Abstract
The crystal structures of the Vβ17+ β chains of two human T cell receptors (TCRs), originally derived from the synovial fluid (SF4) and tissue (C5–1) of a patient with rheumatoid arthritis (RA), have been determined in native (SF4) and mutant (C5–1F104→Y/C187→S) forms, respectively. These TCR β chains form homo-dimers in solution and in crystals. Structural comparison reveals that the main-chain conformations in the CDR regions of the C5–1 and SF4 Vβ17 closely resemble those of a Vβ17 JM22 in a bound form; however, the CDR3 region shows different conformations among these three Vβ17 structures. At the side-chain level, conformational differences were observed at the CDR2 regions between our two ligand-free forms and the bound JM22 form. Other significant differences were observed at the Vβ regions 8–12, 40–44, and 82–88 between C5–1/SF4 and JM22 Vβ17, implying that there is considerable variability in the structures of very similar β chains. Structural alignments also reveal a considerable variation in the Vβ–Cβ associations, and this may affect ligand recognition. The crystal structures also provide insights into the structure basis of T cell recognition of Mycoplasma arthritidis mitogen (MAM), a superantigen that may be implicated in the development of human RA. Structural comparisons of the Vβ domains of known TCR structures indicate that there are significant similarities among Vβ regions that are MAM-reactive, whereas there appear to be significant structural differences among those Vβ regions that lack MAM-reactivity. It further reveals that CDR2 and framework region (FR) 3 are likely to account for the binding of TCR to MAM.
Keywords: TCR, crystal structure, superantigen, rheumatoid arthritis, Mycoplasma arthritidis mitogen, structure/function studies, proteins of the immune system, crystallography, protein crystallization, mutagenesis (site-directed and general), protein structures—new, sedimentation
Rheumatoid arthritis (RA) is a chronic inflammatory disease resulting in radiographic joint destruction, severe functional deterioration, and work disability. Although the causes of RA remain unclear, it is thought to involve autoimmune recognition of self-antigens by autoreactive T lymphocytes (Van Boxel and Paget 1975; Gregersen et al. 1987; Nepom et al. 1989; Panayi et al. 1992). The cause of this breakdown of tolerance to self-antigens is not known, but both genetic and environmental factors are probably required to trigger the onset of RA. Genetic risk for RA has been strongly associated with certain major histocompatibility complex (MHC) class II alleles, in particular those in the HLA-DRB1 locus (Nepom et al. 1989; Nepom and Erlich 1991; Lanchbury and Hall 1995). A recognition mechanism involving a “shared epitope” of the QKRAA motif encoded by the RA-related DRB1 genes has been proposed (Gregersen et al. 1987; Nepom et al. 1989; Hiraiwa et al. 1990; Dessen et al. 1997). However, the autoantigens responsible for the induction of autoimmunity in RA have not been determined. Type II collagen, as the predominant protein of joint cartilage, has been proposed as a candidate antigen (Banerjee et al. 1988b; Fugger et al. 1994; Khare et al. 1995; Rosloniec et al. 1997).
RA is a multifactorial disease. In addition to genetic factors, environmental factors are thought to be of importance in the disease process. Microbial agents have long been thought to trigger autoimmune disease via their possession of epitopes that resemble self-peptides on target organs (molecular mimicry) (Oldstone 1987). In addition, it was proposed that microbial superantigens (SAgs) play a role in the initiation of the autoimmune diseases through activation of whole populations of specific Vβ-bearing T cells, including pathogenic anti-self T cell clones (Howell et al. 1991; Paliard et al. 1991; Conrad et al. 1994; Kotzin 1994; VanderBorght et al. 1999; Chini et al. 2002; Zhang et al. 2002). In collagen II-induced arthritis (CIA), an animal model of human RA, T cells expressing Vβ8.2 and Vβ6 are found to be selectively expanded in the synovial tissue, compared to the levels in the peripheral blood of the same individuals (Banerjee et al. 1988a; Spinella et al. 1991; Haqqi et al. 1992, 1995a, Haqqi et al. b; Moder et al. 1993; Chini et al. 2002). In human RA, T cells expressing Vβ3, Vβ14, and Vβ17 are most frequently expanded (Howell et al. 1991; Grom et al. 1993; Li et al. 1994; Zagon et al. 1994; Alam et al. 1995; Moreland et al. 1996, 1998; Cuesta et al. 1997; Goronzy et al. 1998; Yen et al. 1998; Zhang et al. 2002), although conflicting results have also been reported (Uematsu et al. 1991; Van Laar et al. 1991; Struyk et al. 1995). It has been speculated that a SAg produced by Mycoplasma arthritidis (MAM) is involved in the pathogenesis of human RA (Emery et al. 1985; Cole and Griffiths 1993; Knudtson et al. 1997b; Mu et al. 2000; Sawitzke et al. 2000). MAM has been shown capable of triggering and exacerbating autoimmune arthritis in the CIA mouse model. In fact, administration of MAM has been shown to markedly exacerbate arthritis in mice that were convalescent from CIA, or to trigger arthritis in animals that had previously been immunized with collagen II but that had failed to develop clinical disease (Cole and Griffiths 1993). It has also been reported that MAM can induce human lymphocytes to secrete rheumatoid factor (Emery et al. 1985), a typical feature of RA. Moreover, antibodies against MAM were found to be selectively elevated in sera from patients with RA, compared to sera from control patients, whereas elevation of antibodies to Staphylococcus enterotoxins SEA and SEB in sera from patients with rheumatic diseases was less specific (Sawitzke et al. 2000). This suggests that, although M. arthritidis commonly infects rodents, its product, MAM or a MAM-like molecule, may play a role in human RA.
While the importance of TCR in RA appears clear, no crystal structure of a TCR implicated in human RA has yet been determined. Several TCRs derived from the synovial fluid and tissue of a RA patient with “classic” RF+ polyarticular, symmetrical inflammation, and a DR4,7 haplotype have been cloned (Li et al. 1994). Among these TCRs, two (SF4 and C5–1)Vβ17 TCRs are particularly interesting because they respectively represent a majority TCR population (C5–1) and a minority one (SF4) in the synovial fluid and tissue of the RA patient. SF4 Vβ17 was isolated from the synovial fluid of the patient, 4 mo before synovectomy, while C5–1 Vβ17 was obtained upon activation of T cells from the synovial tissue by the SAg MAM (Li et al. 1994). The CDR3 sequence of the C5–1 Vβ17 is highly homologous (86.4%) to that of a dominant Vβ17, representing more than 40% of the Vβ17 TCR population that was present in tissue of the patient (Li et al. 1994). The antigen specificities of the SF4 and C5–1 TCRs are unknown. However, the T cell clone expressing C5–1 TCR (Vα2.3Vβ17) could be selectively proliferated by EBV-transformed lymphoblastoid B cell lines expressing RA-associated HLA-DR alleles of DR4, Dw4, and Dw14 (Li et al. 1994).
In the current study, we describe the crystal structures of the two Vβ17+ β chains related to human RA. We expressed, purified, and crystallized the native SF4 Vβ17 and a F104Y/C187S double mutant of the C5–1 Vβ17 (designated as C5–1YS) in unbound states (see Materials and Methods). The crystal structures were determined by the multiple anomalous diffraction (MAD) method. Structural comparisons with the TCR β chains of known structures revealed that the overall topology of the two Vβ17 TCR β chains derived from the RA patient is very similar to that of most other known TCR β chains. The conformations in the complementarity determining regions (CDRs) of the RA-related Vβ17 β chains in the absence of contacts with antigens are very close to the conformation in a JM22 TCR Vβ17Vα10 recognizing the matrix protein 58–66 of influenza virus presented by HLA-A2 (Stewart-Jones et al. 2003), although they were originated from CD4+ (SF4 and C5–1 Vβ17) and CD8+ (JM22 TCR) T cells, respectively. The individual Vβ and Cβ domains of these two classes of Vβ17 β chains can be readily superimposed onto the counterparts of other TCR β chains; however, structure alignments of the complete β chains resulted in large root-mean-square deviations (RMSDs), implying considerable variation in the Vβ–Cβ associations. The crystal structures also provide insights into the structure basis of T cell recognition of MAM, a superantigen that may be implicated in the development of human RA. Structure comparison of the Vβ17 domains with Vβ domains of known structures indicates that there are significant similarities among Vβ regions that are MAM-reactive whereas there appear to be significant structural differences among those Vβ regions that lack MAM-reactivity. Detailed structural comparison further reveals that CDR2 and framework region (FR) 3 may account for the TCR binding to MAM.
Results and Discussion
Overall structure
We have expressed, purified, and crystallized two human RA-related TCR β chains, designated as Vβ17SF4 and Vβ17C5–1, respectively. The SF4 and C5–1 Vβ17 β chains possess different CDR3 sequences (Fig. 1 ▶). The crystal structures of the RA-associated Vβ17 β chains, native SF4 and mutant C5–1YS (see Materials and Methods), were determined to about 2.7 Å resolution (Table 1; Fig. 2 ▶). For C5–1, a mutation of C187→S was introduced to eliminate the free cysteine, while a mutation of F104→Y was introduced to generate a TCR Vβ17 molecule with high affinity to MAM (Hodtsev et al. 1998). The electron-density maps were of good quality for both β chains (Fig. 2 ▶). Most parts of the structures were well ordered except for the CDR3 regions, where electron densities were not observed for some side chains, including Arg96 and Arg98 of both β chains in SF4 Vβ17, and Gln99 (chain A) and Met100 (chain B) in C5–1 Vβ17 (Fig. 2A,B ▶).
Figure 1.
Primary sequence alignment of the CDR3 regions (underlined) of three human Vβ17 TCR β chains: Vβ17 SF4, C5–1, and JM22 (Stewart-Jones et al. 2003). Identical CDR3 residues are shaded.
Table 1.
Data collection, phasing, and refinement statistics
| Vβ17C5-1YS | ||||
| Vβ17SF4 Native | λ1 | λ2 | λ3 | |
| Data collection | ||||
| Wavelength (Å) | 1.10 | 0.9794 | 0.9798 | 0.93 |
| Resolution (Å) | 2.65 | 2.7 | 2.7 | 2.8 |
| No. of unique reflections | 25,627 | 24,387 | 24,442 | 22,072 |
| Completeness (%) | 96.4 | 97.7 | 97.5 | 98.1 |
| Rsym (%) | 7.5 | 7.5 | 7.8 | 7.9 |
| I/σ(I) | 18 | 35 | 32 | 30 |
| MAD phasing | ||||
| No. of sites | 2 | |||
| Resolution range (Å) | 20–2.8 | |||
| Figure of merit | 0.37 | |||
| Refinement | ||||
| Resolution range (Å) | 42–2.65 | 39–2.7 | ||
| Rcryst (%) | 23.3 | 23.7 | ||
| Rfree (%) | 28.6 | 28.0 | ||
| No. of non-H atoms in the model | ||||
| Protein atoms | 3886 | 3884 | ||
| Solvent molecules | 93 | 90 | ||
| RMSD value from ideality | ||||
| Bond length (Å) | 0.008 | 0.007 | ||
| Bond angle (o) | 1.3 | 1.3 | ||
| Average B factor (Å2) | ||||
| Protein atoms | 60.1 | 58.9 | ||
| Solvent molecules | 47.4 | 53.2 | ||
Figure 2.
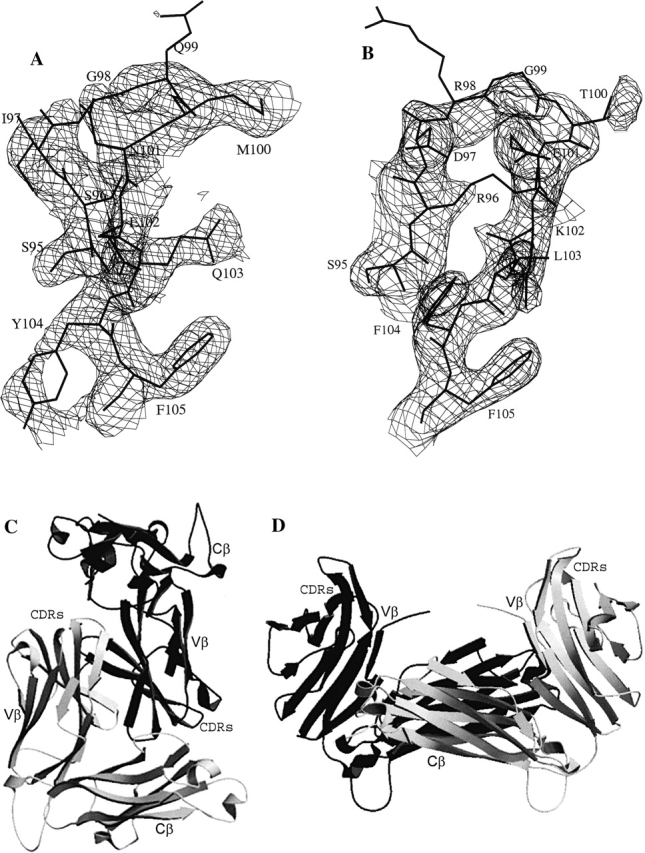
The structures of Vβ17 TCR β chains. (A,B) Representative (2Fo − Fc) electron density maps, contoured at 1.0σ, of the CDR3 regions of Vβ17C5–1YS (A) and Vβ17SF4 (B). (C,D) Ribbon diagram of the Vβ17C5–1YS homodimers in two possible associations. (C) Dimerization via Vβ domains; (D) dimerization via Cβ domains. The TCR Vβ, Cβ and CDR loops are labeled.
Sedimentation characterization of soluble Vβ17 TCR β chains
Although the profile of gel filtration chromatography indicated that the refolded Vβ17 proteins were mainly monomeric, the unit cell parameters imply that the TCR Vβ17 forms a dimer in the crystal.
To characterize the oligomerization state of the re-folded Vβ17 molecules, we performed sedimentation-velocity (SV) experiments using the analytical ultracentrifugation technique. As shown in Figure 3A ▶, the sedimentation coefficient (S value) distribution of Vβ17SF4 at a concentration of 14 μ M (~0.39 mg/mL) displayed two distinct peaks, at 2.2 S and 3.2 S, corresponding respectively to the monomeric and dimeric forms of Vβ17SF4. Such a dimer formation is noncovalent, because mutation of the free cysteine (C187) to serine does not affect the dimerization of the Vβ17 C5–1, showing both monomeric and dimeric peaks in the SV experiment (Fig. 3A ▶). This demonstrated that noncovalently associated Vβ17 dimer exists in solution, as well as in the crystal structures. At moderate protein concentrations, it is apparent that the majority of Vβ17SF4 and Vβ17C5–1YS mutant proteins exist as monomers with a peak at 2.2 S. Higher protein concentration promoted dimer formation, resulting in an overall increase of the proportion of the Vβ17 dimer (Fig. 3A ▶). This shift strongly implies that dimer formation for Vβ17 is concentration-dependent, which is consistent with a self-association behavior.
Figure 3.
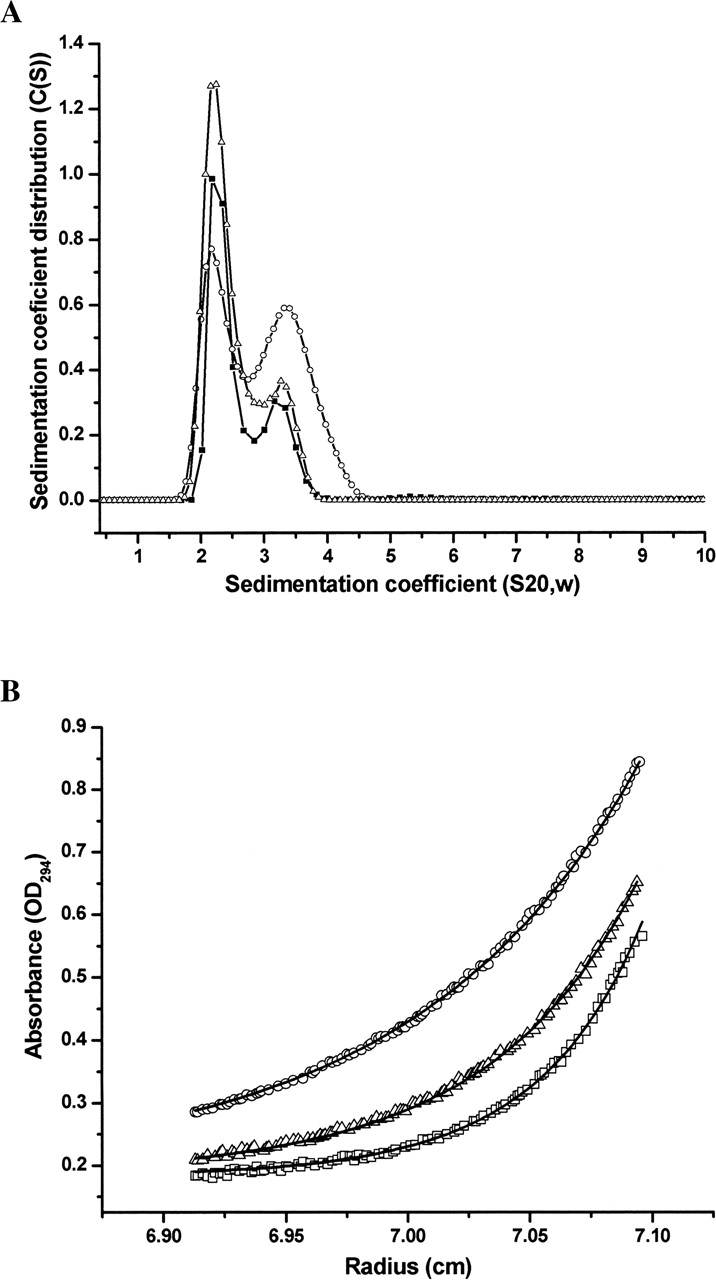
Sedimentation velocity and equilibrium analysis of Vβ17C5– 1YS and Vβ17SF4 dimerization. (A) Sedimentation coefficient distributions of Vβ17SF4 at 14 μ M (▪) and 140 μ M (○), and of Vβ17C5–1YS at 42 μ M (▴) concentration. (B) Absorbance distributions for the sedimentation of Vβ17SF4 at 22.3 μ M at 20° C at rotor speeds of 20,000 rpm (○), 25,000 rpm (▵), and 30,000 rpm (▪). For clarity, only every second data point is shown. Distributions were analyzed as part of a global fitting to the absorbance data at multiple loading concentrations. Solid lines are the global best-fit distributions using a reversible monomer–dimer model with an equilibrium constant of KD = 380 μ M.
To further investigate the self-association of Vβ17, we performed sedimentation equilibrium studies for the wild-type Vβ17SF4 at three concentrations (8.7, 14, and 22.3 μ M) with a Beckman Optima XL-I analytical ultracentrifuge running at three rotor speeds (Fig. 3B ▶). An equilibrium constant KD of 380 μ M was estimated using a reversible monomer–dimer model by global analysis of the equilibrium data. The sedimentation data clearly demonstrated that the TCR Vβ17 β chains can form a dimer in solution.
The homodimer of the Vβ17 TCR β chain
The crystals of the SF4 and C5–1YS Vβ17 β chains are isomorphous. The Vβ17 β chains form homodimers in the crystals. Because noncrystallographic symmetric (NCS) restraints were used throughout the structure refinement, the main-chain conformations of the two monomers of the dimer are nearly identical, with respective RMSD values of 0.05 Å and 0.06 Å for Vβ17C5–1YS and Vβ17SF4.
The packing in the crystal structure suggests two possible ways in which the homodimer in solution could form (Fig. 2C,D ▶). A Vβ17 homodimer could be formed through interactions between the TCR Cβ domains (Fig. 2D ▶). Alternatively, a homodimer could be formed through interactions between the TCR Vβ domains (Fig. 2C ▶). In the latter way, the dimer is formed in a head-to-tail manner, with the CDR loops pointing in opposite directions. Similar arrangements of homodimers have been found in the crystal structures of the 14.3.d TCR β chain and of its complexes with SAgs (Bentley et al. 1995; Fields et al. 1996; Li et al. 1998b; Sundberg et al. 2002), as well as in the structure of the ES204 TCR Vα3 (Li et al. 1998a). This type of dimer arrangement is not analogous to that in the TCR heterodimer; the physiological, active form of TCRs. In the TCR heterodimer, the TCR Vα chain pairs with the TCR Vβ using the same buried surface of the TCR Vβ domain as is used in the Vβ homodimer studied here, but in such a way that all of the CDR loops of the TCR Vα and Vβ chains are oriented in the same direction. In this manner, an antigen-combining site is formed (Garcia et al. 1999).
Similar to the authentic TCR αβ heterodimers, the Vβ17 β chains in the crystals form homodimers mainly through hydrophobic interactions. In both forms of homodimers, considerable amounts of the solvent-accessible surfaces of the Vβ17 β chains are buried (data not shown). It has also been reported that the TCR Vα domains can form homodimers in the absence of TCR Vβ domains (Fields et al. 1995; Li et al. 1997; Plaksin et al. 1999). Although the homodimer arrangement does not have relevance to the physiological Vα:Vβ dimers, the dimerization of TCR V domains may involve a general mechanism by which the hydrophobic surface is shielded so that each TCR Vα or Vβ can be efficiently folded, regardless of the presence of their partners (Rudolph et al. 2001). Therefore, a cross-linking disulfide bond is required for the native, heterodimeric αβ TCRs to ensure their correct chain pairing so as to form the antigen recognition site (Rudolph et al. 2001). In addition, the self-association mechanism of TCR V domains may be important for T cell development and differentiation. Skewed expression of TCR Vα gene families into CD4+ or CD8+ subsets has been reported, apparently occurring independently of the TCR Vβ pairing (Utsunomiya et al. 1989). The unpaired TCR Vα chains might be protected through a self-association mechanism during the early stage of T cell development.
The conformations of the CDR regions in ligand-free and ligand-bound states
To date, only a limited number of crystal structures of TCRs have been determined in both a ligand-free and a bound form. These include TCRs 2C (Garcia et al. 1996, 1998), D10 (Hare et al. 1999; Reinherz et al. 1999), Kb5-C20 (Housset et al. 1997; Reiser et al. 2002), and Lc13 (Kjer-Nielsen et al. 2002, 2003). Structural superimposition of the liganded and unliganded forms of each TCR revealed that no major domain rearrangements are associated with ligand binding. However, it has been proposed that TCRs recognize pMHC molecules through an induced-fit mechanism involving conformational changes at CDR regions (Garcia et al. 1998, 1999; Reiser et al. 2002; Kjer-Nielsen et al. 2003).
To date, the crystal structures of three Vβ17 β chains of human TCR, namely the Vβ17SF4 and Vβ17C5–1YS β chains in this paper, and a Vβ17 β chain of the JM22 TCR (Stewart-Jones et al. 2003), have been determined. The structure of JM22 TCR was determined in a ligand-bound form, whereas the structures of SF4 and C5–1YS β chains were determined in a ligand-free form. Structural comparison between the C5–1YS/SF4 β chains and the JM22 TCR β chain may allow us to determine the range of structural changes that occurs upon complex formation, particularly in the CDR1, CDR2, and hypervariable 4 (HV4) regions, in which the sequences are identical. The variable domains of the RA-related Vβ17 C5–1YS and SF4 have nearly identical conformation with an overall RMSD of 0.19 Å for the Cα positions. The largest structural differences are in the CDR3 regions, which have an RMSD of 0.74 Å. Because C5–1YS and SF4 β chains have nearly identical structures, and because JM22 Vβ17 is more similar to C5–1YS in sequence than it is to SF4 Vβ17, the structure of C5–1YS Vβ17 was used for interpretation and further discussion, except for the CDR3 regions: In the latter, the conformations of C5–1YS and SF4 Vβ17 differ significantly. However, the conclusions are broadly applicable to SF4 Vβ17 as well.
The CDR1, CDR2, and HV4 regions of Vβ17 C5– 1YS adopt canonical CDR conformations (Al-Lazikani et al. 2000; Fig. 4A,B ▶). In the crystal, Vβ17 CDR1 residues are not involved in crystal packing (except for Asn28, which makes one contact with Ser168 of the NCS-mate). In contrast, both CDR2β and HV4β regions participate in crystal contacts, including both van der Waals contacts and hydrogen bonds, with symmetry-related molecules. Nevertheless, regardless of crystal contacts, the conformations of the CDR1, CDR2, and HV4 regions of Vβ17 C5–1YS closely resemble those seen for JM22 Vβ17 in its liganded form (Stewart-Jones et al. 2003; Fig. 4A,B ▶). Structural superimposition of the Vβ domains resulted in RMSD values of 0.5 Å (CDR1), 0.6 Å (CDR2), and 0.3 Å (HV4) in Cα positions between C5–1YS and JM22 Vβ17 molecules. These values are smaller than the RMSD (~0.7 Å) for the entire Vβ domain of the two molecules, implying that the conformations of these loops are well conserved upon complex formation. Although no major conformational differences are observed between the ligand-free C5–1YS and liganded JM22 Vβ17, subtle conformational differences can be seen at the side-chain level. The largest conformational differences occur at residues Tyr50 and Asn55, with positional differences of the tips of Tyr50 and Asn55 as large as 6 Å and 4 Å, respectively (Fig. 4B ▶). In the crystal structure of JM22-HLA-A2/MP complex, Asn55β is involved in recognition of the HLA molecule (Stewart-Jones et al. 2003). In the structure of C5–1YS Vβ17, Asn55β makes a number of crystal contacts, including three hydrogen bonds. Nevertheless, these differences are within the natural range of variations seen for TCRs (Wang et al. 1998).
Figure 4.
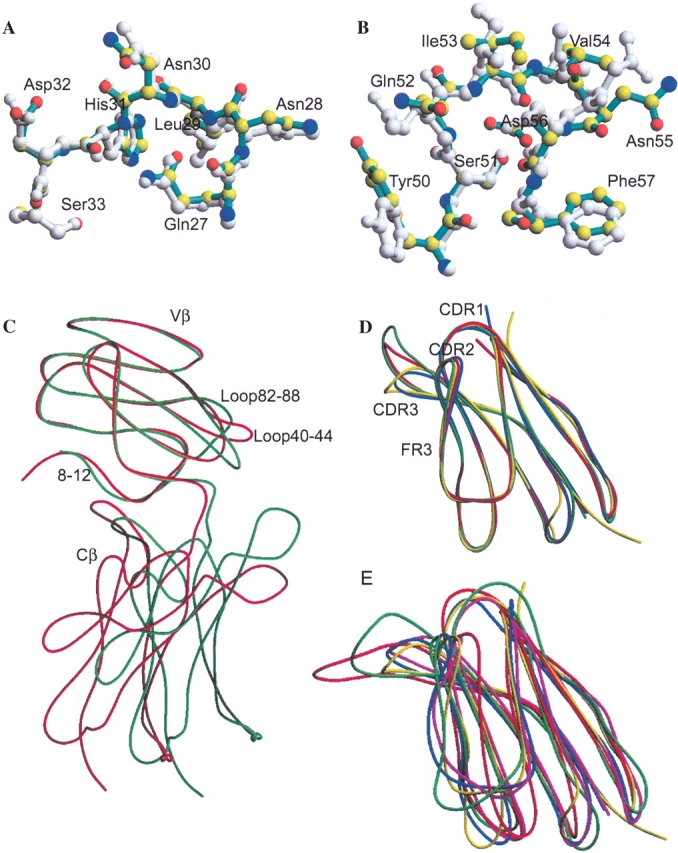
Structural comparisons of Vβ17C5–1YS with the known three-dimensional structures of TCR β chains. (A) Structural comparison of the CDR1 regions of C5–1YS (cyan) and JM22 Vβ17 (PDB code 1OGA) (gray). Atoms for the C5–1YS Vβ17 residues are colored as follows: nitrogen, blue; oxygen, red; carbon, yellow. (B) Structural comparison of the CDR2 regions of C5–1YS (cyan) and JM22 Vβ17 (gray). Atoms for the C5–1YS Vβ17 residues are colored as in A. (C) Structural comparison of the complete β chains of Vβ17C5–1YS (red) and JM22 Vβ17 (green). The TCR Vβ domains have been optimally superimposed. (D) Structural comparison of the variable domain of human Vβ17C5–1YS (yellow) with three MAM-reactive TCR β chains: mouse Vβ8.2 (blue), human Vβ3 (green), and Vβ12.3 (red). (E) Structural comparison of the variable domain of human Vβ17C5–1YS (yellow) with four MAM-nonreactive TCR β chains: mouse Vβ2 (red) and Vβ5.2 (blue), and human Vβ2.1 (green), and Vβ6.3 (magenta). The PDB codes for the TCR β chains used here are as follows: mouse Vβ2, 1KB5 (Housset et al. 1997); Vβ5.2, 1NFD (Wang et al. 1998); Vβ8.2, 1TCR (Garcia et al. 1996); human Vβ2.1, 1KTK (Sundberg et al. 2002); Vβ3, 1FYT (Hennecke et al. 2000); Vβ6.3, 1KGC (Kjer-Nielsen et al. 2002); Vβ12.3, 1AO7 (Garboczi et al. 1996a); and Vβ17, 1OGA (Stewart-Jones et al. 2003).
In contrast to the CDR1, CDR2, and HV4 regions, the TCR CDR3 regions are highly flexible. In the absence of antigen and TCR Vα, CDR3 regions of C5–1YS and SF4 Vβ17 are less well ordered than the other parts of the structure, as reflected by the high temperature factors and lack of electron densities corresponding to some side-chain atoms in these regions. Similar flexibility at CDR3 has been observed in the structures of other unpaired TCR components (Bentley et al. 1995; Fields et al. 1995; Li et al. 1997). The C5–1YS and SF4 Vβ17 molecules adopt very different conformations in CDR3, with an RMSD of 0.74 Å that is much greater than the overall RMSD (0.2 Å) between the two molecules. Therefore, the CDR3 structures analyzed here may be displaying their native conformations, rather than conformations induced by the crystal packing. This is because similar crystal contacts are expected for C5–1YS and SF4 due to the isomorphism of their crystals. In both molecules, the conformation of Vβ17 CDR3 is stabilized by a number of hydrogen bonds between CDR3 and CDR1 residues, as well as by contacts with symmetry-mates.
The C5–1YS, SF4, and JM22 Vβ domains encode different sequences with C5–1YS more similar to JM22 than is SF4. The structural differences in CDR3 between C5–1YS and JM22 (RMSD 1.8 Å) and between SF4 and JM22 (RMSD 1.3 Å) are greater than the RMSD for the CDR3 regions between C5–1YS and SF4 (0.74 Å ). Although the CDR3 regions have different conformations, they all contain β-turns at the tip of CDR3. JM22 Vβ17 forms a type II β-turn composed of residues 98–101, while these residues in C5–1YS and SF4 form type I β-turns. In addition, regardless of their sequence differences and ligand states, the CDR3 loops of the three molecules are positioned similarly; each fold back toward CDR1 and CDR2. Moreover, several hydrogen bonds, including the one between His31 Nδ1 and conserved Ser95 Oγ, are well conserved. The large structural differences in CDR3 seem not to have a large impact on other CDR regions, as only small RMSD values are observed for CDR1 and CDR2. These data could imply that Vβ17 molecules are required to undergo only small conformational changes upon complex formation.
Comparison of the Vβ17 domains of MHC class I and class II-restricted TCRs
The TCR Vβ gene products are known to interact with both class I and class II MHC molecules (Acuto et al. 1985). Although the structures of many TCRs have been determined, structural determinations for those TCRs that use the same TCR Vβ to recognize both class I and II peptide/MHC molecules are thus far limited solely to murine Vβ8.2. TCR 2C (Vα3Vβ8.2) recognizes class I MHC H-2Kb (Garcia et al. 1996, 1998), while TCR D10 (Vα2Vβ8.2) and 172.10 (Vα2.3Vβ8.2) are respectively specific for class II MHCs I-Ak (Reinherz et al. 1999) and I-Au (Maynard et al. 2005). Structural comparisons indicate that the overall conformation of Vβ8.2 in 2C, D10, and 172.10 TCRs is very well conserved, with the exception of Vβ CDR3 regions (Garcia et al. 1999; Hare et al. 1999; Maynard et al. 2005). It is not currently known whether other TCRs maintain a single Vβ conformation in the recognition of different classes of MHC molecules.
Structural determination of SF4 and C5–1YS Vβ17 of class II MHC-restricted TCRs, together with the structure of JM22 Vβ17 of a class-I MHC restricted TCR, provided additional support for the hypothesis that the TCR Vβ domains maintain their main-chain conformation regardless of their CD4+ or CD8+ origins. TCRs encoding Vβ17 SF4 and C5–1 were derived from CD4+ T cells with unknown epitopes, while the JM22 TCR originated from CD8+ T cells, recognizing class I MHC HLA-A2 in the context of a matrix protein peptide (MP58–66) of influenza virus (Stewart-Jones et al. 2003). These three Vβ17 β chains differ significantly in their CDR3 sequences (Fig. 1 ▶). In other regions, the sequences of Vβ17C5–1 and the JM22 Vβ17 are nearly identical, except for amino acid 114. Amino acid 114 is Thr in the JM22 Vβ17 β chain, while it is Leu in the Vβ17 β chains derived from the RA patient. In addition, Vβ17SF4 differs from the other two Vβ17 β chains at positions 107, 110, 112, 118, and 119. The residues at these positions are Ser107, Gln110, Ser112, Asn118, and Lys119 for Vβ17SF4, while they are Pro107, Arg110, Thr112, Lys118, and Asn119 in the other two Vβ17 β chains.
The SF4 and C5–1YS Vβ17 β chains of the class II MHC-restricted TCRs superimpose closely with the JM22 Vβ17 of class I MHC-restricted TCR (Stewart-Jones et al. 2003), with overall RMSD values of 0.67 Å and 0.74 Å, respectively (Fig. 4C ▶). Regions 8–12, 40–44, 82–88, and 97–101 showed structure differences larger than 1.5 Å between the β chains of MHC class I and II-restricted TCRs. The first three among the four regions are within the Vβ framework (FR) regions, with identical sequences. Analysis of the structural differences in these regions indicated that the structural differences in loop 40–44 between C5–1YS and JM22 Vβ17 are likely due to crystal packing. Residues 40–44 form a loop that is located at the Vβ17 dimer interface in the Vβ17 C5–1YS crystals (Fig. 4C ▶). Loop 40–44, which lies at the interface of the authentic αβ TCR heterodimer, showed various conformations when paired with the TCR α chains in TCRs of known structures (data not shown). A conformation different from that in JM22 Vβ17 is necessary, in order for loop 40–44 to avoid steric conflict with the other monomer of the C5–1YS Vβ17 dimer. In all TCR β chains, residues 8–12 form two consecutive turns linking the interrupted TCR A- and A′-strands that form β-sheets with the B- and G-strands, respectively (Bentley et al. 1995). Structural comparison indicates that the conformation of this linker region is not conserved among the known TCR structures. Nevertheless, superimposition of the JM22 Vβ17 onto the C5–1YS Vβ17 in the crystal structures resulted in only minor close contact between the JM22 residue S9β and N162 of the NCS-mate of C5–1YS Vβ17. Therefore, it is possible that the structural difference at this region is due to crystal packing forces. On the other hand, this structural difference could be due to the TCR β-chain’s intrinsic nature to recognize different classes of MHC molecules. Notably, structural comparison of a class II-restricted D10 TCR with a class I-restricted 2C TCR revealed similar structural differences at this region (Hare et al. 1999).
In contrast to loops 8–12 and 40–44, loop 82–88 is not involved in crystal packing contacts in the crystal structures of either JM22 or C5–1YS Vβ17. Residues 82–88 lie at the opposite ends of the CDR loops, and are close but not in the TCR Vβ–Cβ interface. When the JM22 Vβ17 was superimposed on the C5–1YS Vβ17 or vice versa, the conformation of residues 82–88 from one molecule is not such that it would interfere with the crystal packing of the other. Therefore, the structural differences may not be due to differences in crystal packing, although the possibility can not be formally ruled out. Further structural comparisons revealed that the conformations of this loop in the JM22 and C5–1YS Vβ17 domains are different from those of all other TCR Vβ domains whose structures have been determined. With the exception of murine Vβ2, which shows a different conformation due to a unique conformation of the adjacent C″ β-strand (Reiser et al. 2000, 2003), the loops 82–88 of most other TCR Vβ domains with RMSD values within 2.0 Å adopt similar conformations that are composed of one 310 helix from residues 83 to 87 (nomenclature from the 2C TCR structure). In contrast, loops 82–88 of human JM22 and C5–1 Vβ17 display completely different conformations that lack obvious secondary structure elements (Fig. 4C ▶). The three human Vβ17 β chains, JM22, C5–1YS, and SF4, differ from all other TCR β chains at the loop region (amino acids 82–88) with RMSD values larger than 3.1 Å. The Vβ17 β chains of class I and II MHC-restricted TCRs also differ from one another with an RMSD of 3.2 Å. Although the biological significance of this loop is unknown, it is clear that loop 82–88 is flexible and capable of adopting multiple conformations.
Comparison of the TCR Vβ–Cβ associations
The overall topologies of the Vβ17 TCR β chains derived from the RA patient are very similar to those of most other TCR β chains (Fig. 4 ▶). Although the individual Vβ and Cβ domains of these two Vβ17 β chains can be readily superimposed onto their counterparts in other TCR β chains, structural alignments of the complete β chains result in large RMSD values, indicating considerable variation in the Vβ–Cβ associations. Interestingly, although the residues at the Vβ–Cβ interface are identical in the JM22 Vβ17 β chain (Stewart-Jones et al. 2003) and in the two Vβ17 β chains derived from the RA patient, a rotation angle as large as 23.4° is required for the best alignment of the Cβ domains, after optimal alignment of the Vβ domains (Fig. 4C ▶). Structural comparison with other TCR β chains revealed that the Vβ–Cβ associations of Vβ17 SF4 and C5–1 are most similar to that of 2C TCR (PDB code 1MWA) in the crystal structure of a TCR-MHC complex (Luz et al. 2002), with a 15.3° rotation required to optimally align these Cβ domains after optimal superimposition of the Vβ domains. Comparison with other TCR Vβ–Cβ associations resulted in rotation angles ranging from 15.3° to 25.6° (data not shown). This finding implies that the TCR β chains can tolerate quite large inter-domain movements, although the overall structures of individual domains of the TCR β chains are relatively rigid (Bentley et al. 1995).
As discussed earlier, the crystal structures of Vβ17 SF4 and C5–1YS are in a nonliganded form, while the structure of the JM22 TCR is in a liganded form (Stewart-Jones et al. 2003). Therefore, it is possible that the difference in Vβ–Cβ domain arrangement between C5–1YS and JM22 Vβ17 represents a conformational change upon ligand binding, although crystal packing effects can not be ruled out. The structural differences in Vβ–Cβ domain arrangement observed here may have functional significance. A difference in Vβ–Cβ arrangement may affect the elbow angle of a TCR. Elbow angle is a term coined to define the angle between the VL/VH and CL/CH1 pseudo dyads of a Fab fragment of an antibody. Due to the structural similarity between TCR and Fab, the elbow angle terminology can also be used for TCR, to define the relative orientation between the TCR V and C modules. Although there is no solid evidence for any signaling function through the elbow region of the TCR, the elbow angle of an antibody is thought to be essential for molecular signaling upon complexing with the antigen (Colman 1988; Lesk and Chothia 1988; Guddat et al. 1994, 1995; Landolfi et al. 2001). Nevertheless, a difference in Vβ–Cβ arrangement will affect the relative orientations of Vα and Vβ domains of the TCR, which in turn influence the peptide/MHC specificity (Garcia et al. 1999). It is well known that the relative orientations of the TCR Vα/Vβ domains vary significantly (Li et al. 1997; Garcia et al. 1999). Although this has not been extensively studied, due to the limited number of crystal structures of TCRs available, the impact of Vα/Vβ pairing variability on the TCR’s ligand specificity has been demonstrated in a recent study (Maynard et al. 2005). Both TCRs D10 and 172.10 use Vβ8.2. Their Vα domains, Vα2 in D10 and Vα2.3 in 172.10, are also highly similar, with 84% sequence identity. These two TCRs recognize different peptide/MHC molecules (Reinherz et al. 1999; Maynard et al. 2005). However, structural comparison showed that the Vα–Vβ association in the two TCRs differs by a rotational angle of 15°, resulting in TCR footprints on different regions of the peptide/MHC molecules. Clearly, differences in TCR Vβ–Cβ associations will ultimately affect the relative Vα–Vβ orientations, which may modulate the TCR’s peptide/MHC specificity (Garcia et al. 1999; Maynard et al. 2005).
Possible binding site for the superantigen MAM
The SAg MAM has been proposed to be involved in the pathogenesis of human RA (Emery et al. 1985; Cole and Griffiths 1993; Knudtson et al. 1997b; Mu et al. 2000; Sawitzke et al. 2000). The crystal structure of a MAM-MHC complex (Zhao et al. 2004) reveals that MAM, like other SAgs, binds very well to class II MHC molecules. In addition, MAM interacts with TCRs in a Vβ-restricted fashion, as occurs for other SAgs. It has been demonstrated that MAM stimulates T cells expressing murine Vβ6, 7, and 8.1–3. Although human T cells are less responsive to MAM than are murine ones, T cells bearing human Vβ 3.1, 5.1, 7.1, 8.1, 10, 11.1, 12, 13.1, 14, 17, or 20 can be polyclonally expanded to various degrees during MAM induction (Knudtson et al. 1997a). Structure determination of human Vβ17 may allow us to investigate the molecular mechanism by which human T cells are activated by MAM, a superantigen that may be implicated in the development of human RA.
Although the crystal structure of a MAM-TCR complex has not been determined, crystal structures of several staphylococcal and streptococcal pyrogenic SAgs complexed with TCR β chains have been reported (Fields et al. 1996; Li et al. 1998b; Sundberg et al. 2002). In these pyrogenic SAg-TCR complexes, SAgs bind uniformly to the CDR2 and FR3 regions of the TCR β chains, although the Vβ CDR1, CDR3, and HV4 regions contribute to a certain extent toward the binding to various SAgs (Fields et al. 1996; Li et al. 1998b; Li et al. 1999; Sundberg et al. 2002). A SAg-TCR recognition mechanism, which is highly dependent on the main-chain conformations of the TCR Vβ CDR2 and FR3 regions, has been proposed (Fields et al. 1996; Li et al. 1998b, 1999). The SAgs MAM, SEB, and SEC1–3 have nearly identical TCR Vβ specificities (Fields et al. 1996; Knudtson et al. 1997a). Possibly, the binding site on the TCR Vβ domains for MAM is similar to the binding sites for SEB and SEC1–3. As indirect evidence, MAM, SEB, and SEC3 bind to class II MHC at nearly identical sites (Jardetzky et al. 1994; Sundberg et al. 2003; Zhao et al. 2004), although MAM does not show any structural similarity to SEB or SEC1–3 (Zhao et al. 2004).
Nevertheless, crystal structures have been determined for both MAM-reactive and nonreactive TCR β chains, and comparison of these structures may reveal critical information about the MAM binding sites on TCR β chains. Although there are about 37 PDB entries related to TCRs, including TCRs (Garcia et al. 1996; Housset et al. 1997; Wang et al. 1998; Hare et al. 1999; Allison and Garboczi 2002; Kjer-Nielsen et al. 2002), TCR components (Bentley et al. 1995; Fields et al. 1995; Li et al. 1997, 1998a; Plaksin et al. 1999; Machius et al. 2001; Rudolph et al. 2001), TCRs complexed with class I or class II MHC/peptide complex molecules (Garboczi et al. 1996a; Ding et al. 1998, 1999; Garcia et al. 1998; Reinherz et al. 1999; Degano et al. 2000; Hennecke et al. 2000; Reiser et al. 2000, 2002, 2003; Hennecke and Wiley 2002; Luz et al. 2002; Buslepp et al. 2003; Kjer-Nielsen et al. 2003; Stewart-Jones et al. 2003; Hahn et al. 2005; Maynard et al. 2005), and TCR components complexed with bacterial superantigens (Fields et al. 1996; Li et al. 1998b; Sundberg et al. 2002), only eight TCR Vβ families have been structurally characterized. Three are of mouse origin (Vβ2, Vβ5.2, and Vβ8.2) and five are from humans (Vβ2, Vβ3, Vβ6.3, Vβ12.3, and Vβ17). Structural superposition of the human Vβ17C5–1YS Vβ domain in our study onto other TCR Vβ domains resulted in RMSD values that fell into two distinct groups. The Vβ17C5–1 Vβ domain superimposed well onto murine Vβ8.2, and onto human Vβ3 and Vβ12.3, with overall RMSD values ranging from 0.73 Å to 0.98 Å (Fig. 4D ▶). It is notable that these four TCR β chains, murine Vβ8.2 and human Vβ3, Vβ12.3, and Vβ17, are the primary ones used by MAM (Knudtson et al. 1997b). On the other hand, the RMSD values between Vβ17C5–1 and murine Vβ2 and Vβ5.2, and between C5–1YS and human Vβ2.1 and Vβ6.3, are much larger, ranging from 1.24 Å to 1.67 Å (Fig. 4E ▶). T cells bearing these β chains (murine Vβ2 and Vβ5.2, and human Vβ2.1 and Vβ6.3) are known to be nonreactive to MAM stimulation (Knudtson et al. 1997a).
Because MAM can stimulate T cells bearing β chains of a number of Vβ families, regions that show large structural differences among various MAM-reactive β chains probably do not contribute greatly to the binding to MAM. Structural alignments of the MAM-reactive TCR Vβ domains reveal that these Vβ domains superimpose well onto one another, with exceptions at regions 8–12, 40–44, 82–88, CDR3, and HV4; in these regions, relatively large structural differences are observed (Fig. 4D ▶). In addition to the regions where large structural differences were observed among MAM-reactive Vβ domains, significant structural differences between MAM-reactive and nonreactive TCR Vβ domains were observed at the CDR1, CDR2, and FR3 regions (Fig. 4E ▶). It should be noted that the latter two regions contribute most strongly to the binding to the pyrogenic SAgs (Fields et al. 1996; Li et al. 1998b, 1999; Sundberg et al. 2002). Because MAM is known to activate T cells bearing murine Vβ8.2, and human Vβ3, Vβ12.3, and Vβ17, but not murine Vβ2, Vβ5.2, or human Vβ2.1, Vβ6.3, the structural differences in the CDR2 and FR3 regions indicate that these portions of the TCR β chains account for the binding to MAM. Since the TCR β chains recognized by various SAgs differ significantly in the CDR2 and FR3 regions, we could extend the hypothesis to all SAgs: The CDR2 and FR3 regions of TCR β chains may determine the SAg specificity. In other words, the SAgs may uniformly bind to the CDR2 and FR3 regions of TCR β chains. Structural studies of TCR β-chain complexed with MAM or other SAgs are needed before we can determine whether a common recognition mechanism operates to permit MAM and other SAgs to interact with various Vβ families.
Materials and methods
Expression plasmid constructions
The cDNA plasmids encoding the β chains (Vβ17) of T cell receptors (C5–1 and SF4) derived from the synovial fluid and tissue of a RA patient (Li et al. 1994) were used as the source for the molecular cloning. The primers used to generate TCR β chains (residues 1–242) were 5′-GGAATTCCATATGGATG GTGGAATCACTCAGTCC-3′ (upstream primer) and 5′-CC GCTCGAGTCAGTCTGCTCTACCCCAGGCCTC-3′ (downstream primer), and they contained the NdeI and XhoI restriction sites (underlined), respectively. The PCR products were subcloned into the TA cloning vector (Invitrogen). The resulting positive plasmids were confirmed by restriction mapping and DNA sequencing analysis. The plasmids were double-digested with NdeI and XhoI, and the extracted inserts were ligated into the pET26b(+) vector (Novagen), to generate the expression plasmid. For Vβ17C5–1 (Vβ17–Dβ2–Jβ2.1–Cβ2), further mutagenesis was carried out to change the free cysteine, Cys187, to a serine. In order to generate a TCR Vβ17 molecule with high affinity to the SAg MAM (Hodtsev et al. 1998), we introduced a F104Y mutation into Vβ17 C5–1 (designated as C5–1YS). Pairs of primers (C187S: 5′-AATGACTCCAGATAC TCCCTGAGCAGCCGCCTGAG-3′ and 5′-CTCAGGCGGC TGCTCAGGGAGTATCTGGAGTCATT-3′; F104Y: 5′-CAG ATGAATGAGCAGTACTTCGGGCCAGGGACACG-3′ and 5′-CGTGTCCCTGGCCCGAAGTACTGCTCATTC ATCTG) were used with the expression plasmid pET26b-Vβ17C5–1 as the template for site-directed mutagenesis (Stratagene). The correct mutation of the resulting mutant plasmid was confirmed by DNA sequencing analysis.
Protein expression, purification, and refolding
Native proteins of Vβ17C5–1YS and Vβ17SF4 were expressed as inclusion bodies in Escherichia coli BL21 (DE3)pLysS (Novagen). To produce selenium-methionine (Se-Met)-substituted Vβ17C5–1YS, the expression plasmid was transformed into the methionine auxotrophic E. coli strain B834 (Novagen). Cells were grown in minimal medium containing Se-Met at 37°C. Soluble native and Se-Met-substituted Vβ17 proteins were refolded from purified inclusion bodies, using the protocol as described (Garboczi et al. 1996b). The refolded proteins were concentrated in an Amicon stirred cell unit (Millipore), filtered to remove precipitates, and subjected to gel filtration chromatography on a Superdex-200 column (Pharmacia). The peak fractions were pooled together and dialyzed at 4° against 20 mM Tris buffer (pH 8.0). Further purification was carried out on a Pharmacia MonoQ anion-exchange column with a linear NaCl gradient. Fractions containing the Vβ17 proteins were recovered, buffer-exchanged, and concentrated to about 5 mg/mL in the buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM DTT, for further analysis. For Se-Met-substituted protein, 5 mM DTT was added, to prevent oxidization of the Se-Met.
Crystallization, diffraction data collection, structure determination, and refinement
Using the hanging drop evaporation method, we obtained small crystals, under condition 16 of the Hampton Crystallization Screen (1.5 M lithium sulfate as the precipitate) within 30 min after the start of the crystallization experiment. After optimization, the best crystals were obtained at room temperature by mixing 2 μ L of protein solution with an equal volume of 0.6–0.7 M sodium citrate, 0.1 M HEPES (pH 7.5). Eye-shaped crystals grew to dimensions up to 0.3 × 0.3 × 0.1 mm3 within 1 wk. The crystals belong to space group P6322, with cell dimensions a = b = 187.5 Å, c = 86.7 Å for native Vβ17SF4, and a = b = 187.3 Å, c = 86.3 Å for Se-Met-substituted Vβ17C5–1YS. There are two TCR β chains per asymmetric unit.
Prior to data collection, all crystals were transferred to a reservoir solution containing 20% glycerol, and then flash-cooled under a nitrogen stream at 100 K. Data for the native Vβ17SF4 crystals were collected to 2.65 Å resolution at 100 K at beamline X25 of the National Synchrotron Light Source (NSLS) of the Brookhaven National Laboratory (BNL) (Table 1). All of the diffraction data were processed and scaled using HKL2000 (Otwinowski and Minor 1997), followed by truncation with programs in the CCP4 suite (CCP4 1994).
Attempts to determine the crystal structures of Vβ17SF4 by molecular replacement with known structures of the TCR Vβ domains, including Vβ17 (Stewart-Jones et al. 2003), as the search models failed. We therefore determined the crystal structures using the MAD phasing method. Vβ17C5–1YS was selected for the production of Se-Met-substituted protein, because Vβ17C5–1YS has two nonterminal Met residues, as compared to one in Vβ17SF4. A set of three-wavelength MAD data was collected to 2.7 Å for the Se-Met-substituted Vβ17C5–1YS crystals at beamline X25 of the NSLS (Table 1). MAD calculation using the program SOLVE (Terwilliger and Berendzen 1999) clearly defined the positions of the two Se atoms. Further search for additional sites failed to generate obvious solutions, presumably because other Met residues were not well-ordered in the crystal. Nevertheless, an interpretable electron density map could be generated after density modification using the program RESOLVE (Terwilliger 2001). Polyalanine fragments containing about 60% of the Vβ17C5–1YS residues of one molecule and 40% of the residues of the second molecule could be automatically traced. The partial structure showed β-strand structure, which is typical for a TCR. Therefore, the variable and constant domains of the known Vβ17 structure of a JM22 TCR (Stewart-Jones et al. 2003) were manually aligned with this partial structure using the program TURBO-FRODO (Roussel and Cambillau 1989). The structure refinement was completed using CNS (Brunger et al. 1998). After the rigid-body refinement, iterative cycles of simulated annealing and positional, torsion angle, and temperature factor (B) refinements, were carried out with noncrystallographic symmetry (NCS) restrains; these cycles were interspersed with model rebuilding into σA-weighted (Fo − Fc) and (2Fo − Fc) electron density maps. At 2.7 Å resolution, the final Rcryst is 23.7%, with an Rfree of 28.0%.
The structure of Vβ17SF4 was determined by the difference Fourier method, using the partially refined structure of Vβ17C5–1YS as a starting model. The structure refinement was carried out similarly to that done for Vβ17C5–1YS using CNS. Omit maps for groups of Vβ17 CDR3 residues were frequently calculated, as a check of the correct tracing and conformations. At 2.65 Å resolution, the final Rcryst is 23.3% with an Rfree of 28.6%.
For the nonglycine residues, the main-chain torsion angles of all residues lie in the most favored or allowed regions of the Ramachandran plot (data not shown). The refinement statistics are summarized in Table 1. Atomic coordinates for Vβ17C5–1YS and Vβ17SF4 have been deposited in the Protein Data Bank (PDB) as entries 2AXH and 2AXJ.
Analytical ultracentrifugation
Sedimentation-velocity experiments were done at 20°C in a Beckman Optima XL-I analytical ultracentrifuge at a rotor speed of 55,000 rpm. (An50Ti rotor). Double-sector cells were loaded with 400 μ L of proteins at a concentration of 14 μ M (Vβ17SF4), 42 μ M (Vβ17C5–1YS), or 140 μ M (Vβ17SF4), in 2 mM DTT, 0.1 M NaCl, 10 mM Hepes buffer (pH 7.5). Data were recorded with absorbance detection at wavelengths of 280, 294, and 300 nm for low, moderate, and high concentrations of protein, respectively. Absorbance fringe displacement profiles were analyzed with the software SEDFIT (http://www.analyticalultracentrifugation.com) (Vistica et al. 2004), using a model for continuous sedimentation coefficient distributions c(s) (Schuck et al. 2002). Distributions were calculated with maximum entropy regularization at a predetermined confidence level of 1 standard deviation. In further analysis, the differential S value distribution was integrated, to determine weight-average sedimentation coefficients.
Sedimentation equilibrium studies were conducted at a temperature of 20°C and at three rotor speeds of 20,000 rpm, 25,000 rpm, and 30,000 rpm. 110 μ L of proteins were respectively loaded into Epon double-sector centerpieces, at a concentration of 8.7 μ M, 14 μ M, or 22.3 μ M, in 2 mM DTT, 0.1 M NaCl, 10 mM Hepes buffer (pH 7.5). Equilibrium absorbance profiles were acquired at 294-nm wavelength. The equilibrium sedimentation data were analyzed using the software SEDPHAT (http://www.analyticalultracentrifugation.com) (Vistica et al. 2004). Data analysis was performed by global least-squares analysis of the data from multiple concentrations and multiple rotor speeds, based on the well-known superposition of the Boltzmann distributions of ideal species in the centrifugal field, using conservation of mass constraints (Vistica et al. 2004).
Acknowledgments
This research is supported by grant AI50628 from the NIH (to H.L.). We thank the Biochemistry and Macromolecular Crystallography Core facilities at the Wadsworth Center for assistance with the sedimentation and crystallography experiments, and the Molecular Genetics Core facility for DNA sequencing. We also thank A. Verschoor at the Wadsworth Center for critical review of the manuscript and R. Sweet and M. Becker at NSLS for assistance in X-ray data collection. The X-ray diffraction facilities at the NSLS are supported by the Department of Energy and by grants from the NIH.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051748305.
References
- Acuto, O., Hussey, R.E., and Reinherz, E.L. 1985. Multiple class I and class II major histocompatibility complex allospecificities are generated with T cell receptor variable (V) domains created by a single Ti β V gene family. J. Exp. Med. 162 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam, S.M., Sim, B.C., and Gascoigne, N.R. 1995. Selection of TCR V α by MHC class II predicts superantigen reactivity. Int. Immunol. 7 1311–1318. [DOI] [PubMed] [Google Scholar]
- Al-Lazikani, B., Lesk, A.M., and Chothia, C. 2000. Canonical structures for the hypervariable regions of T cell αβ receptors. J. Mol. Biol. 295 979–995. [DOI] [PubMed] [Google Scholar]
- Allison, T.J. and Garboczi, D.N. 2002. Structure of γδ T cell receptors and their recognition of non-peptide antigens. Mol. Immunol. 38 1051–1061. [DOI] [PubMed] [Google Scholar]
- Banerjee, S., Haqqi, T.M., Luthra, H.S., Stuart, J.M., and David, C.S. 1988a. Possible role of V β T cell receptor genes in susceptibility to collagen-induced arthritis in mice. J. Exp. Med. 167 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S., Luthra, H.S., Moore, S.B., and O’Fallon, W.M. 1988b. Serum IgG anti-native type II collagen antibodies in rheumatoid arthritis: Association with HLA DR4 and lack of clinical correlation. Clin. Exp. Rheumatol. 6 373–380. [PubMed] [Google Scholar]
- Bentley, G.A., Boulot, G., Karjalainen, K., and Mariuzza, R.A. 1995. Crystal structure of the β chain of a T cell antigen receptor. Science 267 1984–1987. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Buslepp, J., Wang, H., Biddison, W.E., Appella, E., and Collins, E.J. 2003. A correlation between TCR Vα docking on MHC and CD8 dependence: Implications for T cell selection. Immunity 19 595–606. [DOI] [PubMed] [Google Scholar]
- Chini, L., Bardare, M., Cancrini, C., Angelini, F., Mancia, L., Cortis, E., Finocchi, A., Riccardi, C., and Rossi, P. 2002. Evidence of clonotypic pattern of T-cell repertoire in synovial fluid of children with juvenile rheumatoid arthritis at the onset of the disease. Scand. J. Immunol. 56 512–517. [DOI] [PubMed] [Google Scholar]
- Cole, B.C. and Griffiths, M.M. 1993. Triggering and exacerbation of autoimmune arthritis by the Mycoplasma arthritidis superantigen MAM. Arthritis Rheum. 36 994–1002. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project, Number 4 (CCP4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Colman, P.M. 1988. Structure of antibody-antigen complexes: Implications for immune recognition. Adv. Immunol. 43 99–132. [DOI] [PubMed] [Google Scholar]
- Conrad, B., Weidmann, E., Trucco, G., Rudert, W.A., Behboo, R., Ricordi, C., Rodriquez-Rilo, H., Finegold, D., and Trucco, M. 1994. Evidence for superantigen involvement in insulin-dependent diabetes mellitus aetiology. Nature 371 351–355. [DOI] [PubMed] [Google Scholar]
- Cuesta, I.A., Sud, S., Song, Z., Affholter, J.A., Karvonen, R.L., Fernandez- Madrid, F., and Wooley, P.H. 1997. T cell receptor (V β) bias in the response of rheumatoid arthritis synovial fluid T cells to connective tissue antigens. Scand. J. Rheumatol. 26 166–173. [DOI] [PubMed] [Google Scholar]
- Degano, M., Garcia, K.C., Apostolopoulos, V., Rudolph, M.G., Teyton, L., and Wilson, I.A. 2000. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity 12 251–261. [DOI] [PubMed] [Google Scholar]
- Dessen, A., Lawrence, C.M., Cupo, S., Zaller, D.M., and Wiley, D.C. 1997. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity 7 473– 481. [DOI] [PubMed] [Google Scholar]
- Ding, Y.H., Smith, K.J., Garboczi, D.N., Utz, U., Biddison, W.E., and Wiley, D.C. 1998. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity 8 403–411. [DOI] [PubMed] [Google Scholar]
- Ding, Y.H., Baker, B.M., Garboczi, D.N., Biddison, W.E., and Wiley, D.C. 1999. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11 45–56. [DOI] [PubMed] [Google Scholar]
- Emery, P., Panayi, G.S., Welsh, K.I., and Cole, B.C. 1985. Rheumatoid factors and HLA-DR4 in RA. J. Rheumatol. 12 217–222. [PubMed] [Google Scholar]
- Fields, B.A., Ober, B., Malchiodi, E.L., Lebedeva, M.I., Braden, B.C., Ysern, X., Kim, J.K., Shao, X., Ward, E.S., and Mariuzza, R.A. 1995. Crystal structure of the V α domain of a T cell antigen receptor. Science 270 1821–1824. [DOI] [PubMed] [Google Scholar]
- Fields, B.A., Malchiodi, E.L., Li, H., Ysern, X., Stauffacher, C.V., Schlievert, P.M., Karjalainen, K., and Mariuzza, R.A. 1996. Crystal structure of a T-cell receptor β-chain complexed with a superantigen. Nature 384 188–192. [DOI] [PubMed] [Google Scholar]
- Fugger, L., Michie, S.A., Rulifson, I., Lock, C.B., and McDevitt, G.S. 1994. Expression of HLA-DR4 and human CD4 transgenes in mice determines the variable region β-chain T-cell repertoire and mediates an HLA-DR-restricted immune response. Proc. Natl. Acad. Sci. 91 6151–6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi, D.N., Ghosh, P., Utz, U., Fan, Q., Biddison, W., and Wiley, D.C. 1996a. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384 134–141. [DOI] [PubMed] [Google Scholar]
- Garboczi, D.N., Utz, U., Ghosh, P., Seth, A., Kim, J., VanTienhoven, E.A., Biddison, W.E., and Wiley, D.C. 1996b. Assembly, specific binding, and crystallization of a human TCR-αβ with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J. Immunol. 157 5403–5410. [PubMed] [Google Scholar]
- Garcia, K.C., Degano, M., Stanfield, R.L., Brunmark, A., Jackson, M.R., Peterson, P.A., Teyton, L., and Wilson, I.A. 1996. An αβ T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science 274 209–219. [DOI] [PubMed] [Google Scholar]
- Garcia, K.C., Degano, M., Pease, L.R., Huang, M., Peterson, P.A., Teyton, L., and Wilson, I.A. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279 1166– 1172. [DOI] [PubMed] [Google Scholar]
- Garcia, K.C., Teyton, L., and Wilson, I.A. 1999. Structural basis of T cell recognition. Annu. Rev. Immunol. 17 369–397. [DOI] [PubMed] [Google Scholar]
- Goronzy, J.J., Zettl, A., and Weyand, C.M. 1998. T cell receptor repertoire in rheumatoid arthritis. Int. Rev. Immunol. 17 339–363. [DOI] [PubMed] [Google Scholar]
- Gregersen, P.K., Silver, J., and Winchester, R.J. 1987. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30 1205–1213. [DOI] [PubMed] [Google Scholar]
- Grom, A.A., Thompson, S.D., Luyrink, L., Passo, M., Choi, E., and Glass, D.N. 1993. Dominant T-cell-receptor β chain variable region Vβ14+ clones in juvenile rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 90 11104–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guddat, L.W., Shan, L., Anchin, J.M., Linthicum, D.S., and Edmundson, A.B. 1994. Local and transmitted conformational changes on complexation of an anti-sweetener Fab. J. Mol. Biol. 236 247–274. [DOI] [PubMed] [Google Scholar]
- Guddat, L.W., Shan, L., Fan, Z.C., Andersen, K.N., Rosauer, R., Linthicum, D.S., and Edmundson, A.B. 1995. Intramolecular signaling upon complexation. FASEB J. 9 101–106. [DOI] [PubMed] [Google Scholar]
- Hahn, M., Nicholson, M.J., Pyrdol, J., and Wucherpfennig, K.W. 2005. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 6 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqqi, T.M., Anderson, G.D., Banerjee, S., and David, C.S. 1992. Restricted heterogeneity in T-cell antigen receptor V β gene usage in the lymph nodes and arthritic joints of mice. Proc. Natl. Acad. Sci. 89 1253–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqqi, T.M., Qu, X.M., and Banerjee, S. 1995a. Limited heterogeneity in T-cell receptor V β chain gene expression in arthritic joints of BUB/BnJ (H-2q) mice—a T-cell receptor V β a strain. Ann. N. Y. Acad. Sci. 756 221–224. [DOI] [PubMed] [Google Scholar]
- Haqqi, T.M., Qu, X.M., Sy, M.S., and Banerjee, S. 1995b. Restricted expression of T cell receptor V β and lymphokine genes in arthritic joints of a TCR V β a (H-2q) mouse strain-BUB/BnJ-with collagen-induced arthritis. Autoimmunity 20 163–170. [DOI] [PubMed] [Google Scholar]
- Hare, B.J., Wyss, D.F., Osburne, M.S., Kern, P.S., Reinherz, E.L., and Wagner, G. 1999. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat. Struct. Biol. 6 574–581. [DOI] [PubMed] [Google Scholar]
- Hennecke, J. and Wiley, D.C. 2002. Structure of a complex of the human αβ T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): Insight into TCR cross-restriction and alloreactivity. J. Exp. Med. 195 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke, J., Carfi, A., and Wiley, D.C. 2000. Structure of a covalently stabilized complex of a human αβ T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 19 5611– 5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraiwa, A., Yamanaka, K., Kwok, W.W., Mickelson, E.M., Masewicz, S., Hansen, J.A., Radka, S.F., and Nepom, G.T. 1990. Structural requirements for recognition of the HLA-Dw14 class II epitope: A key HLA determinant associated with rheumatoid arthritis. Proc. Natl. Acad. Sci. 87 8051–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodtsev, A.S., Choi, Y., Spanopoulou, E., and Posnett, D.N. 1998. Mycoplasma superantigen is a CDR3-dependent ligand for the T cell antigen receptor. J. Exp. Med. 187 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset, D., Mazza, G., Gregoire, C., Piras, C., Malissen, B., and Fontecilla- Camps, J.C. 1997. The three-dimensional structure of a T-cell antigen receptor V α V β heterodimer reveals a novel arrangement of the V β domain. EMBO J. 16 4205–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, M.D., Diveley, J.P., Lundeen, K.A., Esty, A., Winters, S.T., Carlo, D.J., and Brostoff, S.W. 1991. Limited T-cell receptor β-chain heterogeneity among interleukin 2 receptor-positive synovial T cells suggests a role for superantigen in rheumatoid arthritis. Proc. Natl. Acad. Sci. 88 10921–10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky, T.S., Brown, J.H., Gorga, J.C., Stern, L.J., Urban, R.G., Chi, Y.I., Stauffacher, C., Strominger, J.L., and Wiley, D.C. 1994. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature 368 711–718. [DOI] [PubMed] [Google Scholar]
- Khare, S.D., Krco, C.J., Griffiths, M.M., Luthra, H.S., and David, C.S. 1995. Oral administration of an immunodominant human collagen peptide modulates collagen-induced arthritis. J. Immunol. 155 3653– 3659. [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Clements, C.S., Brooks, A.G., Purcell, A.W., McCluskey, J., and Rossjohn, J. 2002. The 1.5 Å crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure (Camb.) 10 1521–1532. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Clements, C.S., Purcell, A.W., Brooks, A.G., Whisstock, J.C., Burrows, S.R., McCluskey, J., and Rossjohn, J. 2003. A structural basis for the selection of dominant αβ T cell receptors in antiviral immunity. Immunity 18 53–64. [DOI] [PubMed] [Google Scholar]
- Knudtson, K., Sawitzke, A., and Cole, B. 1997a. The superantigens Mycoplasma arthritidis mitogen (MAM): Physical properties and immunology. In Superantigens: Molecular biology and relavence to human diseases (eds. D. Leung et al.), pp. 339–368. Marcel Dekker, New York.
- Knudtson, K.L., Manohar, M., Joyner, D.E., Ahmed, E.A., and Cole, B.C. 1997b. Expression of the superantigen Mycoplasma arthritidis mitogen in Escherichia coli and characterization of the recombinant protein. Infect. Immun. 65 4965–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin, B.L. 1994. Superantigens and their role in disease. Hosp. Pract. (Off. Ed.) 29 59–63, 68–70. [DOI] [PubMed] [Google Scholar]
- Lanchbury, J.S. and Hall, M.A. 1995. T cell receptor usage in rheumatoid arthritis. Br. Med. Bull. 51 346–358. [DOI] [PubMed] [Google Scholar]
- Landolfi, N.F., Thakur, A.B., Fu, H., Vasquez, M., Queen, C., and Tsurushita, N. 2001. The integrity of the ball-and-socket joint between V and C domains is essential for complete activity of a humanized anti-body. J. Immunol. 166 1748–1754. [DOI] [PubMed] [Google Scholar]
- Lesk, A.M. and Chothia, C. 1988. Elbow motion in the immunoglobulins involves a molecular ball-and-socket joint. Nature 335 188–190. [DOI] [PubMed] [Google Scholar]
- Li, Y., Sun, G.R., Tumang, J.R., Crow, M.K., and Friedman, S.M. 1994. CDR3 sequence motifs shared by oligoclonal rheumatoid arthritis synovial T cells. Evidence for an antigen-driven response. J. Clin. Invest. 94 2525–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lebedeva, M.I., Ward, E.S., and Mariuzza, R.A. 1997. Dual conformations of a T cell receptor V α homodimer: Implications for variability in V α V β domain association. J. Mol. Biol. 269 385– 394. [DOI] [PubMed] [Google Scholar]
- Li, H., Lebedeva, M.I., Llera, A.S., Fields, B.A., Brenner, M.B., and Mariuzza, R.A. 1998a. Structure of the Vδ domain of a human γδ T-cell antigen receptor. Nature 391 502–506. [DOI] [PubMed] [Google Scholar]
- Li, H., Llera, A., Tsuchiya, D., Leder, L., Ysern, X., Schlievert, P.M., Karjalainen, K., and Mariuzza, R.A. 1998b. Three-dimensional structure of the complex between a T cell receptor β chain and the super-antigen staphylococcal enterotoxin B. Immunity 9 807–816. [DOI] [PubMed] [Google Scholar]
- Li, H., Llera, A., Malchiodi, E.L., and Mariuzza, R.A. 1999. The structural basis of T cell activation by superantigens. Annu. Rev. Immunol. 17 435–466. [DOI] [PubMed] [Google Scholar]
- Luz, J.G., Huang, M., Garcia, K.C., Rudolph, M.G., Apostolopoulos, V., Teyton, L., and Wilson, I.A. 2002. Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: A buried alloreactive mutation subtly alters peptide presentation substantially increasing V(β) interactions. J. Exp. Med. 195 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machius, M., Cianga, P., Deisenhofer, J., and Ward, E.S. 2001. Crystal structure of a T cell receptor Vα11 (AV11S5) domain: New canonical forms for the first and second complementarity determining regions. J. Mol. Biol. 310 689–698. [DOI] [PubMed] [Google Scholar]
- Maynard, J., Petersson, K., Wilson, D.H., Adams, E.J., Blondelle, S.E., Boulanger, M.J., Wilson, D.B., and Garcia, K.C. 2005. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: Insights into MHC bias and antigen specificity. Immunity 22 81–92. [DOI] [PubMed] [Google Scholar]
- Moder, K.G., Luthra, H.S., Griffiths, M., and David, C.S. 1993. Prevention of collagen-induced arthritis in mice by deletion of T cell receptor V β 8 bearing T cells with monoclonal antibodies. Br. J. Rheumatol. 32 26–30. [DOI] [PubMed] [Google Scholar]
- Moreland, L.W., Heck Jr., L.W., Koopman, W.J., Saway, P.A., Adamson, T.C., Fronek, Z., O’Connor, R.D., Morgan, E.E., Diveley, J.P., Richieri, S.P., et al. 1996. V β 17 T cell receptor peptide vaccination in rheumatoid arthritis: Results of phase I dose escalation study. J. Rheumatol. 23 1353–1362. [PubMed] [Google Scholar]
- Moreland, L.W., Morgan, E.E., Adamson 3rd, T.C., Fronek, Z., Calabrese, L.H., Cash, J.M., Markenson, J.A., Matsumoto, A.K., Bathon, J., Matteson, E.L., et al. 1998. T cell receptor peptide vaccination in rheumatoid arthritis: A placebo-controlled trial using a combination of Vβ3, Vβ14, and Vβ17 peptides. Arthritis Rheum. 41 1919–1929. [DOI] [PubMed] [Google Scholar]
- Mu, H.H., Sawitzke, A.D., and Cole, B.C. 2000. Modulation of cytokine profiles by the Mycoplasma superantigen Mycoplasma arthritidis mitogen parallels susceptibility to arthritis induced by M. arthritidis. Infect. Immun. 68 1142–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom, G.T. and Erlich, H. 1991. MHC class-II molecules and auto-immunity. Annu. Rev. Immunol. 9 493–525. [DOI] [PubMed] [Google Scholar]
- Nepom, G.T., Byers, P., Seyfried, C., Healey, L.A., Wilske, K.R., Stage, D., and Nepom, B.S. 1989. HLA genes associated with rheumatoid arthritis. Identification of susceptibility alleles using specific oligo-nucleotide probes. Arthritis Rheum. 32 15–21. [DOI] [PubMed] [Google Scholar]
- Oldstone, M.B. 1987. Molecular mimicry and autoimmune disease. Cell 50 819–820. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Paliard, X., West, S.G., Lafferty, J.A., Clements, J.R., Kappler, J.W., Marrack, P., and Kotzin, B.L. 1991. Evidence for the effects of a superantigen in rheumatoid arthritis. Science 253 325–329. [DOI] [PubMed] [Google Scholar]
- Panayi, G.S., Lanchbury, J.S., and Kingsley, G.H. 1992. The importance of the T cell in initiating and maintaining the chronic synovitis of rheumatoid arthritis. Arthritis Rheum. 35 729–735. [DOI] [PubMed] [Google Scholar]
- Plaksin, D., Chacko, S., Navaza, J., Margulies, D.H., and Padlan, E.A. 1999. The X-ray crystal structure of a Vα2.6Jα38 mouse T cell receptor domain at 2.5 Å resolution: Alternate modes of dimerization and crystal packing. J. Mol. Biol. 289 1153–1161. [DOI] [PubMed] [Google Scholar]
- Reinherz, E.L., Tan, K., Tang, L., Kern, P., Liu, J., Xiong, Y., Hussey, R.E., Smolyar, A., Hare, B., Zhang, R., et al. 1999. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science 286 1913–1921. [DOI] [PubMed] [Google Scholar]
- Reiser, J.B., Darnault, C., Guimezanes, A., Gregoire, C., Mosser, T., Schmitt-Verhulst, A.M., Fontecilla-Camps, J.C., Malissen, B., Housset, D., and Mazza, G. 2000. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 1 291–297. [DOI] [PubMed] [Google Scholar]
- Reiser, J.B., Gregoire, C., Darnault, C., Mosser, T., Guimezanes, A., Schmitt-Verhulst, A.M., Fontecilla-Camps, J.C., Mazza, G., Malissen, B., and Housset, D. 2002. A T cell receptor CDR3β loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity 16 345–354. [DOI] [PubMed] [Google Scholar]
- Reiser, J.B., Darnault, C., Gregoire, C., Mosser, T., Mazza, G., Kearney, A., van der Merwe, P.A., Fontecilla-Camps, J.C., Housset, D., and Malissen, B. 2003. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 4 241–247. [DOI] [PubMed] [Google Scholar]
- Rosloniec, E.F., Brand, D.D., Myers, L.K., Whittington, K.B., Gumanovskaya, M., Zaller, D.M., Woods, A., Altmann, D.M., Stuart, J.M., and Kang, A.H. 1997. An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen. J. Exp. Med. 185 1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, A. and Cambillau, C. 1989. TURBO-FRODO. In Silicon graphics geometry partners directory, pp. 77–78. Silicon Graphics, Mountain View, CA.
- Rudolph, M.G., Huang, M., Teyton, L., and Wilson, I.A. 2001. Crystal structure of an isolated V(α) domain of the 2C T-cell receptor. J. Mol. Biol. 314 1–8. [DOI] [PubMed] [Google Scholar]
- Sawitzke, A., Joyner, D., Knudtson, K., Mu, H.H., and Cole, B. 2000. Anti-MAM antibodies in rheumatic disease: Evidence for a MAM-like superantigen in rheumatoid arthritis? J. Rheumatol. 27 358–364. [PubMed] [Google Scholar]
- Schuck, P., Perugini, M.A., Gonzales, N.R., Howlett, G.J., and Schubert, D. 2002. Size-distribution analysis of proteins by analytical ultracen-trifugation: Strategies and application to model systems. Biophys. J. 82 1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinella, D.G., Jeffers, J.R., Reife, R.A., and Stuart, J.M. 1991. The role of C5 and T-cell receptor Vβ genes in susceptibility to collagen-induced arthritis. Immunogenetics 34 23–27. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones, G.B., McMichael, A.J., Bell, J.I., Stuart, D.I., and Jones, E.Y. 2003. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4 657–663. [DOI] [PubMed] [Google Scholar]
- Struyk, L., Hawes, G.E., Chatila, M.K., Breedveld, F.C., Kurnick, J.T., and van den Elsen, P.J. 1995. T cell receptors in rheumatoid arthritis. Arthritis Rheum. 38 577–589. [DOI] [PubMed] [Google Scholar]
- Sundberg, E.J., Li, H., Llera, A.S., McCormick, J.K., Tormo, J., Schlievert, P.M., Karjalainen, K., and Mariuzza, R.A. 2002. Structures of two streptococcal superantigens bound to TCR β chains reveal diversity in the architecture of T cell signaling complexes. Structure (Camb.) 10 687–699. [DOI] [PubMed] [Google Scholar]
- Sundberg, E.J., Andersen, P.S., Schlievert, P.M., Karjalainen, K., and Mariuzza, R.A. 2003. Structural, energetic, and functional analysis of a protein–protein interface at distinct stages of affinity maturation. Structure (Camb.) 11 1151–1161. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. 2001. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. D Biol. Crystallogr. 57 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated structure solution for MIR and MAD. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu, Y., Wege, H., Straus, A., Ott, M., Bannwarth, W., Lanchbury, J., Panayi, G., and Steinmetz, M. 1991. The T-cell-receptor repertoire in the synovial fluid of a patient with rheumatoid arthritis is polyclonal. Proc. Natl. Acad. Sci. 88 8534–8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya, Y., Bill, J., Palmer, E., Gollob, K., Takagaki, Y., and Kanagawa, O. 1989. Analysis of a monoclonal rat antibody directed to the α-chain variable region (V α 3) of the mouse T cell antigen receptor. J. Immunol. 143 2602–2608. [PubMed] [Google Scholar]
- Van Boxel, J.A. and Paget, S.A. 1975. Predominantly T-cell infiltrate in rheumatoid synovial membranes. N. Engl. J. Med. 293 517–520. [DOI] [PubMed] [Google Scholar]
- VanderBorght, A., van der Aa, A., Geusens, P., Vandevyver, C., Raus, J., and Stinissen, P. 1999. Identification of overrepresented T cell receptor genes in blood and tissue biopsies by PCR-ELISA. J. Immunol. Methods 223 47–61. [DOI] [PubMed] [Google Scholar]
- Van Laar, J.M., Miltenburg, A.M., Verdonk, M.J., Daha, M.R., De Vries, R.R., Van den Elsen, P.J., and Breedveld, F.C. 1991. Lack of T cell oligoclonality in enzyme-digested synovial tissue and in synovial fluid in most patients with rheumatoid arthritis. Clin. Exp. Immunol. 83 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistica, J., Dam, J., Balbo, A., Yikilmaz, E., Mariuzza, R.A., Rouault, T.A., and Schuck, P. 2004. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326 234–256. [DOI] [PubMed] [Google Scholar]
- Wang, J., Lim, K., Smolyar, A., Teng, M., Liu, J., Tse, A.G., Hussey, R.E., Chishti, Y., Thomson, C.T., Sweet, R.M., et al. 1998. Atomic structure of an αβ T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. EMBO J. 17 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, J.H., Tsai, W.C., Tsai, J.J., Chen, C.J., Lin, C.H., Ou, T.T., and Liu, H.W. 1998. T cell receptor gene V α and V β usage in patients with rheumatoid arthritis in Taiwan. Kaohsiung J. Med. Sci. 14 251–257. [PubMed] [Google Scholar]
- Zagon, G., Tumang, J.R., Li, Y., Friedman, S.M., and Crow, M.K. 1994. Increased frequency of V β 17-positive T cells in patients with rheumatoid arthritis. Arthritis Rheum. 37 1431–1440. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Zhang, G., and Dong, Y. 2002. T cell receptor Vβ gene bias in rheumatoid arthritis. Chin. Med. J. (Engl.) 115 856–859. [PubMed] [Google Scholar]
- Zhao, Y., Li, Z., Drozd, S., Guo, Y., Mourad, W., and Li, H. 2004. Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen- MHC complex. Structure 12 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]



