near the site of the P114G mutation in maps calculated with Fo–Fc amplitudes and phases from the refined protein model. The height of this peak was significantly greater than that of the active site sulfate. An anomalous difference map calculated with anomalous difference amplitudes between 8 and 3 Å resolution and using refined model phases, revealed significant density at the same site (12.8σ) and lower density at the sulfate and each disulfide site. Because of the strong anomalous signal, the missing density was attributed to an element with an anomalous scattering f″ component larger than sulfur (f″sulfur = 0.6 e− at CuKα). Consideration of the elements in the crystallization and storage buffers indicated cesium (f″cesium = 8 electrons at CuKα) as the only element that could account for both the normal and anomalous electron density.
The occupancy (Q) and B-factor (B) of the cesium ion were refined as part of the final structure refinement in CNS (Q = 0.46, B = 24.8 Å2). The final model was refined using CNS with all data within 30.0–2.0 Å resolution, using a bulk solvent model. A final round of alternating positional and B-factor refinement (until the Rfree no longer improved) resulted in an R-factor of 19.5% and an Rfree of 21.9%.
Structural analysis
RMS differences between the structures were calculated after superimposing the backbone atoms (N, C, O, and Cα) of the wild-type (high-salt crystal form) (Kim et al. 1992) and P114G mutant using the least-squares methods in CNS (Brünger et al. 1998) and MIDAS (UCSF Graphics Laboratory). Ligands and residues 110–117 were excluded from the superposition. Figures were prepared using the program NEON in the MIDAS package (UCSF Graphics Laboratory). The structure was also analyzed using PROCHECK (Laskowski et al. 1996).
Coordinates
The coordinates and structure factors have been deposited in the Protein Data Bank (accession code 1KH8).
Acknowledgments
This work is dedicated to the memory of Professor Harold W. Wyckoff, our mentor, colleague, and friend. This work was supported by The Robert A. Welch Foundation (H-1345 to R.O.F.), The Sealy and Smith Foundation, NIH grants to R.O.F. (GM51332 and AI23923), and The Howard Hughes Medical Institute (R.O.F.). We thank Drs. Harold Wyckoff, Eunice Kim, Paul Harkins, Jonathan Friedman, Alec Hodel, and Marc Jacobs for helpful discussions. We also thank the staff at the Yale Center for Structural Biology for their efforts and Dr. David Konkel for editing the manuscript. We thank Drs. Harold Wyckoff and Paul Harkins for providing us with the high-salt ribonuclease A coordinates.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051610505.
Supplemental material: see www.proteinscience.org
References
- Adler, M. and Scheraga, H.A. 1990. Identification of a new site of conformational heterogeneity in unfolded ribonuclease A. J. Protein Chem. 9 583–588. [DOI] [PubMed] [Google Scholar]
- Beintema, J.J., Fitch, W.M., and Carsana, A. 1986. Molecular evolution of pancreatic-type ribonucleases. Mol. Biol. Evol. 3 262–275. [DOI] [PubMed] [Google Scholar]
- Bhat, R., Wedemeyer, W.J., and Scheraga, H.A. 2003. Proline isomerization in bovine pancreatic ribonuclease A. 2. Folding conditions. Biochemistry 42 5722–5728. [DOI] [PubMed] [Google Scholar]
- Birolo, L., Malashkevich, V.N., Capitani, G., De Luca, F., Moretta, A., Jansonius, J.N., and Marino, G. 1999. Functional and structural analysis of cis-proline mutants of Escherichia coli aspartate aminotransferase. Biochemistry 38 905–913. [DOI] [PubMed] [Google Scholar]
- Brandts, J.F., Halvorson, H.R., and Brennan, M. 1975. Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14 4953–4963. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1987. X-PLOR version 3.1: A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Brünger, A.T., Krukowski, A., and Erickson, J.W. 1990. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. A 46 585–593. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Dodge, R.W. and Scheraga, H.A. 1996. Folding and unfolding kinetics of the proline-to-alanine mutants of bovine pancreatic ribonuclease A. Biochemistry 35 1548–1559. [DOI] [PubMed] [Google Scholar]
- Drakenberg, T., Dahlqvist, K.I., and Forsen, S. 1972. Barrier to internal rotation in amides. IV. N, N-Dimethylamides. Substituent and solvent effects. J. Phys. Chem. 76 2178–2183. [Google Scholar]
- Evans, P.A., Dobson, C.M., Kautz, R.A., Hatfull, G., and Fox, R.O. 1987. Proline isomerism in staphylococcal nuclease characterized by NMR and site-directed mutagenesis. Nature 329 266–268. [DOI] [PubMed] [Google Scholar]
- Evans, P.A., Kautz, R.A., Fox, R.O., and Dobson, C.M. 1989. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry 28 362–370. [DOI] [PubMed] [Google Scholar]
- Grathwohl, C. and Wüthrich, K. 1976. The X-Pro peptide bond as an NMR probe for conformational studies of flexible linear peptides. Biopolymers 15 2025–2041. [DOI] [PubMed] [Google Scholar]
- Hinck, A.P., Eberhardt, E.S., and Markley, J.L. 1993. NMR strategy for determining Xaa-Pro peptide bond configurations in proteins: Mutants of staphylococcal nuclease with altered configuration at proline-117. Biochemistry 32 11810–11818. [DOI] [PubMed] [Google Scholar]
- Hodel, A., Kim, S.-H., and Brünger, A.T. 1992. Model bias in macromolecular crystal structures. Acta Crystallogr. A 48 851–858. [Google Scholar]
- Hodel, A., Kautz, R.A., Jacobs, M.D., and Fox, R.O. 1993. Stress and strain in staphylococcal nuclease. Protein Sci. 2 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel, A., Kautz, R.A., Adelman, D.M., and Fox, R.O. 1994. The importance of anchorage in determining a strained protein loop conformation. Protein Sci. 3 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel, A., Kautz, R.A., and Fox, R.O. 1995a. Stabilization of a strained protein loop conformation through protein engineering. Protein Sci. 4 484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel, A., Rice, L.M., Simonson, T., Fox, R.O., and Brünger, A.T. 1995b. Proline cis-trans isomerization in staphylococcal nuclease: Multi-substrate free energy perturbation calculations. Protein Sci. 4 636–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry, W.A. and Scheraga, H.A. 1996. Nature of the unfolded state of ribonuclease A: Effect of cis-trans X-Pro peptide bond isomerization. Biochemistry 35 11719–11733. [DOI] [PubMed] [Google Scholar]
- Hynes, T.R., Hodel, A., and Fox, R.O. 1994. Engineering alternative β-turn types in staphylococcal nuclease. Biochemistry 33 5021–5030. [DOI] [PubMed] [Google Scholar]
- Ihara, S. and Ooi, T. 1985. Energy difference associated with proline isomerization in ribonuclease A. Biochim. Biophys. Acta 830 109–112. [DOI] [PubMed] [Google Scholar]
- Jabs, A., Weiss, M.S., and Hilgenfeld, R. 1999. Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286 291–304. [DOI] [PubMed] [Google Scholar]
- Jin, L., Stec, B., and Kantrowitz, E.R. 2000. A cis-proline to alanine mutant of E. coli aspartate transcarbamoylase: Kinetic studies and three-dimensional crystal structures. Biochemistry 39 8058–8066. [DOI] [PubMed] [Google Scholar]
- Jones, T.A. 1985. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 115 157–171. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L. and Gao, J. 1988. Cis-trans energy difference for the peptide bond in the gas phase and in aqueous solution. J. Am. Chem. Soc. 110 4212–4216. [Google Scholar]
- Juminaga, D., Wedemeyer, W.J., Garduno-Juarez, R., McDonald, M.A., and Scheraga, H.A. 1997. Tyrosyl interactions in the folding and unfolding of bovine pancreatic ribonuclease A: A study of tyrosine-to-phenylalanine mutants. Biochemistry 36 10131–10145. [DOI] [PubMed] [Google Scholar]
- Kamen, D.E. and Woody, R.W. 2002. Identification of proline residues responsible for the slow folding kinetics in pectate lyase C by mutagenesis. Biochemistry 41 4724–4732. [DOI] [PubMed] [Google Scholar]
- Kelley, R.F. and Richards, F.M. 1987. Replacement of proline-76 with alanine eliminates the slowest kinetic phase in thioredoxin folding. Biochemistry 26 6765–6774. [DOI] [PubMed] [Google Scholar]
- Kiefhaber, T., Quaas, R., Hahn, U., and Schmid, F.X. 1990a. Folding of ribonuclease T1. 1. Existence of multiple unfolded states created by proline isomerization. Biochemistry 29 3053–3061. [DOI] [PubMed] [Google Scholar]
- ———. 1990b. Folding of ribonuclease T1. 2. Kinetic models for the folding and unfolding reactions. Biochemistry 29 3061–3070. [DOI] [PubMed] [Google Scholar]
- Kim, E.E., Varadarajan, R., Wyckoff, H.W., and Richards, F.M. 1992. Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures. Biochemistry 31 12304–12314. [DOI] [PubMed] [Google Scholar]
- Kundrot, C.E. and Richards, F.M. 1987. Crystal structure of hen egg-white lysozyme at a hydrostatic pressure of 1000 atmospheres. J. Mol. Biol. 193 157–170. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Moss, D.S., and Thornton, J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231 1049–1067. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Luzzati, V. 1952. Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr. 5 802–810. [Google Scholar]
- MacArthur, M.W. and Thornton, J.M. 1991. Influence of proline residues on protein conformation. J. Mol. Biol. 218 397–412. [DOI] [PubMed] [Google Scholar]
- Maki, K., Ikura, T., Hayano, T., Takahashi, N., and Kuwajima, K. 1999. Effects of proline mutations on the folding of staphylococcal nuclease. Biochemistry 38 2213–2223. [DOI] [PubMed] [Google Scholar]
- Mayr, L.M. and Schmid, F.X. 1993. Kinetic models for unfolding and refolding of ribonuclease T1 with substitution of cis-proline 39 by alanine. J. Mol. Biol. 231 913–926. [DOI] [PubMed] [Google Scholar]
- Mayr, L.M., Landt, O., Hahn, U., and Schmid, F.X. 1993. Stability and folding kinetics of ribonuclease T1 are strongly altered by the replacement of cis-proline 39 with alanine. J. Mol. Biol. 231 897–912. [DOI] [PubMed] [Google Scholar]
- Mayr, L.M., Willbold, D., Rosch, P., and Schmid, F.X. 1994. Generation of a non-prolyl cis peptide bond in ribonuclease T1. J. Mol. Biol. 240 288–293. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Odefey, C., Mayr, L.M., and Schmid, F.X. 1995. Non-prolyl cis-trans peptide bond isomerization as a rate-determining step in protein unfolding and refolding. J. Mol. Biol. 245 69–78. [DOI] [PubMed] [Google Scholar]
- Oka, M., Montelione, G.T., and Scheraga, H.A. 1984. Chain-folding initiation structures in ribonuclease A: Conformational free energy calculations on Ac-Asn-Pro-Tyr-NHMe, Ac-Tyr-Pro-Asn-NHMe, and related peptides. J. Am. Chem. Soc. 106 7946–7958. [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. In Macromolecular crystallography, part A (eds. C.W. Carter Jr. and R.M. Sweet), pp. 307–326. Academic Press, New York. [DOI] [PubMed]
- Pal, D. and Chakrabarti, P. 1999. Cis peptide bonds in proteins: Residues involved, their conformations, interactions and locations. J. Mol. Biol. 294 271–288. [DOI] [PubMed] [Google Scholar]
- Pearson, M.A., Karplus, P.A., Dodge, R.W., Laity, J.H., and Scheraga, H.A. 1998. Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A. Protein Sci. 7 1255–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus, M.R., Gerewitz, F., Schwartz, R.H., and Scheraga, H.A. 1983. Correlation between the conformation of cytochrome c peptides and their stimulatory activity in a T-lymphocyte proliferation assay. Proc. Natl. Acad. Sci. 80 3297–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzicka, A., Pedersen, L., and Wolfenden, R. 1988. Influences of solvent water on protein folding: Free energies of solvation of cis and trans peptides are nearly identical. Biochemistry 27 4538–4541. [DOI] [PubMed] [Google Scholar]
- Ramachandran, G.N. and Mitra, A.K. 1976. An explanation for the rare occurrence of cis peptide units in proteins and polypeptides. J. Mol. Biol. 107 85–92. [DOI] [PubMed] [Google Scholar]
- Richards, F.M. and Wyckoff, H.W. 1971. Bovine pancreatic ribonuclease. In The enzymes (ed. P.D. Boyer), pp. 647–806. Academic Press, New York.
- Richardson, J.S. 1981. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34 167–339. [DOI] [PubMed] [Google Scholar]
- Robertson, A.D., Purisima, E.O., Eastman, M.A., and Scheraga, H.A. 1989. Proton NMR assignments and regular backbone structure of bovine pancreatic ribonuclease A in aqueous solution. Biochemistry 28 5930–5938. [DOI] [PubMed] [Google Scholar]
- Santoro, J., Gonzalez, C., Bruix, M., Neira, J.L., Nieto, J.L., Herranz, J., and Rico, M. 1993. High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 229 722–734. [DOI] [PubMed] [Google Scholar]
- Schultz, D.A. and Baldwin, R.L. 1992. Cis proline mutants of ribonuclease A. I. Thermal stability. Protein Sci. 1 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D.A., Schmid, F.X., and Baldwin, R.L. 1992. Cis proline mutants of ribonuclease A. II. Elimination of the slow-folding forms by mutation. Protein Sci. 1 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, L.W., Hargraves, S.R., Klink, T.A., and Raines, R.T. 1998. Structure and stability of the P93G variant of ribonuclease A. Protein Sci. 7 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E.E., White, M.A., He, Y.A., Johnson, E.F., Stout, C.D., and Halpert, J.R. 2004. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-Å resolution: Insight into the range of P450 conformations and the coordination of redox partner binding. J. Biol. Chem. 279 27294–27301. [DOI] [PubMed] [Google Scholar]
- Stewart, D.E., Sarkar, A., and Wampler, J.E. 1990. Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 214 253–260. [DOI] [PubMed] [Google Scholar]
- Stimson, E.R., Montelione, G.T., Meinwald, Y.C., Rudolph, R.K., and Scheraga, H.A. 1982. Equilibrium ratios of cis- and trans-proline conformers in fragments of ribonuclease A from nuclear magnetic resonance spectra of adjacent tyrosine ring resonances. Biochemistry 21 5252–5262. [DOI] [PubMed] [Google Scholar]
- Svensson, L.A., Sjolin, L., Gilliland, G.L., Finzel, B.C., and Wlodawer, A. 1986. Multiple conformations of amino acid residues in ribonuclease A. Proteins 1 370–375. [DOI] [PubMed] [Google Scholar]
- Svensson, L.A., Thulin, E., and Forsen, S. 1992. Proline cis-trans isomers in calbindin D9k observed by X-ray crystallography. J. Mol. Biol. 223 601–606. [DOI] [PubMed] [Google Scholar]
- Tweedy, N.B., Nair, S.K., Paterno, S.A., Fierke, C.A., and Christianson, D.W. 1993. Structure and energetics of a non-proline cis-peptidyl linkage in a proline-202 → alanine carbonic anhydrase II variant. Biochemistry 32 10944–10949. [DOI] [PubMed] [Google Scholar]
- Vanhove, M., Raquet, X., Palzkill, T., Pain, R.H., and Frere, J.M. 1996. The rate-limiting step in the folding of the cis-Pro167Thr mutant of TEM-1 β-lactamase is the trans to cis isomerization of a non-proline peptide bond. Proteins 25 104–111. [DOI] [PubMed] [Google Scholar]
- Wedemeyer, W.J., Welker, E., and Scheraga, H.A. 2002. Proline cis-trans isomerization and protein folding. Biochemistry 41 14637–14644. [DOI] [PubMed] [Google Scholar]
- Weiss, M.S., Jabs, A., and Hilgenfeld, R. 1998. Peptide bonds revisited. Nat. Struct. Biol. 5 676. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A. and Sjolin, L. 1983. Structure of ribonuclease A: Results of joint neutron and X-ray refinement at 2.0-Å resolution. Biochemistry 22 2720–2728. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A., Svensson, L.A., Sjolin, L., and Gilliland, G.L. 1988. Structure of phosphate-free ribonuclease A refined at 1.26 Å. Biochemistry 27 2705–2717. [DOI] [PubMed] [Google Scholar]
- Wu, Y. and Matthews, C.R. 2002. A cis-prolyl peptide bond isomerization dominates the folding of the α subunit of Trp synthase, a TIM barrel protein. J. Mol. Biol. 322 7–13. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., Juminaga, D., Swapna, G.V., Wedemeyer, W.J., Scheraga, H.A., and Montelione, G.T. 2000. Solution NMR evidence for a cis Tyr-Ala peptide group in the structure of [Pro93Ala] bovine pancreatic ribonuclease A. Protein Sci. 9 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W.F., Tung, C.S., Wang, H., and Tasayco, M.L. 2000. NMR analysis of cleaved Escherichia coli thioredoxin (1-73/74-108) and its P76A variant: Cis/trans peptide isomerization. Protein Sci. 9 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]