Abstract
The two active sites of dimeric 5-aminolevulinate synthase (ALAS), a pyridoxal 5′-phosphate (PLP)-dependent enzyme, are located on the subunit interface with contribution of essential amino acids from each subunit. Linking the two subunits into a single polypeptide chain dimer (2XALAS) yielded an enzyme with an approximate sevenfold greater turnover number than that of wild-type ALAS. Spectroscopic and kinetic properties of 2XALAS were investigated to explore the differences in the coenzyme structure and kinetic mechanism relative to those of wild-type ALAS that confer a more active enzyme. The absorption spectra of both ALAS and 2XALAS had maxima at 410 and 330 nm, with a greater A410/A330 ratio at pH ~7.5 for 2XALAS. The 330 nm absorption band showed an intense fluorescence at 385 nm but not at 510 nm, indicating that the 330 nm absorption species is the substituted aldamine rather than the enolimine form of the Schiff base. The 385 nm emission intensity increased with increasing pH with a single pK of ~8.5 for both enzymes, and thus the 410 and 330 nm absorption species were attributed to the ketoenamine and substituted aldamine, respectively. Transient kinetic analysis of the formation and decay of the quinonoid intermediate EQ2 indicated that, although their rates were similar in ALAS and 2XALAS, accumulation of this intermediate was greater in the 2XALAS-catalyzed reaction. Collectively, these results suggest that ketoenamine is the active form of the coenzyme and forms a more prominent coenzyme structure in 2XALAS than in ALAS at pH ~7.5.
Keywords: 5-aminolevulinate synthase, heme biosynthesis, tetrapyrrole, pyridoxal 5′-phosphate, ketoenamine, substituted aldamine
Heme is an essential tetrapyrrole in nearly all living cells. It is the prosthetic group of oxygen carrying molecules (e.g., hemoglobins), gas-sensing receptors (e.g., the bacterial FixL and CooA receptors), electron transport proteins (e.g., cytochromes), drug-metabolizing enzymes (cytochromes P450, peroxidases), and many other enzymes (e.g., NO synthases, guanylyl cyclases, catalases). 5-Aminolevulinate synthase (ALAS) (EC 2.3.1.37) catalyzes the first and regulatory step in heme biosynthesis in mammalian cells. In this step, glycine is condensed with succinyl coenzyme A (CoA) yielding 5-aminolevulinate (ALA), CoA, and carbon dioxide (Ahktar and Jordan 1979; Ferreira and Gong 1995; Ferreira 1999). In humans, of the two genes that encode the two ALAS isoforms, housekeeping or ALAS1 (Bishop et al. 1990) and erythroid-specific or ALAS2 (Cotter et al. 1992), mutations in the ALAS2 gene appear to be responsible for the erythropoietic disorder, X-linked sideroblastic anemia (Bottomley 1999). In addition, reduced hemoglobin synthesis, characteristic of other anemias, may also be associated with decreased ALAS activity, as in chronic anemia of rheumatoid arthritis (Houston et al. 1994).
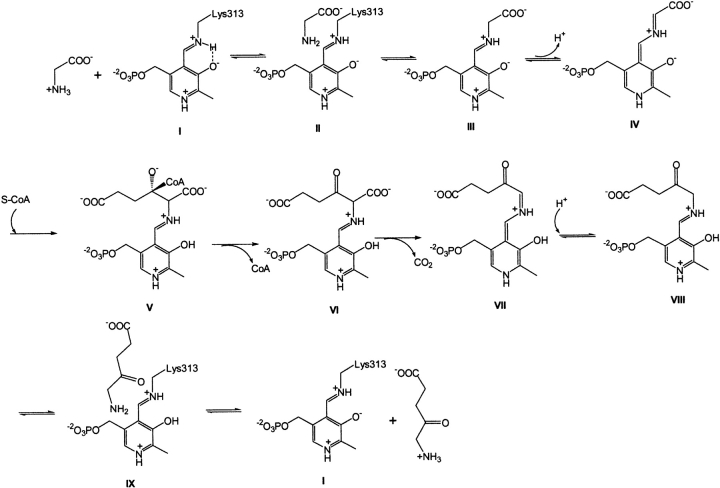
ALAS requires pyridoxal 5′-phosphate (PLP) as an essential cofactor (Ferreira 1999) and has been classified in the α-oxoamine synthase subfamily (Alexeev et al. 1998; Ikushiro et al. 2001; Schmidt et al. 2001) and within the α-family of PLP-dependent enzymes (Alexander et al. 1994) or within class II of fold type I of the PLP-dependent enzyme superfamily (Schneider et al. 2000). The mechanistic information about ALAS indicates that the stereospecific removal of the pro-R proton of the PLP-glycine external aldimine yields a carbanion (i.e., a transient quinonoid intermediate), which then condenses with succinyl-CoA (Zaman et al. 1973; Hunter and Ferreira 1999a,b; Zhang and Ferreira 2002). Briefly, the catalytic pathway of ALAS comprises the following steps (Scheme 1 ▶): (1) association of the glycine substrate with the enzyme, forming the Michaelis complex (II); (2) formation of the external aldimine (III) (i.e., transaldimination); (3) formation of the quinonoid intermediate EQ1 (IV) by abstraction of the α-proton from the PLP-glycine external aldimine; (4) condensation of the succinyl-CoA substrate (V); (5) release of CoA from the tetrahedral intermediate; (6) decarboxylation of the generated α-amino-β-ketoadipate-ALAS aldimine (VI) with formation of a second quinonoid intermediate (EQ2) (VII); (7) protonation of the quinonoid intermediate to form the ALA-ALAS aldimine (VIII); (8) release of ALA and restoration of the internal aldimine (I) (transaldimination) (Hunter and Ferreira 1999b; Zhang and Ferreira 2002) (Scheme 1 ▶).
Scheme 1.
Similar to other fold type I PLP-dependent enzymes, ALAS functions as a homodimer with two active sites per dimer (Eliot and Kirsch 2004). The active sites are located on the dimer interface and comprise residues from each monomer (Tan and Ferreira 1996). K313 (murine ALAS2 numbering) (Schoenhaut and Curtis 1986) forms a Schiff base linkage with the PLP cofactor (Ferreira et al. 1993) and is likely to have a catalytic role (Hunter and Ferreira 1999a). The negatively charged side chain of D279 maintains a hydrogen bond/salt bridge with the protonated pyridine nitrogen of the PLP cofactor (Gong et al. 1998), thus enhancing the electron withdrawing character of the PLP cofactor as observed in other fold type I PLP-dependent enzymes (e.g., aspartate aminotransferase [Yano et al. 1993], tryptophanase [Isupov et al. 1998], 1-aminocyclopropane-1-carboxylate synthase [Eliot and Kirsch 2002], dialkylglycine decarboxylase [Toney et al. 1993], ornithine decarboxylase [Momany et al. 1995], 8-amino-7-oxononanoate synthase [Alexeev et al. 1998], 2-amino-3-ketobutyrate CoA ligase [Schmidt et al. 2001]).
Recently, the construction of a “single-chain dimeric” ALAS (i.e., 2XALAS), which involved the linkage of the two murine erythroid-specific ALAS monomeric units, resulted in a functional, monomeric enzyme with distinct spectroscopic properties and substantially greater enzymatic activity than wild-type murine erythroid-specific ALAS (Cheltsov et al. 2001, 2003). In this study, we examine the differences, at the coenzyme structure and mechanistic levels, between murine erythroid-specific ALAS and 2XALAS that confer an increased turnover to 2XALAS.
Results and Discussion
As with many other PLP-dependent enzymes (e.g., Li et al. 1997; Alexeev et al. 1998; Morollo et al. 1999; Eliot and Kirsch 2004; Paiardini et al. 2004) the two active sites of the ALAS homodimer are located at the subunit interface, and amino acids from both subunits contribute to the architecture of the active sites (Tan and Ferreira 1996). The design of an expression vector with two in-tandem cDNAs of murine erythroid-specific ALAS led to the synthesis of a monomeric protein corresponding to the two linked ALAS subunits, in other words, a single-chain dimer (Cheltsov et al. 2001, 2003). This protein (2XALAS) did not only exhibit ALAS activity but, intriguingly, at 20°C, it also had kcat and kcat/KS–CoA m values about 7- and 117-fold greater, respectively, than those of ALAS (Table 1). A similar trend was observed at 30°C, although with lower enhancements (Table 1). In the present work, we analyzed the spectroscopic and kinetic (transient and steady-state) properties of ALAS and 2XALAS to begin to unravel the possible mechanism whereby enzymatic activity is enhanced by linking the two ALAS subunits.
Table 1.
Steady-state kinetic parameters for reaction of 2XALAS
| Glycine | SCoA | |||||
| Enzyme | Temperaturea (°C) | kcat (sec−1) | Km (mM) | kcat/Km (sec−1 mM−1) | Km (μM) | kcat/Km (sec−1 μM−1) |
| 2XALAS | 20 | 0.11 ± 0.02 | 11.7 ± 0.3 | 0.009 | 0.63 ± 0.03 | 0.175 |
| ALAS | 20 | 0.016 ± 0.002b | 14.0 ± 2.0b | 0.0011b | 11.0 ± 1.0b | 0.0015b |
| 2XALAS | 30 | 0.92 ± 0.02 | 16.7 ± 2.4 | 0.055 | 0.45 ± 0.03 | 2.044 |
| ALAS | 30 | 0.167 ± 0.016c | 23.0 ± 1.0c | 0.007c | 2.30 ± 0.10c | 0.072c |
a Determinations of ALAS activity were conducted at either 20°C or 30°C.
b From Cheltsov et al. (2001).
c From Gong et al. (1998).
Spectroscopic properties of 2XALAS: CD and UV-visible absorption and fluorescence
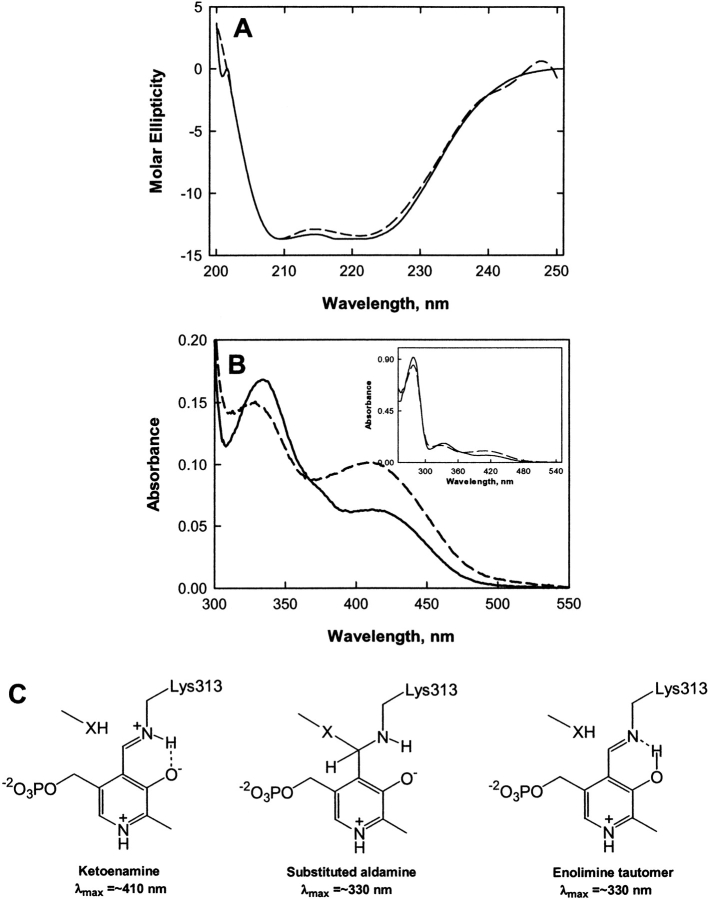
Functional ALAS and 2XALAS have practically the same molecular mass (i.e., ~110 kDa) as the two ALAS subunits (each 56 kDa) are “cross-linked” in 2XALAS. The cross-link resulted from an engineered DNA construct in which two ALAS cDNAs were concatamerized and not from protein chemical cross-linking (Cheltsov et al. 2001, 2003). Thus, the amino acid compositions of ALAS and 2XALAS are essentially the same, except for the two extra amino acids (Gln and Leu), corresponding to the Gln-Leu cross-link-encoding sequence, which were introduced into 2XALAS with the construction of an MfeI site (5′-CAATTG-3′) between the sequences of the two ALAS cDNAS in the 2XALAS-expression plasmid (Cheltsov et al. 2001). Further, while SDS-PAGE indicated a subunit of 56 kDa and ~110 kDa for ALAS and 2XALAS, respectively, gel-filtration chromatography yielded Stokes radii results agreeable with ~112 kDa for both functional ALAS and 2XALAS (Cheltsov et al. 2003). The structural content and overall conformation of 2XALAS, as verified by CD, are identical to those of the wild-type enzyme (Fig. 1A ▶), suggesting that the “linking” of the two subunits did not introduce drastic alterations in the secondary structure of the protein. The ɛ-amino group of K313 of murine erythroid ALAS forms a Schiff base with the PLP carbonyl group yielding the absorption maximum at ~410 nm (Ferreira and Dailey 1993; Ferreira et al. 1993), which is typically observed with PLP-dependent enzymes at neutral pH (Kallen et al. 1985; Li et al. 1997; Ikushiro et al. 1998; Bertoldi et al. 2002). Similar to ALAS, the absorption spectrum of 2XALAS exhibited maxima at 410 and 330 nm (Fig. 1B ▶). However, while the absorbance band at 410 nm was much more pronounced, the absorbance band at 330 nm was less defined in 2XALAS (with A410/A330 of 0.684 and 0.380 for 2XALAS and ALAS at pH 7.5, respectively) (Fig. 1B ▶). With other PLP-dependent enzymes, the 420 nm-absorption species is generally attributed to the ketoenamine form of the internal aldimine (Ahmed et al. 1996; Li et al. 1997; Ikushiro et al. 1998; Zhou and Toney 1999) (Fig. 1C ▶), but the assignment of the 330 nm-absorption species is less straightforward, as several potential structures have the same absorption maximum. Among the potential structures for the 330 nm-absorption species are an aldimine form in which the pyridinium nitrogen is deprotonated, a substituted aldamine (Fig. 1C ▶) and an enolimine tautomer (Fig. 1C ▶).
Figure 1.
Spectroscopic (CD and absorption) properties of ALAS and 2XALAS and possible structures of internal aldimine formed between PLP and the ɛ-amino group of ALAS K313. (A) CD spectra of ALAS (——) and 2XALAS (— —). Both proteins were at 2.0 μM in 20 mM potassium phosphate, pH 7.5, and the spectra were recorded at 25°C. (B) UV-visible absorption spectra (300–550 nm) of ALAS at 15.0 μM (——) and 2XALAS at 7.75 μM (— —). (Inset) UV-visible absorption spectra (250–550 nm) of ALAS at 15.0 μM (——) and 2XALAS at 7.75 μM (— —). Both proteins were in 20 mM HEPES (pH 7.5) containing 10% glycerol. (C) Structures of the ketoenamine, enolimine, and substituted aldamine forms of the internal aldimine.
Deprotonation of the pyridinium nitrogen is an unlikely event for several reasons. If deprotonation were to occur, an absorption maximum at 360 nm should be observed, which is not the case in the ALAS spectra. In ALAS and other fold type I PLP-dependent enzymes (e.g., aspartate aminotransferase [Yano et al. 1993], 8-amino-7-oxononanoate synthase [Toney et al. 1993] and 2-amino-3-ketobutyrate CoA ligase [Schmidt et al. 2001]), a hydrogen bond/salt bridge is maintained between the protonated pyridine nitrogen of the PLP cofactor and the negatively charged side chain of an active-site aspartate residue. The aspartate residue in murine erythroid ALAS is D279, which has been shown to have an essential role in catalysis by stabilizing protonation of the pyridinium ring nitrogen and thus enhancing the electron withdrawing properties of the PLP cofactor (Gong et al. 1998). This interaction is believed to increase dramatically the intrinsic pK value of the ring nitrogen (Gloss and Kirsch 1995), rendering the possibility of deprotonation of the pyridinium nitrogen of ALAS-bound PLP remote. Thus, to elucidate which structure, the enolimine tautomer or the substituted aldamine, is associated with the 330 nm-absorption species, we turned to fluorescence spectroscopy.
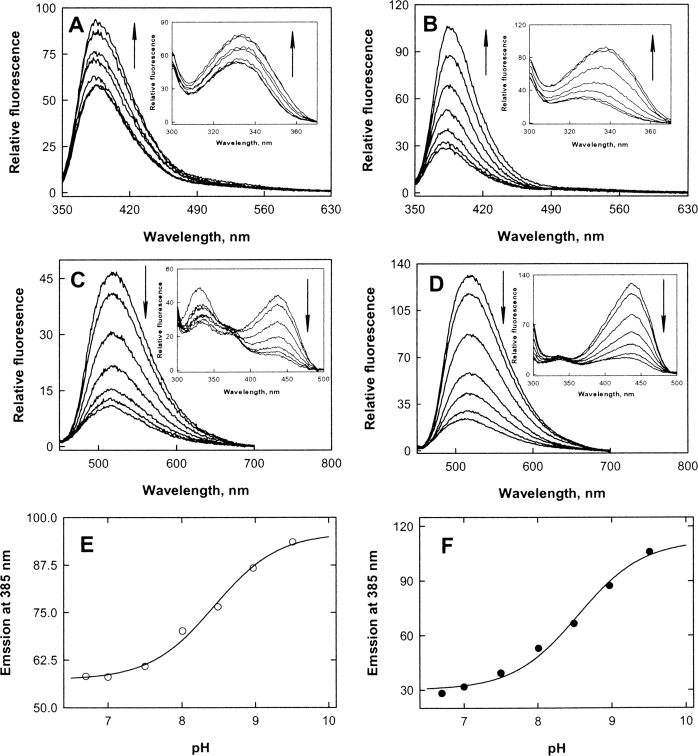
ALAS and 2XALAS emitted fluorescence maxima at 385 and 518 nm upon excitation at 331 and 436 nm, respectively (Fig. 2A–D ▶). (Similar emission spectra, with a maximum at 515 nm, were obtained upon excitation at 420 nm; data not shown.) When 2XALAS fluorescence was monitored at 385 and 518 nm, the excitation spectra exhibited maxima at 331 and 436 nm, respectively, corresponding to the absorption maxima of 2XALAS. However, a second maximum (at 331 nm) in the excitation spectra of ALAS was observed when the fluorescence was monitored at 518 nm. ALAS fluorescence intensity was higher at 385 nm than at 518 nm in the alkaline pH region, although the fluorescence intensity at these wavelengths and pH was similar for 2XALAS (Fig. 2E,F ▶). Emission fluorescence intensity at 518 nm increased and that at 385 nm decreased with decreasing pH for both ALAS and 2XALAS. Indeed, the 385 nm emission fluorescence intensity increased with increasing pH with a single pK of 8.44 ± 0.08 and 8.55 ± 0.10 for ALAS and 2XALAS, respectively (Fig. 2 ▶). It has been shown that a substituted aldamine exhibits a fluorescence maximum intensity at ~390 nm upon excitation at ~330 nm (Hayashi et al. 1993; Ikushiro et al. 1998; Bertoldi et al. 2002), while an enolimine tautomer displays a fluorescence emission maximum at ~515 nm (Ikushiro et al. 1998; Bertoldi et al. 2002). Excitation of a substituted aldamine at 330 nm results in an emission band at ~390 nm (and not at ~515 nm) because of the lack of double bonds conjugated with the pyridinium ring (Ikushiro et al. 1998). These results are consistent with the assignment of a substituted aldamine (and not an enolimine) structure for the 330 nm-absorption band of ALAS (or 2XALAS), although the nature of the nucleophile involved in the formation of the adduct remains to be investigated.
Figure 2.
Fluorescence spectra of ALAS (A,C) and 2XALAS (B,D) and pH profiles for the 385 nm emission fluorescence of ALAS (E) and 2XALAS (F). (A) Fluorescence emission spectra of ALAS at pH 6.7, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5 upon excitation at 331 nm. (Inset) Fluorescence excitation spectra of ALAS at the same pH values with emission wavelength of 385 nm. (B) Fluorescence emission spectra of 2XALAS at pH 6.7, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5 upon excitation at 331 nm. (Inset) Fluorescence excitation spectra of 2XALAS at the same pH values with emission wavelength of 385 nm. (C) Fluorescence emission spectra of ALAS at pH 6.7, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5 upon excitation at 436 nm. (Inset) Fluorescence excitation spectra of ALAS at the same pH values with emission wavelength of 518 nm. (D) Fluorescence emission spectra of 2XALAS at pH 6.7, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5 upon excitation at 436 nm. (Inset) Fluorescence excitation spectra of 2XALAS at the same pH values with emission wavelength of 518 nm. (E) pH dependence of the 385 nm emission intensity of ALAS (excitation at 331 nm). (F) pH dependence of the 385 nm emission intensity of 2XALAS (excitation at 331 nm). The solid lines in E and F represent the theoretical curves from fits of the data to
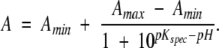 |
Arrows indicate pH increase.
Taken together, the differences between the absorption and fluorescence spectra of ALAS and 2XALAS most likely reflect a slightly different nature of the active sites. It appears that the “linking” of the two ALAS subunits did not affect the global conformation of ALAS but affected the environment of the PLP cofactor—mirrored by the change in the predominant tautomeric form of the internal aldimine.
Steady-state kinetic behavior
The variation of the kinetic parameters of ALAS and 2XALAS with pH is illustrated in Figure 3 ▶. Despite the greater turnover numbers for 2XALAS in relation to those of wild-type ALAS, the pH-rate profiles for the reactions catalyzed by ALAS and 2XALAS are similar (Fig. 3A–C ▶). Log kcat decreases with increasing pH with a pKa of 8.79 ± 0.03 and of 8.65 ± 0.04 for ALAS and 2XALAS, respectively (Fig. 3A ▶). The kcat/Km Gly-pH profile decreased on the acid and basic sides with limiting slopes of 1 and −1, indicating that the ionization of two groups is relevant to the enzymatic reaction, with one being protonated and the other unprotonated (Fig. 3B ▶). However, a fit of the kcat/Km Gly data using Equation 2 indicated that the pKa values were very close to one another (less than 0.16 and 0.69 pH units for ALAS and 2XALAS, respectively) and therefore the same pKa was assumed for both groups, i.e., a pKa value of 8.6 for ALAS and 2XALAS (Fig. 3B ▶). Nevertheless, this pKa value appears to be similar to that controlling the kcat. Variation with pH of pKGlym yielded a pKa value of ~8.6 (Fig. 3C ▶), which likely represents the pKa of a group in the free enzyme, given that glycine does not ionize in the pH range studied and is not a sticky substrate (Cleland 1977).
Figure 3.
pH dependence of logkcat, log kcat/Km Gly and pKm Gly for ALAS (A) 2XALAS (B). The lines represent the nonlinear regression to Equation 2 for logkcat and logkcat/Km Gly vs. pH and Equation 3 for pKm Gly vs. pH. The filled and open symbols are for 2XALAS and ALAS, respectively. The concentrations 2XALAS (or ALAS) and succinyl-CoA were maintained at 1.0 μM (or 4.0 μM) and 20 μM, respectively, while the concentration of glycine covered a range from 0.625 to 200 mM. The buffers for different pH values are described under Materials and Methods.
Besides the involvement in the Schiff base linkage with the PLP cofactor, a catalytic role for K313 was previously advanced (Hunter and Ferreira 1999a). We proposed that K313 could act as a general base catalyst to remove the pro-R proton of glycine and as a general acid to donate a proton to the quinonoid intermediate, EQ2 (Scheme 1 ▶) (Hunter and Ferreira 1999a). In fact, we determined that a pKa of ~8.1 was associated with the titration of EQ2 formed upon the reaction of ALA with ALAS (Hunter and Ferreira 1999a). The kinetic pKas reflected in the kcat/Km versus pH profiles for ALAS (and 2XALAS) (Fig. 3B ▶) are consistent with that determined by titration and with K313 being involved in the abstraction of the proton from the α-position of glycine to afford the quinonoid intermediate, EQ1 (Scheme 1 ▶). On the other hand, although the spectral titrations of the free enzymes, which were the basis for the proposed structures of the coenzyme in ALAS and 2XALAS (ketoenamine and substituted aldamine) (Fig. 1C ▶) yielded pKas of ~8.4 and ~8.5 for ALAS and 2XALAS, respectively (Fig. 2E–F ▶), we believe that this equilibrium does not reflect the protonation state of the iminic nitrogen, but instead it reflects the structural change of the internal aldimine (see below). A second ionizing group could not be distinguished in the kcat/KGlym -pH profiles (Fig. 3B ▶), and while kinetics cannot elucidate how the Michaelis complex is formed, it is expected that, given the pKa of the α-NH3+ of glycine (i.e., 9.6), the amino group of glycine would have to be deprotonated before the transaldimination reaction could occur. The unprotonated phenolate oxygen of the PLP cofactor and the imidazole nitrogen of H282 have been advanced among the possible candidates in the proton abstraction (Zhang and Ferreira 2002).
Kinetics of a pre-steady-state burst of ALA, accumulated EQ2 intermediate and the predominance of the ketoenamine form of the coenzyme
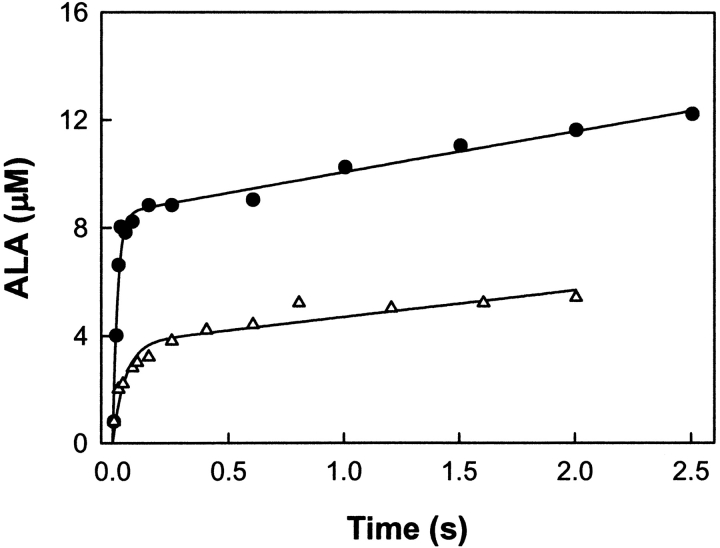
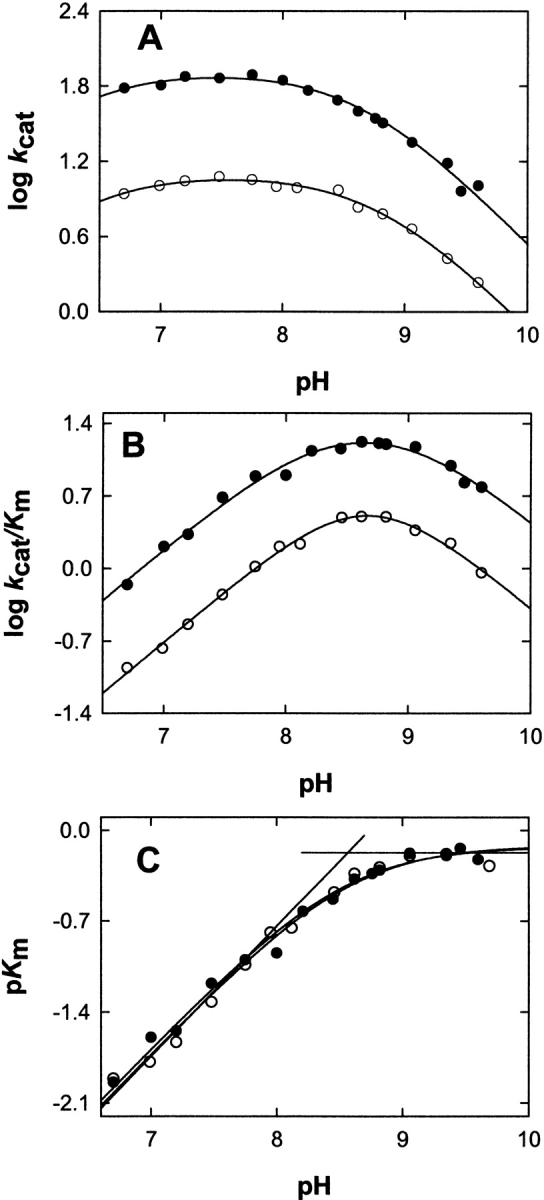
The rate-limiting step of the ALAS-catalyzed reaction has been shown to occur after catalysis, and was proposed to be the ALA release from the enzyme or a protein conformational change associated with it (Hunter and Ferreira 1999b; Zhang and Ferreira 2002). To assess whether (1) the rate-limiting step of the 2XALAS reaction also occurs after the chemical step and (2) the amount of ALA produced in the first turnover correlates with the coenzyme structure, we performed chemical quenched-flow studies and looked for a pre-steady-state burst of ALA formation upon reaction of 2XALAS with saturating concentrations of glycine and succinyl-CoA. Reactions were conducted at 20°C in 25 mM HEPES (pH 7.5) containing 10% glycerol; they were initiated by adding succinyl-CoA (150 μM) to 2XALAS (17.2 μM) preincubated with glycine (200 mM) and stopped at different times with perchloric acid (Fig. 4 ▶). (It was not possible to determine reliably the formation of ALA at 30°C, as the reaction occurred too rapidly and the “burst” could not be clearly defined. Thus, the reactions associated with our chemical quenched-flow studies were performed at 20°C.) The first turnover occurred at a rate of 48.6 ± 6.1 sec−1 and with an amplitude of 0.49/enzyme site, whereas subsequent turnovers were at a rate 0.09 ± 0.01 sec−1, in agreement with the independently determined steady-state rate (kcat = 0.11 ± 0.02) (Table 1). The pre-steady-state burst (Fig. 4 ▶) indicates that the rate-limiting step occurs after the chemical step, which, as with ALAS, we assign to ALA release (Hunter and Ferreira 1999b; Zhang and Ferreira 2002). It appears that while the rate-determining step remained the same in the 2XALAS reaction, the extent of the catalytically competent species, judging from the burst amplitudes, increased in 2XALAS (i.e., burst amplitudes of 0.49 and 0.12/active site [Hunter and Ferreira 1999b; Zhang and Ferreira 2002] for 2XALAS and ALAS, respectively). The amplitude of the burst corresponds to the amount of E • Gly species in the preincubated solution (E + Gly⇌E • Gly) that is catalytically active. Thus, an approximate fourfold increase in the amplitude of the burst suggests a greater proportion of catalytic active species in 2XALAS than in ALAS. This enhancement is consistent with the observed increase in the quinonoid intermediate concentration (Fig. 5 ▶) and correlates with the ketoenamine as the major 2XALAS coenzyme form (Fig. 1B ▶).
Figure 4.
Kinetics of a pre-steady-state burst of ALA product in the 2XALAS reaction. 2XALAS (8.6 μM) (filled circles) preincubated with glycine (200 mM) was quickly reacted with succinyl-CoA (150 μM) at 20°C. The concentrations shown in parentheses are final concentrations after mixing. The reactions were quenched with perchloric acid (0.14 M) at various aging times, and the ALA concentration was determined. The curve represents the best fit to Equation 4 with a burst amplitude of 8.52 ± 0.30 μM, a burst rate of 48.6 ± 6.1 sec−1, and a steady-state rate of 0.09 ± 0.01 sec−1. For comparison, the time course for the reaction of ALAS (30 μM) (open triangles) preincubated with glycine (200 mM) with succinyl-CoA (150 μM) at 20°C is included (from Zhang and Ferreira 2002).
Figure 5.
Reaction of 2XALAS (A) (12.5 μM) or ALAS (B) (50 μM), preincubated with glycine (200 mM), with succinyl-CoA (120 μM) at pH 7.5 and 30°C. (A,B) Selected pre-steady-state spectra from the 2000 spectra collected during the reaction. The concentrations shown in parentheses are final concentrations after mixing. (C) Time courses of the (1) 2XALAS-and (2) ALAS-catalyzed reactions at 510 nm. The time course data were best fitted to Equation 5 for a two-exponential process (see Materials and Methods). An increase, defined by
 |
in the accumulated 510 nm-absorbing species (or EQ2) of 4.1 was associated with the 2XALAS-catalyzed reaction. [E]0 represents the initial enzyme active site concentration.
Moreover, a similar correlation was observed when the formation of the quinonoid intermediate (EQ2) in the ALAS and 2XALAS-catalyzed reactions was monitored using rapid scanning stopped-flow spectroscopy. Previously, using rapid scanning stopped-flow spectroscopy and single turnover conditions, we demonstrated that the catalytic reaction of ALAS involves the formation of two quinonoid reaction intermediates (Scheme 1 ▶); the first one, EQ1, is formed very rapidly (within the dead-time of the instrument, i.e., 2 msec) (Zhang and Ferreira 2002), and the second one, EQ2, a relatively stable quinonoid intermediate, is formed with an estimated rate of ~5 sec−1 (Hunter and Ferreira 1999b; Zhang and Ferreira 2002). When either ALAS (50 μM) or 2XALAS (12.5 μM), preincubated with glycine (200 mM), was reacted with succinyl-CoA (120 μM), a 510-nm absorbing species, corresponding to the quinonoid intermediate EQ2, accumulated (Fig. 5A,B ▶). Although the rates of formation and decay of the intermediate into the steady-state were similar for ALAS and 2XALAS, the concentration of the transiently accumulated quinonoid species was approximately 4.1-fold greater in the 2XALAS-catalyzed reaction (Fig. 5C ▶). This was in agreement with the enhancement observed in the burst amplitude for the 2XALAS reaction.
In several other PLP-dependent enzymes, including tryptophanase (Morino and Snell 1967; Ikushiro et al. 1998) and cystalysin (Bertoldi et al. 2002), the ~420 nm-absorption species, ascribed to the ketoenamine form of the Schiff base of PLP with the active-site lysine residue, has been found to be the active form of the enzyme that reacts with the substrate (Ikushiro et al. 1998). Tryptophanase represents a remarkable example of how, in a given pH range, a PLP-dependent enzyme can be catalytically active in spite of the inactive form of the free enzyme being the major species (Ikushiro et al. 1998). Indeed, tryptophanase is more catalytically competent at alkaline pH (Morino and Snell 1967; Ikushiro et al. 1998), where the 338 nm-absorbing species, a catalytically incompetent, substituted aldamine structure, predominates (Morino and Snell 1967; Ikushiro et al. 1998). The enzyme overcomes this apparent conundrum by converting the structure of its coenzyme from the 338 nm-absorbing species to the ~420-absorbing species in an early step of the reaction cycle (Ikushiro et al. 1998). The binding of the substrate to tryptophanase has been postulated to promote the conversion of the substituted aldamine structure of the coenzyme into the ketoenamine structure and active form of the enzyme. This active form can then undergo transaldimination with the substrate amino group to give the external aldimine necessary for subsequent catalysis (Ikushiro et al. 1998). Specifically, the electrostatic interaction between the coenzyme and the ligand was proposed to induce the removal of the nucleophilic group from the tryptophanase tetrahedral adduct (Ikushiro et al. 1998). Although the mechanism involved in the activation of the cystalysin coenzyme remains to be elucidated, Bertoldi et al. (2002) proposed that a small and localized ligand-induced conformational change of the enzyme could be another plausible explanation to account for the transition between the inactive, substituted aldamine and the active, ketoenamine structure. Whether binding of glycine to ALAS and 2XALAS causes a shift between two (active and inactive) forms of the coenzyme and even if observed, whether the degree of coenzyme conversion differs in ALAS and 2XALAS, remain to be elucidated. However, it seems clear that the rate of the reaction is dependent on the proportion of active sites initially in the ketoenamine form, and this is greater in 2XALAS at pH values ~7.5. If it is a conformational change of the enzyme associated with ALA release that limits turnover, then linking the N and C termini in 2XALAS may result in strain that lowers the energy barrier for interconversion of the enzyme between different conformations required for ALA formation and release. It is also conceivable that the linked structure of 2XALAS better accommodates the succinyl-CoA substrate, as reflected by the 5- to 17.5-fold lower KmS–CoA value for 2XALAS than ALAS (i.e., determinations at 30°C and 20°C, Table 1). Questions related to the nuances of the succinyl-CoA-binding site in ALAS and 2XALAS and the nature of the nucleophile proposed to form an adduct with the PLP-Lys313 aldimine will require the determination of the three-dimensional structures of ALAS and 2XALAS, particularly at high pH for the identification of the structure of the nucleophile. Crystallization studies of ALAS and 2XALAS are in progress. Enhancing the specific activity of ALAS will be relevant in the development of novel ALAS variants of biotechnological interest, as ALAS controls the rate-limiting step of heme biosynthesis. To our knowledge, this is the first example of creating a more active enzyme by just linking its monomeric subunits and raises the possibility of enhancing the activity of enzymes with product release as the rate-limiting step by simply controlling the degree of protein domain movement.
In summary, our data provide evidence that, at neutral pH, the ketoenamine is a more predominant coenzyme structure in the free 2XALAS than in ALAS. Although the rates associated with the formation and decay of the quinonoid intermediate EQ2 are similar for ALAS and 2XALAS, the amplitude of this intermediate in the steady-state increased and an enhanced burst of ALA formation and ALA release were observed in the 2XALAS-catalyzed reaction. These factors probably account for the increased turnover number of 2XALAS.
Materials and methods
Reagents
The following reagents were from Sigma Chemical Co.: DEAE-sephacel, β-mercaptoethanol, p-dimethylamino-benzaldehyde, acetylacetone, PLP, bovine serum albumin, α-ketoglutarate dehydrogenase, α-ketoglutarate, NAD+, thiamin pyrophosphate, succinyl-CoA, HEPES-free acid, AMPSO-free acid, CAPS-free acid, MOPS, tricine, aprotinin, pepstatin, leupeptin, phenylmethylsulfonyl fluoride (PMSF), and the bicinchoninic acid protein concentration determination kit. Glycerol, mono- and dibasic potassium phosphate, sodium acetate, perchloric acid, acetic acid, and disodium ethylenediamine tetraacetic acid dihydrate were provided from Fisher Scientific. Ultrogel AcA-44 was obtained from IBF Biotechnics. Sodium dodecyl sulfate polyacrylamide gel electrophoresis reagents were supplied by Bio-Rad. The Superdex 200 gel-filtration resin and PD-10 gel filtration columns were purchased from Amersham Pharmacia Biotech.
Overexpression, purification, and steady-state kinetic analyses of ALAS and 2XALAS
Recombinant wild-type murine erythroid ALAS and 2XALAS were overproduced and purified from Escherichia coli DH5α and BL21(DE3) cells harboring pGF23 (Ferreira and Dailey 1993) and pAC1 (Cheltsov et al. 2003), ALAS- and 2XALAS-overexpression plasmids, respectively, as previously described (Cheltsov et al. 2003). (The sequence for 2XALAS corresponds to that of two tandem ALAS cDNAs linked through an MfeI site [Cheltsov et al. 2001]). The purified proteins (either ALAS or 2XALAS) were concentrated by pressurized dialysis in an Amicon 8050 stir cell equipped with a YM30 membrane. The concentrated, purified proteins were stored under liquid nitrogen until needed.
Protein concentration determination and SDS-PAGE
Protein concentration was determined by the bicinchoninic acid assay, according to the instructions supplied with the protein concentration determination kit (Sigma Chemical Co.), and using bovine serum albumin as standard. ALAS and 2XALAS concentrations are reported based on a subunit molecular mass of 56,000 Da and 112,000 Da, respectively. Protein purity was assessed by sodium dodecyl sulfate-polyacylamide gel electrophoresis (Laemmli 1970).
UV/visible absorption and fluorescence spectroscopic measurements
All UV/vis absorption spectra were obtained with a Shimadzu UV2100U UV/vis dual beam spectrophotometer. This spectrophotometer is equipped with thermostatically controlled cell holders and allows the exporting of data as ASCII files through the RS232 interface. ALAS activity was measured using a continuous spectrophotometric assay according to a previously described method (Hunter and Ferreira 1995). Fluorescence spectra were collected on a Shimadzu RF-5301 PC spectrofluorophotometer. Spectra of blanks, i.e., of samples containing all component except ALAS (or 2XALAS), were taken immediately prior to measurements of samples containing protein. Blank spectra were subtracted from spectra of samples containing enzyme. Before spectra were acquired, the enzymes were passed through a PD-10 (Pharmacia) gel filtration column equilibrated with 20 mM potassium phosphate (pH 7.5), containing 10% glycerol, to remove free PLP.
Enzymatic assay and determination of steady-state kinetic parameters
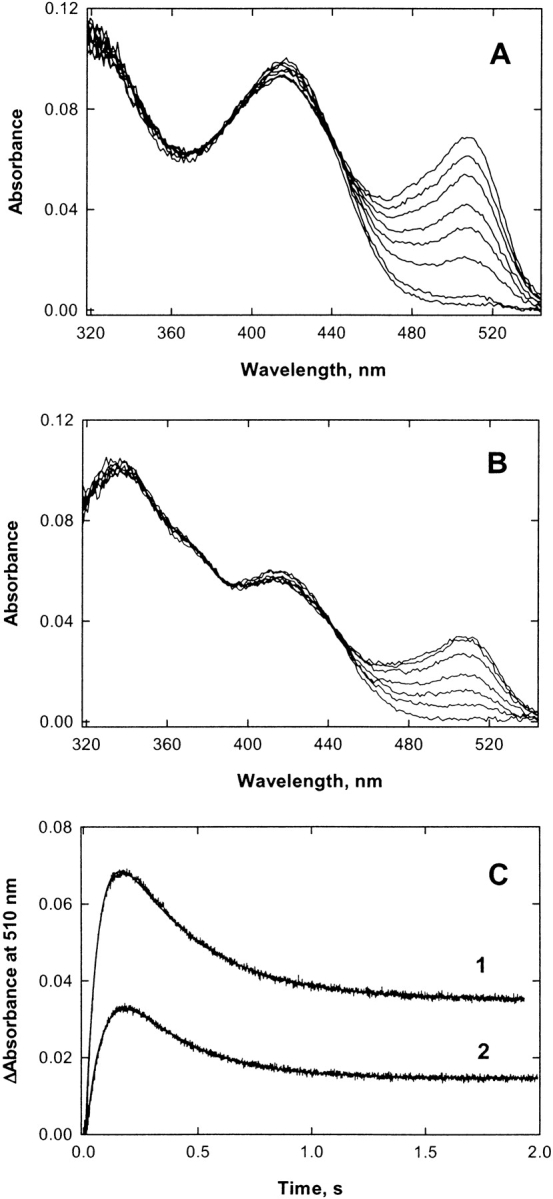
The steady-state kinetic parameters KmGly, KmS–CoA and kcat of ALAS and 2XALAS were determined at pH 7.5 and either 20°C or 30°C, using a continuous spectrophotometric assay (Hunter and Ferreira 1995). To determine Km and Vm values, the concentrations of the substrates were varied (in matrices of six glycine and six succinyl-CoA concentrations), and the observed rates were fitted to the hyperbolic form of the Michaelis-Menten equation (Equation 1) with SigmaPlot (version 7.0):
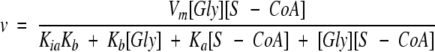 |
(1) |
where Ka is the limiting Michaelis constant for glycine when the succinyl-CoA concentration is saturating, Kb is the limiting Michaelis constant for succinyl-CoA when the glycine concentration is saturating and Kia is the limiting value of the Michaelis constant for glycine when the succinyl-CoA concentration approaches zero. Values of kcat, kcat/KmGly, and kcat/KmS–CoA were calculated by dividing the fitted values of Vm, Vm/KmGly and Vm/KmS–CoA by the concentration of enzyme, using a molecular mass of 56 and 112 kDa for ALAS and 2XALAS, respectively.
pH dependence of the kinetic parameters ALAS and 2XALAS
The enzymatic assays were performed in MOPS (pH 6.7), HEPES (pH 7.0–8.0), or AMPSO (pH 8.2–9.6) at the pH value indicated and at a buffer concentration of 20 mM. Following the reactions, which were run at 30°C and with 4.0 μM of ALAS or 2.0 μM of 2XALAS, the pH of the reaction mixtures was measured. The pH dependencies of log kcat and log kcat/Km and −logKGly m (or pKGly m ) for ALAS and 2XALAS enzymes were fitted to Equations 2 and 3, respectively.
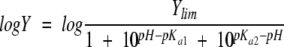 |
(2) |
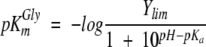 |
(3) |
Rapid chemical quenched-flow experiments and data analysis
Rapid chemical quenched flow experiments were performed using a SFM-400/Q mode quenched-flow apparatus (BioLogic Science Instrument), equipped with a circulating water bath to control the temperature of the reactants essentially as described in Zhang and Ferreira (2002). ALA concentration in the quenched samples was also determined as previously described (Zhang and Ferreira 2002). ALA produced at different reaction times were plotted against time and fitted to Equation 4 (Johnson 1992), using the nonlinear least-squares regression analysis program SigmaPlot, where Pt represents the product concentration at an aging time t, A is the amplitude of the burst phase, kb is the burst rate constant, kss is the steady-state rate constant, and E0 is the total enzyme concentration (Zhang and Ferreira 2002).
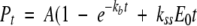 |
(4) |
Rapid scanning stopped-flow spectroscopy and pre-steady-state kinetic data analysis
Rapid scanning stopped-flow kinetic measurements were performed using a model RSM-1000 stopped-flow spectrophotometer (OLIS Inc.). This instrument has a 2 msec dead time, a 4.0-mm path length, and a temperature-controlled observation chamber. In general, scan spectra covering the wavelength range of 317–544 nm were collected at a rate of 1000 spectra per second. A fixed 0.6-mm slit and a 16 × 0.2-mm scandisk were used to collect data. A circulating water bath, controlled thermostatically at 30°C, was used to maintain the temperature of the loading syringes containing the reactants and the stopped-flow cell compartment. Reactant concentrations in the two loading syringes were twofold greater than the final concentrations in the observation chamber. The reaction buffer for the experiments was 50 mM HEPES (pH 7.5), containing 10% glycerol. Time course data were fitted using single-wavelength analysis to the minimum number of exponentials required to obtain a good fit, i.e., the data were fitted to Equation 5 for one to three exponentials using the program provided by OLIS, where At is the absorbance at time t, a is the amplitude of each phase, k is the observed rate for each phase, and c is the final absorbance.
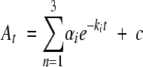 |
(5) |
Acknowledgments
We thank Dr. Gregory A. Hunter for critically reading the manuscript and Mr. W. Christopher Adams for some of the purified 2XALAS batches. This work was supported by the NIH (grant no. DK63191) and the Chiles Endowment Biomedical Research Program of the Florida Department of Health (grant no. BM036). A.V.C. was recipient of an American Heart Association/Florida Division Predoctoral Fellowship.
The opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the Biomedical Research Program of the Florida Department of Health.
Abbreviations
ALA, 5-aminolevulinate
ALAS, 5-aminolevulinate synthase
ALAS2, erythroid-specific ALAS isoform
AMPSO, 3([1,1-dimethyl-2-hydroxyethyl]-amino)-2-hydroxypropane sulfonic acid
AONS, 7-amino-8-oxanonaoate synthase
CAPS, 3(cyclohexylamino)-1-propane sulfonic acid
CD, circular dichroism
DEAE, diethylaminoethyl
EDTA, ethylenediamine tetraacetate
HEPES, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
MOPS, 3-morpholinopropane sulfonic acid
PLP, pyri-doxal 5′phosphate
PMSF, phenylmethylsulfonyl fluoride
SDS, sodium dodecyl sulfate
SDS-PAGE, SDS-polyacrylamide gel electrophoresis
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041258305.
References
- Ahktar, M. and Jordan, P.M. 1979. In Comprehensive organic chemistry: The synthesis and reaction of organic compounds (ed. E. Haslam), 1st ed., pp. 1121–1144. Pergamon Press, New York.
- Ahmed, S.A., McPhie, P., and Miles, E.W. 1996. A thermally induced reversible conformational transition of the tryptophan synthase b2 subunit probed by the spectroscopic properties of pyridoxal phosphate and by enzymatic activity. J. Biol. Chem. 271 8612–8617. [DOI] [PubMed] [Google Scholar]
- Alexander, F.W., Sandmeier, E., Mehta, P.K., and Christen, P. 1994. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific α, β and γ families. Eur. J. Biochem. 219 953–960. [DOI] [PubMed] [Google Scholar]
- Alexeev, D., Alexeeva, M., Baxter, R.L., Campopiano, D.J., Webster, S.P., and Sawyer, L. 1998. The crystal structure of 8-amino-7-oxononanoate synthase: A bacterial PLP-dependent, acyl-CoA-condensing enzyme. J. Mol. Biol. 284 401–419. [DOI] [PubMed] [Google Scholar]
- Bertoldi, M., Cellini, B., Clausen, T., and Voltattorni, C.B. 2002. Spectroscopic and kinetic analyses reveal the pyridoxal 5′-phosphate binding mode and the catalytic features of Treponema denticola cystalysin. Biochemistry 41 9153–9164. [DOI] [PubMed] [Google Scholar]
- Bishop, D.F., Henderson, A.S., and Astrin, K.H. 1990. Human δ-aminolevulinate synthase: Assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics 7 207–214. [DOI] [PubMed] [Google Scholar]
- Bottomley, S.S. 1999. Sideroblastic anemias. In Wintrobe’s clinical hematology (ed. G.R. Lee), pp. 1022–1045. Lippincott Williams & Wilkins, Baltimore, MD.
- Cheltsov, A.V., Barber, M.J., and Ferreira, G.C. 2001. Circular permutation of 5-aminolevulinate synthase. Mapping the polypeptide chain to its function. J. Biol. Chem. 276 19141–19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheltsov, A.V., Guida, W.C., and Ferreira, G.C. 2003. Circular permutation of 5-aminolevulinate synthase: Effect on folding, conformational stability and structure. J. Biol. Chem. 278 27945–27955. [DOI] [PubMed] [Google Scholar]
- Cleland, W.W. 1977. Determining the chemical mechanisms of enzyme-catalyzed reactions by kinetic studies. Adv. Enzymol. Relat. Areas Mol. Biol. 45 273–387. [DOI] [PubMed] [Google Scholar]
- Cotter, P.D., Willard, H.F., Gorski, J.L., and Bishop, D.F. 1992. Assignment of human erythroid δ-aminolevulinate synthase (ALAS2) to a distal subregion of band Xp11.21 by PCR analysis of somatic cell hybrids containing X; autosome translocations. Genomics 13 211–212. [DOI] [PubMed] [Google Scholar]
- Eliot, A.C. and Kirsch, J.F. 2002. Modulation of the internal aldimine pK(a)’s of 1-aminocyclopropane-1-carboxylate synthase and aspartate aminotransferase by specific active site residues. Biochemistry 41 3836–3842. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73 383–415. [DOI] [PubMed] [Google Scholar]
- Ferreira, G.C. 1999. In Iron metabolism. Inorganic biochemistry and regulatory mechanism (eds. G.C. Ferreira et al.), pp. 15–34. Wiley-VCH, Weinheim, Germany.
- Ferreira, G.C. and Dailey, H.A. 1993. Expression of mammalian 5-aminolevulinate synthase in Escherichia coli. Overproduction, purification, and characterization. J. Biol. Chem. 268 584–590. [PubMed] [Google Scholar]
- Ferreira, G.C. and Gong, J. 1995. 5-Aminolevulinate synthase and the first step of heme biosynthesis. J. Bioenergetics Biomembr. 27 151–159. [DOI] [PubMed] [Google Scholar]
- Ferreira, G.C., Neame, P.J., and Dailey, H.A. 1993. Heme biosynthesis in mammalian systems: Evidence of a Schiff base linkage between the pyridoxal 5′-phosphate cofactor and a lysine residue in 5-aminolevulinate synthase. Protein Sci. 2 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss, L.M. and Kirsch, J.F. 1995. Decreasing the basicity of the active site base, Lys-258, of Escherichia coli aspartate aminotransferase by replacement with γ-thialysine. Biochemistry. 34 3990–3998. [DOI] [PubMed] [Google Scholar]
- Gong, J., Hunter, G.A., and Ferreira, G.C. 1998. Aspartate-279 in aminolevulinate synthase affects enzyme catalysis through enhancing the function of the pyridoxal 5′-phosphate cofactor. Biochemistry 37 3509–3517. [DOI] [PubMed] [Google Scholar]
- Hayashi, H., Mizuguchi, H., and Kagamiyama, H. 1993. Rat liver aromatic L-amino acid decarboxylase: Spectroscopic and kinetic analysis of the co-enzyme and reaction intermediates. Biochemistry 32 812–818. [DOI] [PubMed] [Google Scholar]
- Houston, T., Moore, M., Porter, D., Sturrock, R., and Fitzsimons, E. 1994. Abnormal haem biosynthesis in the chronic anaemia of rheumatoid arthritis. Ann. Rheum. Dis. 53 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, G.A. and Ferreira, G.C. 1995. A continuous spectrophotometric assay for 5-aminolevulinate synthase that utilizes substrate cycling. Anal. Biochem. 226 221–224. [DOI] [PubMed] [Google Scholar]
- ———. 1999a. Lysine-313 of 5-aminolevulinate synthase acts as a general base during formation of the quinonoid reaction intermediates. Biochemistry 38 3711–3718. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Pre-steady-state reaction of 5-aminolevulinate synthase. Evidence for a rate-determining product release. J. Biol. Chem. 274 12222–12228. [DOI] [PubMed] [Google Scholar]
- Ikushiro, H., Hayashi, H., Kawata, Y., and Kagamiyama, H. 1998. Analysis of the pH- and ligand-induced spectral transitions of tryptophanase: Activation of the coenzyme at the early steps of the catalytic cycle. Biochemistry 37 3043–3052. [DOI] [PubMed] [Google Scholar]
- Ikushiro, H., Hayashi, H., and Kagamiyama, H. 2001. A water-soluble homodimeric serine palmitoyltransferase from Sphingomonas paucimobilis EY2395T strain. Purification, characterization, cloning, and overproduction. J. Biol. Chem. 276 18249–18256. [DOI] [PubMed] [Google Scholar]
- Isupov, M.N., Antson, A.A., Dodson, E.J., Dodson, G.G., Dementieva, I.S., Zakomirdina, L.N., Wilson, K.S., Dauter, Z., Lebedev, A.A., and Harutyunyan, E.H. 1998. Crystal structure of tryptophanase. J. Mol. Biol. 276 603–623. [DOI] [PubMed] [Google Scholar]
- Johnson, K.A. 1992. Transient-state kinetic analysis of enzyme reaction pathways. Enzymes 20 1–61. [Google Scholar]
- Kallen, R.G., Korpela, T., Martell, A.E., Matsushima, Y., Metzler, C.M., Metzler, D.E., Morozov, Y.V., Ralston, I.M., Savin, F.A., Torchinsky, Y.M., et al. 1985. Chemical and spectroscopic properties of pyridoxal and pyridoxamine phosphates. In Transaminases (eds. P. Christen and D.E. Metzler), pp. 37–107. John Wiley & Sons, New York.
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Li, Y., Feng, L., and Kirsch, J.F. 1997. Kinetic and spectroscopic investigations of wild-type and mutant forms of apple 1-aminocyclopropane-1-carboxylate synthase. Biochemistry 36 15477–15488. [DOI] [PubMed] [Google Scholar]
- Momany, C., Ernst, S., Ghosh, R., Chang, N.L., and Hackert, M.L. 1995. Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30 Å to 3.0 Å resolution. J. Mol. Biol. 252 643–655. [DOI] [PubMed] [Google Scholar]
- Morino, Y. and Snell, E.E. 1967. The relation of spectral changes and tritium exchange reactions to the mechanism of tryptophanase-catalyzed reactions. J. Biol. Chem. 242 2800–2809. [PubMed] [Google Scholar]
- Morollo, A.A., Petsko, G.A., and Ringe, D. 1999. Structure of a Michaelis complex analogue: Propionate binds in the substrate carboxylate site of alanine racemase. Biochemistry 38 3293–3301. [DOI] [PubMed] [Google Scholar]
- Paiardini, A., Bossa, F., and Pascarella, S. 2004. Evolutionarily conserved regions and hydrophobic contacts at the superfamily level: The case of the fold-type I, pyridoxal-5′-phosphate-dependent enzymes. Protein Sci. 13 2992–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., Sivaraman, J., Li, Y., Larocque, R., Barbosa, J.A., Smith, C., Matte, A., Schrag, J.D., and Cygler, M. 2001. Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: Inferred reaction mechanism. Biochemistry 40 5151–5160. [DOI] [PubMed] [Google Scholar]
- Schneider, G., Kack, H., and Lindqvist, Y. 2000. The manifold of vitamin B6 dependent enzymes. Struct. Fold. Des. 8 R1–R6. [DOI] [PubMed] [Google Scholar]
- Schoenhaut, D.S. and Curtis, P.J. 1986. Nucleotide sequence of mouse 5-aminolevulinic acid synthase cDNA and expression of its gene in hepatic and erythroid tissues. Gene 48 55–63. [DOI] [PubMed] [Google Scholar]
- Tan, D. and Ferreira, G.C. 1996. Active site of 5-aminolevulinate synthase resides at the subunit interface. Evidence from in vivo heterodimer formation. Biochemistry 35 8934–8941. [DOI] [PubMed] [Google Scholar]
- Toney, M.D., Hohenester, E., Cowan, S.W., and Jansonius, J.N. 1993. Dialkylglycine decarboxylase structure: Bifunctional active site and alkali metal sites. Science 261 756–759. [DOI] [PubMed] [Google Scholar]
- Yano, T., Hinoue, Y., Chen, V.J., Metzler, D.E., Miyahara, I., Hirotsu, K., and Kagamiyama, H. 1993. Role of an active site residue analyzed by combination of mutagenesis and coenzyme analog. J. Mol. Biol. 234 1218–1229. [DOI] [PubMed] [Google Scholar]
- Zaman, Z., Jordan, P.M., and Akhtar, M. 1973. Mechanism and stereochemistry of the 5-aminolaevulinate synthetase reaction. Biochem. J. 135 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. and Ferreira, G.C. 2002. Transient-state kinetic investigation of 5-aminolevulinate synthase reaction mechanism. J. Biol. Chem. 277 44660–44669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. and Toney, M.D. 1999. pH studies on the mechanism of the pyridoxal phosphate-dependent dialkylglycine decarboxylase. Biochemistry 38 311–320. [DOI] [PubMed] [Google Scholar]