Abstract
The new antigen receptor (IgNAR) antibodies from sharks are disulphide bonded dimers of two protein chains, each containing one variable and five constant domains. Three types of IgNAR variable domains have been discovered, with Type 3 appearing early in shark development and being overtaken by the antigen-driven affinity-matured Type 1 and 2 response. Here, we have determined the first structure of a naturally occurring Type 2 IgNAR variable domain, and identified the disulphide bond that links and stabilizes the CDR1 and CDR3 loops. This disulphide bridge locks the CDR3 loop in an “upright” conformation in contrast to other shark antibody structures, where a more lateral configuration is observed. Further, we sought to model the Type 3 isotype based on the crystallographic structure reported here. This modeling indicates (1) that internal Type 3-specific residues combine to pack into a compact immunoglobulin core that supports the CDR loop regions, and (2) that despite apparent low-sequence variability, there is sufficient plasticity in the CDR3 loop to form a conformationally diverse antigen-binding surface.
Keywords: immunoglobulin new antigen receptor, single variable domain, antibody, paratope, shark, cell adhesion molecule
Immunoglobulin new antigen receptors (IgNARs) are a unique antibody isotype found in the serum of sharks (Greenberg et al. 1995; Nuttall et al. 2001). They are bivalent, but target antigen through a single immunoglobulin variable domain (~13 kDa) displaying two complementarity determining region (CDR) loops (Roux et al. 1998; Nuttall et al. 2003). In contrast, conventional antibodies have a variable heavy (VH) + variable light (VL) domain format (~26 kDa) and bind antigen through up to six CDRs (Chothia et al. 1989; Padlan 1994). To compensate for their reduced size, IgNARs encode unusually long and structurally complex CDR3s, which display a high degree of variability (Greenberg et al. 1995; Nuttall et al. 2004).
To date, three IgNAR isotypes have been identified, which vary in the number and configuration of their framework cysteine residues, and time of appearance in shark development (Rumfelt et al. 2002). Type 3 IgNARs, the last discovered, display limited diversity in both the size and composition of their CDR loop regions (Diaz et al. 2002). They appear early in development and are hypothesized to form an early defense against infection prior to maturation of the full adaptive immune response. Both Type 1 and 2 IgNAR levels increase as the shark immune system is exposed to exogenous antigen, and show significant diversity consistent with extensive antibody affinity maturation (Diaz et al. 1999; Dooley et al. 2003).
Recently, both our laboratory (Streltsov et al. 2004) and Stanfield et al. (2004), have reported three-dimensional crystallographic structures for IgNAR variable domains, which provide significant insight into their evolutionary origin and antigen-binding strategy. Interestingly, the IgNAR immunoglobulin fold resembles I-set proteins (e.g., cell adhesion molecules) as much as it does conventional V-set immunoglobulins (e.g., VH/VL antibodies; T-cell receptors), suggesting an early divergence among the molecules of the shark immune system (Streltsov and Nuttall 2005). The crystallographic structures also clearly delineate the Type 1 and Type 2 isotypes. For Type 2, a disulphide bridge usually, though not in our first structures, links the CDR1 and CDR3 regions producing a loop structure extending high above the immunoglobulin framework. In contrast, for Type 1, two conserved framework cysteine residues form disulphide bridges with matching residues within the extended CDR3, distending the loop laterally. These appear to be two related strategies to enhance stability, and concurrently position the extended loop allowing access to cleft-like epitopes, such as the lysozyme active site in one of the reported structures (Stanfield et al. 2004), in a manner similar to that observed in camelid VHHs, the only other naturally occurring single domain antibodies (Muyldermans 2001; Desmyter et al. 2002).
Now, we have solved the first structure of a fully natural Type 2 IgNAR variable domain, and one which possesses a disulphide bridge linking the CDR1 and 3 loops. In addition, the fortuitous close sequence homology to the Type 3 IgNARs has allowed us to model the antigen-binding paratope of this early developmental isotype, and address the question of how limited sequence diversity can still accommodate a wide range of antigen-binding paratopes.
Results
The 12A-9 crystal structure
Protein 12A-9 is an IgNAR single variable domain antibody specific for the Gingipain K protease from Porphyromonas gingivalis (Nuttall et al. 2002). It was originally isolated from a combinatorial library of naturally occurring Type 2 VNAR antibody fragments derived from the wobbegong shark (Orectolobus maculatus) immune repertoire. In common with many IgNAR variable domains, a disulphide bridge links and stabilizes the CDR1 and CDR3 loop regions, in this case connecting residues Cys29 and Cys89.
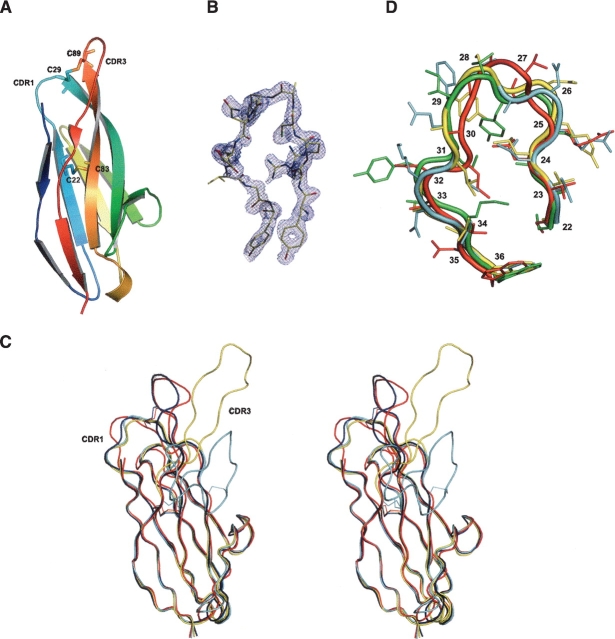
Recombinant 12A-9 protein was purified from the Escherichia coli periplasmic space and placed into a 960-condition robotic crystallization trial. Successful conditions were scaled up and final crystallization conditions were 0.1 M CHES (pH 9.5)/50% PEG200. Diffraction quality crystals (space group P21212) were obtained after 40 d, and the structure of 12A-9 was determined by molecular replacement. The search model for 12A-9 was the previously determined Type 2 IgNAR 12Y-1 (PDB: 1VER) without the CDR3 loop. In the final 12A-9 structure (Fig. 1A,B ▶) 88.4% of the residues are in the most favored regions of the Ramachandran plot, with one residue in the generously allowed region. Details of the diffraction data and refinement statistics are presented in Table 1. The characteristic CDR1-CDR3 disulphide linkage is clearly visible as a disulphide bond in the electron density.
Figure 1.
Structure of the 12A-9 Type 2 IgNAR antibody variable domain. (A) PyMOL image of the 12A-9 molecule in cartoon representation and colored in rainbow fashion (blue N terminus through to red C terminus). Cysteine residues and disulfide bonds are shown as sticks. (B) XtalView/Raster3D image of σA-weighted (2m|Fo|-D|Fc|) electron density of the CDR3 loop of the 12A-9 molecule. The contour level is 1σ. (C) Stereo pair for superimposed structures of IgNAR Type 1, (1SQ2) (cyan); Type 2, 12A-9 (red) and 12Y-2 (1VES) (yellow); and Type 3, modeled (blue) in ribbon representation. Disulphide bonds are shown. (D) PyMol image of superimposed CDR1 loops (22–36) for Type 2 IgNAR’s 12A-9 (red) and 12Y-2 (1VES) (cyan), Type 1 IgNAR (1SQ2) (yellow), and camel cAb-Lys3 (1MEL) (green).
Table 1.
Crystallographic statistics
| 12A-9 | |
| Diffraction data | |
| Space group | P21212 |
| Unit cell (Å) (a,b,c) | 38.27,68.32,39.51 |
| Resolution (Å) (outer shell) | 34.2–2.1 (2.15–2.1) |
| Wavelength (Å) | 1.000 |
| Measured reflections | 43,653 |
| Unique reflections | 6048 |
| Multiplicity | 7.2 (6.3) |
| Completeness (%) (outer shell) | 98.6 (100) |
| I/σ (I) (outer shell) | 15.2 (2.0) |
| χ2 (outer shell) | 1.03 (0.97) |
| Rmerge (%) | 12.6 |
| Refinement | |
| Resolution range (Å) | 34.2–2.10 |
| R % (outer shell) | 21.7 (29.5) |
| Rfree % (outer shell) | 28.0 (39.9) |
| RMS deviations: | |
| Bond length (Å) | 0.012 |
| Bond angles (deg.) | 1.494 |
| Average B values (Å2) | 49.9 |
The structure has an Ig superfamily β-sandwich fold consisting of two β-sheets with seven major β-strands characteristic of IgNAR variable domain antibodies (RMSD of 0.83 Å compared to 1VES for 94 atoms and of 0.72 Å compared to 1SQ2 for 95 atoms) (Fig. 1C ▶). As for other IgNAR’s (Stanfield et al. 2004; Streltsov et al. 2004) the 12A-9 framework exhibits a folding topology which resembles both the intermediate (I-set) and variable (V-set) folds. The similarity to I-set domains is heavily biased by the absence of the extended C′ and C″ strands or conventional CDR2 region, in marked contrast to T-cell receptor and conventional VH/VL antibody V-set domains. The VH/VL “interface” in VNAR Type 2 domains is dominated by the charged/polar residues such as Tyr37, Glu46, Lys82, Lys84, Tyr97, and Glu99 in the 12A-9 structure, which form a charged hydrophilic patch ringed by water molecules. This patch is altered in Type 1 (Stanfield et al. 2004) because the positions 84 and 102 (101 in Type 2) are “covered” by the slightly lateral position of the CDR3, and are therefore replaced with the hydrophobic residues Gly84 and Ala102. However, this CDR3 “cover” now forms another layer, with selection presumably operating at residues such as Tyr91 and Aps93 to reestablish the charged/polar surface typical for IgNAR’s and IgSF I-set domains (Streltsov et al. 2004).
The 12A-9 CDR loop structures
Antibody CDR1s are classically divided into a number of canonical forms describing the structure of this loop (Al-Lazikani et al. 1997). For our previous shark antibody structures analysis of CDR1 residues 28–33 showed similarity to the well-established canonical structures L1 and L2 (Streltsov et al. 2004). However, upon examination of the 12A-9 structure in the context of a more extended CDR1 region (residues 22–36) deviations from this pattern become apparent, revealing a conformation closer to a Type 4 topology. A similar topology is observed in single-domain VHH antibodies and shark Type 1 IgNARs (Decanniere et al. 2000; Stanfield et al. 2004). In Figure 1D ▶ we compare the 12A-9 CDR1 loop with our previously determined Type 2 structure (1VES), with a Type 1 IgNAR (1SQ2), and with the camelid single-domain antibody cAb-Lys3 (1MEL). The alignment of these extended CDR1 regions indicates their similarity, as do the low deviations (12A-9/1VES RMSD = 1.9 Å; 12A-9/1SQ2 RMSD = 2.1 Å; 12A-9/cAb-Lys3 RMSD =1.9 Å, for 15 Cα atoms). In comparison, RMSDs of 12A-9 from canonical Type L1 (1FBJ) and Type L2 (1ZHZ) are 2.9 Å and 3.6 Å, respectively.
The alignment of the 12A-9 CDR1 deviates somewhat from that of the other structures shown, due to the involvement of the key residue Cys29 in binding to the CDR3 loop (Fig. 1D ▶). This impacts on residue Thr27, where the side chain is now rotated outward and is hence more accessible for antigen binding. The CDR1 inside pocket is filled by the bulky hydrophobic residues Leu24 and Leu31, well-conserved in all IgNAR types. This supports the hypothesis (Decanniere et al. 2000) that the combined effect of residues at positions 27, 29, and 31 is critical in formation of the archaetypal Type 4 canonical structure. This structure may be intrinsic to single domain antibodies as it was observed in the camelid and Type 1 structures obtained in complex with antigen, as well as in the uncomplexed Type 2 IgNARs, and is thus unlikely to be induced by antigen binding.
In contrast to the canonical similarity and standard length of the IgNAR CDR1 loop, the CDR3 region exhibits huge sequence and length variability (Supplementary Fig. 1). Structurally, this variability is enhanced by the different strategies of disulphide bond patterns utilized by shark Type 1 and Type 2 antibodies to stabilize the extended CDR3 loops. The 12A-9 structure exhibits the most “upright” CDR3 loop, locked into position by the disulphide bridge with the CDR1 loop (Fig. 1C ▶) and with additional stabilization from H-bonding between the Tyr86 phenolic O-atom and the Asn35 ammonium N-atom (3.4 Å), and between the Tyr86 and Tyr37 phenolic O-atoms (3.6 Å). In contrast, for the Type 1 structures the additional framework residues pull the loop laterally. Where none of these linkages occur, an intermediate position is adopted, for example, for 1VES, where stability is generated by the intraloop hydrogen bond pattern. Thus, of the solved structures, there is continuity between CDR loop positions from “upright” to laterally extended, as dictated by the presence, absence, and position of the stabilizing disulphide bridges (Fig. 1C ▶).
Modeling of the Type 3 IgNAR isotype using the 12A-9 template
Next, we sought to model the IgNAR Type 3 isotype based on the 12A-9 Type 2 structure. Fortuitously, the 12A-9 CDR3 loop is exactly the same length as that found in all but one of the Type 3 IgNARs (Supplementary Fig. 1), and the position of the CDR1-CDR3 disulphide linkage is identical. No other reported IgNAR structures have this particular disulphide bond, and their CDR3 lengths are widely variant (Fig. 2A ▶). Thus, we modeled a typical Type 3 IgNAR (GenBank AAM77191) using the Type 1 and 2 IgNAR structures listed in Figure 2A ▶, with particular emphasis on the core packing and CDR loops.
Figure 2.
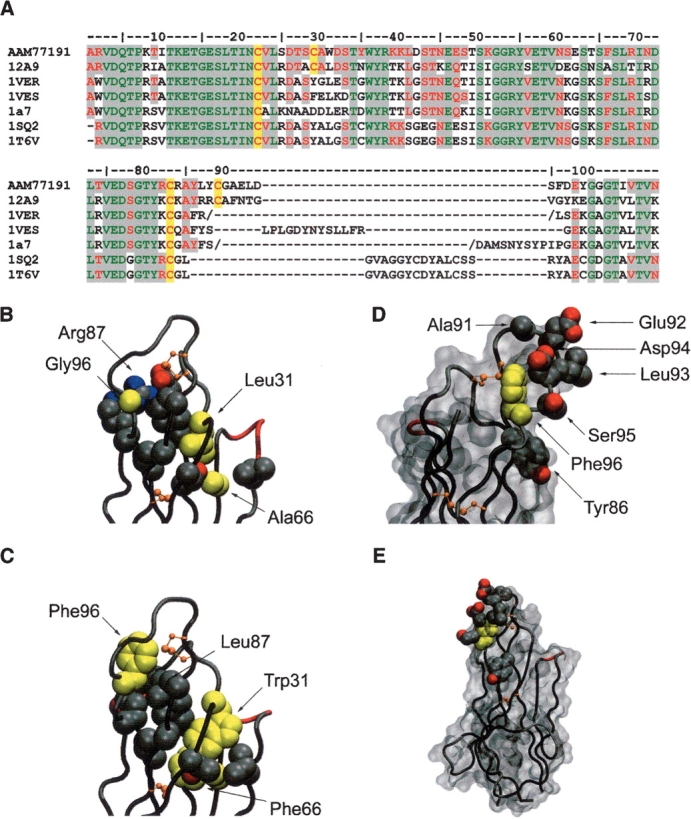
Modeling of the Type 3 IgNAR antibody isotype. (A) Sequence alignment of Type 3 VNAR AAM77191 with structural templates used for modeling. The conserved Ig superfamily framework cysteine residues and those found in the Type 2 and Type 3 CDR3 loop regions are highlighted in yellow. Regions of sequence homology are highlighted in gray (green, residues identical in all known VNAR structures and AAM77191; red, residues conserved in all known VNAR structures and AAM77191). (B) The 12A-9 CDR loops shown in Cα tube representation. Side-chain atoms of residues involved in packing of the hydrophobic core supporting these loops are shown as cpk balls. Modeled residue side chains for Leu31/Trp31 and Gly96/Phe96 (as well as Ala66/Phe66) are shown in yellow. (C) Predicted model of the Type 3 loop in the same representation as for B. (D) Predicted model of the Type 3 CDR3 loop showing the conserved central hydrophobic Phe96 residue in yellow, surrounded by variable residues putatively involved in antigen binding shown as side-chain atoms rendered as cpk balls. A solvent accessible surface is also shown in gray, orientation ~170° rotation in the vertical axis. (E) Same as for D, but in the same orientation as C, and illustrating the relatively small surface area occupied by the hypervariable residues in the context of the complete single domain antibody. Diagrams were constructed in VMD (Humphrey et al. 1996).
CDR stabilizing aromatic residues
Type 3 IgNARs are characterized by an invariant tryptophan (Trp31) within the CDR1 loop, which is not present in Type 1 or Type 2 forms, and which is thought to pack within the Ig framework (Diaz et al. 2002), or alternatively contribute to the CDR1 binding loop. In the Type 2 crystal structures solved to date this position is occupied by Leu31 and this comparatively smaller side chain rotamer is directed toward the Ig framework core, presenting a solvent-exposed surface of ~85.9 Å2 located at the C-terminal end of CDR1 (Fig. 2B ▶). This orientation is positionally and rotamerically conserved, with the largest positional difference being for the 12A-9 structure where there is a backbone Cα shift ~1 Å toward the core.
Detailed modeling of the larger hydrophobic Trp31 side chain in general led to an expected increase in side chain solvent accessible surface area, over a number of different modeling runs (Table 2). However, the increase in solvent-exposed surface area was somewhat less than expected in the top 10 models in each run (as selected by modeler objective score), suggesting that these side chains would be at least partially buried within the existing Type 3 framework. For example, 14/40 of the selected models had the Trp31 side chain rotamer directed into the core of the protein reducing solvent exposure to ~112 Å2, while 6/40 models exhibited the opposite rotamer with the side chain directed toward the solvent but a slight rotation of the backbone leading to greater burial, ameliorating the effects of hydrophobic exposure to solvent (~117 Å2 exposed). While 20/40 models showed the Trp31 side chain rotamers directed toward the solvent, these produced larger solvent-exposed areas (~125 Å2). Given the relative invariance of the position 31 rotamers in the known Type 2 structures, such as 12A-9, and the proximity to the structurally rigid disulphide, we consider these solvent exposed rotamers to be unfavorable and a poor model of the Type 3 Trp31.
Table 2.
Modeler objective scores for Type 3 IgNAR positions Trp31 and Phe96
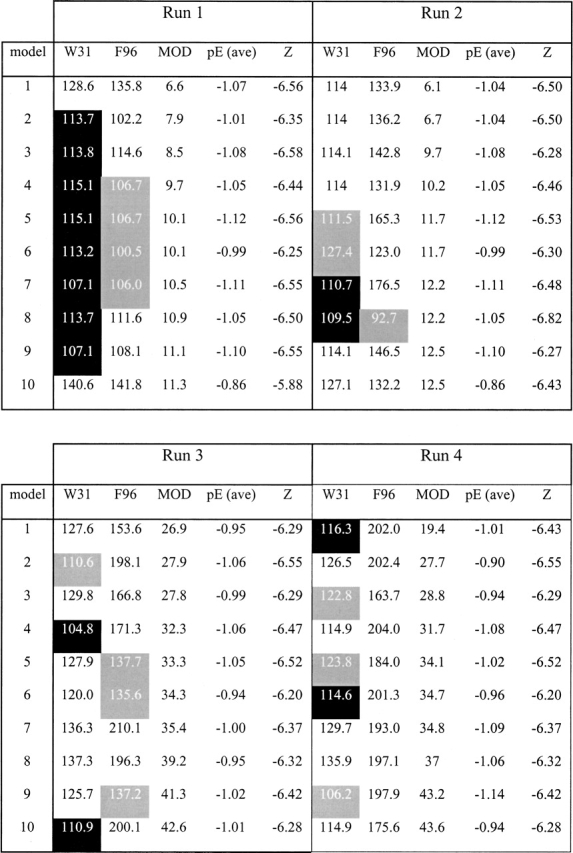
The 10 best scoring models for each modeler run, as described in Materials and Methods. White cells with black text have the side chain directed out away from protein core; black cells with white text have the side chain directed into protein core; gray cells with white text have median exposure to solvent by visual inspection. Numbers are the calculated approximate exposure (Å2) of the side chain atoms to solvent in Å2. MOD = modeler objective energy score. pE (ave) is the average pseudoenergy (~ΔG) and Z is the Z-score of the model calculated in Prosa 3003 for model validation and assessment.
Thus, for Type 3 IgNARs, Trp31 appears to be involved in stabilizing the core of the protein. This was particularly evident when the models were examined for the positions of adjacent side chains. The backbone shift observed in 12A-9 is now reversed (to accommodate the larger Trp31 side chain) and allow it to pack in nearly edge-to-face stacking geometry against the highly conserved Phe66 (Fig. 2C ▶). The “conserved rotamer” Trp31 models show this side chain surrounded by hydrophobic amino acids (Val3, Leu24, Val59, Ala85, and Leu87) allowing formation of hydrophobic contacts. Additionally, Trp31 participates in polar interactions, for example, through (1) the π-electron system of the aromatic rings with the backbone atoms O of Thr64 and N of Ser65 (4.1–4.9 Å), and (2) the H-bond interaction (3.6 Å) of the phenolic O-atoms of Tyr37 and Tyr86, and through the interaction of the phenolic O-atom of Tyr86 with the π-system of the Tyr35 aromatic ring (4.4 Å). These aromatic residues (Tyr35, Tyr37, and Tyr86) are highly to moderately conserved in Type 3 IgNARs (Supplementary Fig. 1). Overall, this tighter core packing (cf. Fig. 2B ▶) appears to reflect a generally stable core that, along with the CDR1-CDR3 disulphide, may further restrict the CDR3 backbone conformations possible in the Type 3 VNARs (Fig. 2C ▶).
We next turned our attention to residue Phe96, a large hydrophobic aromatic residue proximal to the CDRs in Type 3 IgNARs and which appeared to continue a belt of stabilizing framework-CDR hydrophobic residues (Fig. 2C ▶). Again, this residue is invariant in Type 3 IgNARs, despite being within the CDR3 loop region. The equivalent residue in 12A-9 is Gly96 (Fig. 2B ▶), where the lack of a side chain means the Cα has an approximate solvent exposure of ~60.9 Å2 at the end of CDR3 in an otherwise more structurally diverse region (for Type 2 IgNARs). Substitution with the larger aromatic Phe96 in the Type 3 models again gives an unfavorable increase in solvent accessible surface area for the side chain (Table 2); however, Phe96 appears to adopt a number of rotamers in the models, suggesting that it could adopt a number of slightly varying conformations within an otherwise fairly rigid and invariant CDR3 loop structure. Specifically the pairing for the invariant Phe96 and Leu87 in the Type 3 IgNARs is of interest, compared to residues in this position in Type 1/2 IgNARs, where the smaller and more buried Leu87 appears to allow Phe96 to bury itself by adopting an alternative rotamer in high solvent/electrostatic environments. This, combined with the ability of the aromatic Phe96 to participate in both polar and nonpolar interactions, means that it may be exposed, in different rotamers to different interactions with antigen. This significant flexibility has important implications for the diversity of the Type 3 IgNAR as discussed below.
The Type 3 IgNAR antigen-binding interface
Finally, we asked whether our modeling of the Type3 IgNAR could be correlated with its hypothesized role as an early-developmental low-affinity/low-diversity antibody. CDR variability maps almost exclusively to a surprisingly small region of the CDR3 loop, encompassing residues Tyr86, Ala91, Glu92, Leu93, Asp94, and Ser95 (Supplementary Fig. 1). When combined with the adjacent conserved acidic amino acids Asp97 and Glu98, these residues form a large acidic patch surrounding the Phe96. This is not observed in the 12A-9 and the other Type 2 IgNAR structures. In addition, there is a surprisingly high representation of aromatic residues per length of the Type 3 CDR3 region (Tyr86 moderately, and Tyr88, Phe96, and Tyr99 highly conserved). We hypothesize that the structural plasticity of these aromatic residues, in particular Phe96, and the variability of other residues, combine to present a single binding face (Fig. 2D,E ▶). Because of the many conformations of Phe96, the large and charged residues in particular may assume many different orientations in respect to antigen, forming an antigen-binding paratope of far greater diversity than a simple examination of the conserved CDR3 region would suggest.
Discussion
While conventional antibodies combine a vast array of loop conformations to form their antigen-binding paratopes, the shape of these molecular surfaces tend, in general, toward flat or slightly concave topologies (MacCallum et al. 1996). Only rarely do murine and human antibodies break the constraints imposed by the combination of six relatively short CDR loops forming a planar binding surface, usually as the result of powerful selection pressures (Nuttall et al. 2000; Saphire et al. 2001). However, this situation is reversed for the two known examples of single domain antibodies: the IgNARs from sharks, and VHH domains from camelids. With extended CDR3 loops, no associated VL domain, and only two loops to contribute a binding surface, the IgNARs have evolved a radically different antigen-binding paratope with which to target pathogens. This response is most probably complementary to rather than a replacement for the shark IgM response (Dooley and Flajnik 2005). In much the same way camelids encode and utilize both IgG and VHH isotypes (Hamers-Casterman et al. 1993).
However, a problem associated with the IgNAR CDR3s is maintenance of loop stability while retaining both diversity (to target a wide range of antigens) and an extended conformation (suitable for penetrating narrow, cleft-like epitopes). In this report, we describe the structure of a Type 2 IgNAR variable domain antibody, which solves these problems by encoding a disulphide bridge between the CDR1 and CDR3 regions. This effectively constrains the CDR3 in an upright configuration. Similarly, disulphide bonds are a common structural element reinforcing Type 2 and 3 IgNAR variable domains, and the fact that convergent evolution has apparently selected the same solution in camels (CDR1-CDR3 linkages) and llamas (CDR2-CDR3 linkages) confirms this as a powerful structural solution to the problem of loop stability.
The Type 3 IgNARs are hypothesized to form an early line of defense against infection, produced in highest amounts before the antigen-driven antibody response has had time to mature (Diaz et al. 2002). As such they would seem to be required to encode broad binding affinity, however, sequence alignments suggest that they display only minimalist diversity; for example, the CDR3 displays almost zero length variability, due in part to the D1 and D2 diversity elements being germ-line fused (Diaz et al. 2002). Our modeling of the Type 3 isotype suggests that the relative overrepresentation of aromatic residues and the ability of pivotal CDR3 residue Phe96 to adopt a number of different conformations may resolve this apparent contradiction. The importance of aromatic residues in the CDR regions has already been emphasized in several studies of antibody-antigen interfaces (e.g., Hofstädter et al. 1999), and we suggest that the Phe96 is the cornerstone of a diverse antigen-binding face formed by a combination of just a few variable amino acids. While this theory clearly requires validation at the protein chemistry and immunological levels, it is difficult to conceive how Type 3 IgNARs could otherwise perform their role as molecules of immune surveillance. Certainly, more conventional shark antibody isotypes, albeit with greater diversity, have been shown to target an incredibly extensive range of antigens (Adelman et al. 2004).
Alternatively, the possibility exists that Type 3 IgNARs have evolved to target specific classes of pathogenic antigens, which represent particular threats to the embryonic shark. Such a hypothesis takes us firmly into the realm of the innate immunity; and there is a growing awareness of the continuity between the innate and adaptive immune systems (van den Berg et al. 2004). Shark Type 3 IgNARs may combine features of both pathogen recognition systems, providing further evidence of a distinct immunological role for this unique antibody isotype.
Materials and methods
Protein preparation
Recombinant 12A-9 protein was produced as previously described (Nuttall et al. 2002). Briefly, the 12A-9 variable domain coding sequence (GenBank AF466395) was cloned into expression vector pGC (Coia et al. 1997) in-frame with an N-terminal periplasmic targeting peptide, and dual C-terminal FLAG epitope tags. Recombinant protein was expressed into the E. coli TG1 (K12 supE Δ(lac-proAB) thi hsdΔ5 F′ {traD36 proAB+ lacIq lacZΔM15} periplasm as follows. Starter cultures were grown overnight in 2YT medium/ampicillin (100 μg/mL)/glucose (2.0% w/v), diluted 1/100 into fresh 2YT/100 μg/mL ampicillin/glucose (0.1% w/v) and then grown at 37°C/200 rpm to an OD550 nm = 0.2–0.4. Cultures were then induced with IPTG (1 mM final), grown for a further 16 h at 28°C, and harvested by centrifugation (Beckman JA-14/6 K/10 min/4°C). Periplasmic fractions were isolated by the method of Minsky et al. (1986) and recombinant protein purified by affinity chromatography using an anti-FLAG antibody-Sepharose column (10 × 1 cm). The affinity column was equilibrated in PBS (pH 7.4) and bound protein eluted with ImmunoPure gentle elution buffer (Pierce). Eluted proteins were dialysed against two changes of 20 mM Tris HCl (pH 8.0), and concentrated by ultrafiltration over a 3 kDa cutoff membrane (YM3, Diaflo).
Crystallization and data collection
Protein 12A-9 (6 mg/mL) was set up as 0.2 μL sitting drops using a Cartesian Honey Bee robot. Plates were incubated at 25°C. Successful conditions were scaled up to 2 μL hanging drops. Final crystallization conditions were 0.1 M CHES (pH 9.5)/50% PEG200. Crystals appeared after 40 d as small rods with brunched start-like ends (clusters). An isolated piece of the cluster exhibiting diffraction quality was used for data collection.
X-ray diffraction data for 12A-9 crystal were collected at the multipole wiggler beamline BL5 at the Photon Factory, Japan. BL5 is equipped with collimating mirror, double-crystal Si(111) monochromator and focusing mirror which focuses X-rays to the ADSC Quantum 315 CCD detector. Data were collected at −160°C; the crystals required no added cryoprotectant. The data processing was carried out using the DENZO/SCALEPACK suite (Otwinowski and Minor 1997). Diffraction data statistics are summarized in Table 1.
Structure determination
The structure of 12A-9 was determined by molecular replacement using MOLREP (Vagin and Teplyakov 1997) of the CCP4 package (CCP4 1994). The search model for 12A-9 was the 12Y-1 structure (Streltsov et al. 2004) without the CDR3 loop. One molecule of 12A-9 was identified in the asymmetric unit of the P21212 space group. The model was manually revised and adjusted using XTALVIEW (McRee 1999) with the electron density maps produced by REFMAC5 (Murshudov et al. 1997). The electron density was traceable in the CDR3 loop, however, with a somewhat diffuse 92–95 region. Water molecules were added automatically with the program ARP (Lamzin et al. 2001). Progress of the refinement was monitored using the Rfree statistic based on a set encompassing 5% of the observed diffraction amplitudes. The final refinement included the TLS parameters for whole molecule and converged to R and Rfree values of 0.217 and 0.280, respectively, for the full 39.5–2.1 Å range of experimental data. The libration tensor was significant. The final model comprises residues 1 to 108 of 12A-9 and 140 water molecules. In total, 88.4% of residues are in the most favored regions of the Ramachandran plot, with one residue in the generously allowed regions. Further details are in Table 1. The coordinates have been deposited in the Protein Data Bank under the accession code 2COQ.
Modeling
The Type 3 VNAR domain was modeled using the program Modeler 6v2 (Sanchez and Sali 2000). Previously solved Type 2 VNAR structures, namely two nurse shark IgNAR structures (1SQ2, 1T6V), four wobbegong shark IgNAR structures (1VER, 1VES), an unpublished structure from our laboratory (1A7), and the structure revealed here, were used as templates. The structure presented here was the only template used for modeling of the CDR3 loop as it was identical in length to the Type 3 VNAR AAM77191 and all other templates exhibited longer loops by three to five residues. The template alignment used in the modeling input is shown in Figure 2A ▶.
Four modeling runs were undertaken with differing input parameters namely: Run1 with no declaration of loop residues or cis-peptide bonds; Run2 with no declaration of loop residues but declaration of a cis-proline at residue 7; Run3 with declaration of loop residues as 25–28 and 90–98 as well as residue Trp31 and no declaration of cis-peptide bonds; Run4 with declaration of loop residues as 25–28 and 90–98 as well as residue Trp31 and with declaration of a cis-proline at residue 7.
The models were initially ranked using the “modeler objective score’ before subjecting each of the top 10 models from each run to a further round of Cartesian dynamics in CNS (Brünger et al. 1998). In this step the modeled residues containing disulphide bonds (C22–C83 and C29–C89) and framework residues (1–7, 11–21, 23–24, 32–41, 52–59, 65–82, 84–86, and 100–107) were fixed and the remaining mainly loop residues allowed to refine. Post dynamics the selection of the best solutions was based on the modeler objective scores, the relative area exposed to solvent of the side chains of two of the large aromatic substitutions (Trp31 and Phe96) in the Type 3 VNAR models and pseudo energy and Z-scores. The side chain solvent exposure was derived by first calculating the solvent-accessible surface of the side chain in isolation (A), the solvent accessible surface of the model without the side chain and the surface area buried by returning the side chain to its modeled position (B) in CNS. An estimate of the solvent exposure for a side chain was then determined by A − 0.5B. Prosa 2003 (Sippl 1993) was used to calculate the pseudo energies (~ΔG) of the models and the Z-scores. Models with low selected side chain solvent exposure, low modeler objective scores, showing burial of the Trp31 and Phe96 side chains by visual inspection, more negative pseudo-energies and Z-scores were designated the best models for the Type 3 VNAR.
Electronic supplemental material
Supplementary Figure 1: Protein sequence alignment of 40 IgNAR variable domains, representing Type 3 (16 sequences), Type 1 (8 sequences), and Type 2 (16 sequences).
Acknowledgments
We thank Ms. Pat Pilling for assistance with protein crystallization. Financial support from the Australian Synchrotron Research Program funded by the Commonwealth of Australia is also acknowledged.
Abbreviations
IgNAR, new antigen receptor antibody from sharks
VNAR, single variable domain of the IgNAR antibody
CDR, complementarity determining region
kDa, kilodalton
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051709505.
Supplemental material: see www.proteinscience.org
References
- Adelman, M.K., Schluter, S.F., and Marchalonis, J.J. 2004. The natural antibody repertoire of sharks and humans recognizes the potential universe of antigens. Protein J. 23 103–118. [DOI] [PubMed] [Google Scholar]
- Al-Lazikani, B., Lesk, A.M., and Chothia, C. 1997. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 273 927–948. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Chothia, C., Lesk, A.M., Tramontano, A., Levitt, M., Smith-Gill, S.J., Air, G., Sheriff, S., Padlan, E.A., Davies, D., Tulip, W.R., et al. 1989. Conformations of immunoglobulin hypervariable regions. Nature 28 877–883. [DOI] [PubMed] [Google Scholar]
- Coia, G., Ayres, A., Lilley, G.G., Hudson, P.J., and Irving, R.A. 1997. Use of mutator cells as a means for increasing production levels of a recombinant antibody directed against Hepatitis B. Gene 201 203–209. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (CCP4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Decanniere, K., Muyldermans, S., and Wyns, L. 2000. Canonical antigen-binding loop structures in immunoglobulins: More structures, more canonical classes? J. Mol. Biol. 300 83–91. [DOI] [PubMed] [Google Scholar]
- Desmyter, A., Spinelli, S., Payan, F., Lauwereys, M., Wyns, L., Muyldermans, S., and Cambillau, C.J. 2002. Three camelid VHH domains in complex with porcine pancreatic α-amylase. Inhibition and versatility of binding topology. J. Biol. Chem. 277 23645–23650. [DOI] [PubMed] [Google Scholar]
- Diaz, M., Velez, J., Singh, M., Cerny, J., and Flajnik, M.F. 1999. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: The translesion synthesis model of somatic hypermutation. Int. Immunol. 11 825–833. [DOI] [PubMed] [Google Scholar]
- Diaz, M., Stanfield, R.L., Greenberg, A.S., and Flajnik, M.F. 2002. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): Identification of a new locus preferentially expressed in early development. Immunogenetics 54 501–512. [DOI] [PubMed] [Google Scholar]
- Dooley, H. and Flajnik, M.F. 2005. Shark immunity bites back: Affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol. 35 936–945. [DOI] [PubMed] [Google Scholar]
- Dooley, H., Flajnik, M.F., and Porter, A.J. 2003. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol. Immunol. 40 25–33. [DOI] [PubMed] [Google Scholar]
- Greenberg, A.S., Avila, D., Hughes, M., Hughes, A., McKinney, E.C., and Flajnik, M.F. 1995. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374 168–173. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E.B., Bendahman, N., and Hamers, R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363 446–448. [DOI] [PubMed] [Google Scholar]
- Hofstädter, K., Stuart, F., Jiang, L., Vrijbloed, J. W., and Robinson, J.A. 1999. On importance of being aromatic at an antibody-protein antigen interface: Mutagenesis of the extracellular interferon γ receptor and recognition by the neutralizing antibody A6. J. Mol. Biol. 285 805–815. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. 1996. VMD—Visual molecular dynamics. J. Mol. Graph. 14 33–38. [DOI] [PubMed] [Google Scholar]
- Lamzin, V.S., Perrakis, A., and Wilson, K.S. 2001. The ARP/WARP suite for automated construction and refinement of protein models. In International tables for crystallography. Vol. F: Crystallography of biological macromolecules (eds. M.G. Rossmann and E. Arnold), pp. 720–722. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- MacCallum, R.M., Martin, A.C., and Thornton, J.M. 1996. Antibody-antigen interactions: Contact analysis and binding site topography. J. Mol. Biol. 262 732–745. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Minsky, A., Summers, R.G., and Knowles, J.R. 1986. Secretion of β-lactamase into the periplasm of Escherichia coli: Evidence for a distinct release step associated with a conformational change. Proc. Natl. Acad. Sci. 83 4180–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Muyldermans, S. 2001. Single domain camel antibodies: Current status. J. Biotechnol. 74 277–302. [DOI] [PubMed] [Google Scholar]
- Nuttall, S.D., Irving, R.A., and Hudson, P.J. 2000. Immunoglobulin VH domains and beyond: Design and selection of single-domain binding and targeting reagents. Curr. Pharmaceut. Biotechnol. 1 253–263. [DOI] [PubMed] [Google Scholar]
- Nuttall, S.D., Krishnan, U.V., Hattarki, M., De Gori, R., Irving, R.A., and Hudson, P.J. 2001. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 38 313–326. [DOI] [PubMed] [Google Scholar]
- Nuttall, S.D., Krishnan, U.V., Doughty, L., Alley, N., Hudson, P.J., Pike, R.N., Kortt, A.A., and Irving, R.A. 2002. A naturally occurring NAR variable domain against the Gingipain K protease from Porphyromonas gingivalis. FEBS Lett. 516 80–86. [DOI] [PubMed] [Google Scholar]
- Nuttall, S.D., Krishnan, U.V., Doughty, L., Pearson, K., Ryan, M.T., Hoogenraad, N.J., Hattarki, M., Carmichael, J.A., Irving, R.A., and Hudson, P.J. 2003. Isolation and characterisation of an IgNAR variable domain specific for the human mitochondrial translocase receptor Tom70. Eur. J. Biochem. 270 3543–3554. [DOI] [PubMed] [Google Scholar]
- Nuttall, S.D., Humberstone, K.S., Krishnan, U.V., Carmichael, J.A., Doughty, L., Hattarki, M., Coley, A.M., Casey, J.L., Anders, R.F., Foley, M., et al. 2004. Selection and affinity maturation of IgNAR variable domains targeting Plasmodium falciparum AMA1. Proteins 55 187–197. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Padlan, E.A. 1994. Anatomy of the antibody molecule. Mol. Immunol. 31 169–217. [DOI] [PubMed] [Google Scholar]
- Roux, K.H., Greenberg, A.S., Greene, L., Strelets, L., Avila, D., McKinney, E.C., and Flajnik, M.F. 1998. Structural analysis of the nurse shark (new) antigen receptor (NAR): Molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. 95 11804–11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumfelt, L.L., McKinney, E.C., Taylor, E., and Flajnik, M.F. 2002. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 56 130–148. [DOI] [PubMed] [Google Scholar]
- Sanchez, R. and Sali, A. 2000. Comparative protein structure modelling. Introduction and practical examples with modeller. Methods Mol. Biol. 143 97–129. [DOI] [PubMed] [Google Scholar]
- Saphire, E.O., Parren, P.W., Pantophlet, R., Zwick, M.B., Morris, G.M., Rudd, P.M., Dwek, R.A., Stanfield, R.L., Burton, D.R., and Wilson, I.A. 2001. Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science 293 1155–1159. [DOI] [PubMed] [Google Scholar]
- Sippl, M.J. 1993. Recognition of errors in three-dimensional structures of proteins. Proteins 17 355–362. [DOI] [PubMed] [Google Scholar]
- Stanfield, R.L., Dooley, H., Flajnik, M.F., and Wilson, I.A. 2004. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 305 1770–1773. [DOI] [PubMed] [Google Scholar]
- Streltsov, V.A. and Nuttall, S.D. 2005. Do sharks have a new antibody lineage? Immunol. Lett. 97 159–160. [DOI] [PubMed] [Google Scholar]
- Streltsov, V.A., Varghese, J.N., Carmichael, J.A., Irving, R.A., Hudson P.J., and Nuttall, S.D. 2004. Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc. Natl. Acad. Sci. 101 12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A. and Teplyakov, A. 1997. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30 1022–1025. [Google Scholar]
- van den Berg, T.K., Yoder, J.A., and Litman, G.W. 2004. On the origins of adaptive immunity: Innate immune receptors join the tale. Trends Immunol. 25 11–16. [DOI] [PubMed] [Google Scholar]