Abstract
Using immunological approaches and mass spectrometry, five proteins associated with metallothionein-3 in mouse brains have been identified. Metallothionein-3 and associated proteins were isolated using immunoaffinity chromatography over immobilized anti-mouse brain MT3 antibody. Proteins in the recovered pool were separated by SDS-polyacrylamide gel electrophoresis, and distinct bands were excised and the proteins digested using trypsin. Peptides were extracted and analyzed using electrospray ionization mass spectrometry. Initial identification was done comparing the identified peptide mass:charge ratios to the MASCOT database. Confirmation of proteins was accomplished by sequencing of selected peptides using tandem mass spectrometry and comparison to the MASCOT database. The proteins were heat-shock protein 84 (mouse variant of heat-shock protein 90), heat-shock protein 70, dihydropyrimidinase-like protein 2, creatine kinase, and β actin. Independently using antibodies against metallothionein-3, creatine kinase, and heat-shock protein 84 showed that all three proteins were coimmunoprecipitated from whole mouse brain homogenates with each of the three antibodies. Mixing purified samples of metallothionein and human brain creatine kinase also generated a complex that could be immunoprecipitated either by anti-metallothionein-3 or anticreatine kinase antibody. These data are consistent with metallothionein-3 being present in the mouse brain as part of a multiprotein complex providing new functional information for understanding the role of metallothionein-3 in neuronal physiology.
Keywords: metallothionein-3, partner proteins, mass spectrometry, proteomics, mouse brain
Metal ions are both essential constituents to cell growth and development and toxic elements when present in an excess, unchaperoned form. Organisms have developed metal-regulating networks to provide the control of the concentrations and availability of different metal ions. A major constituent of this network is the family of low-molecular weight, cysteine rich, inducible metal-binding proteins called metallothioneins (MTs) (Kägi and Schäffer 1988; Kägi 1993; Klaassen 1999). The metal ions are coordinated to Cys sulfur atoms in metal-thiolate clusters (Otvos and Armitage 1980; Boulanger et al. 1983). Multiple isoforms of MTs are known. MT1 and MT2 are the better characterized proteins, and are induced by increased concentrations of heavy metals, such as Cd2+, among others. Although MT3 is often considered to be specific to brain tissue, it also is reported to be found in human kidney (Garrett et al. 1999a) and numerous cancers (Garrett et al. 1999b; Sens et al. 2000; Dutta et al. 2002). Unlike MT1 and MT2, MT3 is not induced by heavy metals, indicating that the amounts of MT isoforms are regulated differently in cells.
Metallothionein-3 was originally isolated from brain as a growth inhibitory factor (GIF) using bioassays demonstrating the inhibition of neuronal cell growth, a property not exhibited by MT1 and MT2. Many reports indicate that mRNA for MT3 is downregulated in tissue from Alzheimer’s patients (Tsuji et al. 1992; Naruse et al. 1994; Carrasco et al. 1999; Yu et al. 2001), although other studies have not confirmed this result (Erickson et al. 1994; Amoureux et al. 1997). The growth-inhibiting activity was established to reside in the amino terminal β-domain (Sewell et al. 1995; Uchida and Ihara 1995), with mutations at proline residues (positions 7 and 9) found to abolish the inhibitory activity of MT3 (Sewell et al. 1995). Although the inhibitory activity was reportedly independent of metal ion binding (Sewell et al. 1995), inhibitory activity was reported to decrease in zinc-deficient mice (Palmiter 1995). MT3 and Zn2+ were co-localized in neurons of mice, which overexpress MT3 (Masters et al. 1994; Erickson et al. 1995). These studies support unique functions for MT3 attributable to the unique structural features of the protein.
The unique properties of MT3 support the hypothesis that there are specific interacting proteins which mediate the functionality of MT3. In this paper, immunological approaches have been used to identify specific MT3 partner proteins using mass spectrometry.
Experimental procedures
Materials
An antibody preparation against heat-shock protein 84 was purchased from Affinity Bioreagent. Purified human brain creatine kinase (isoform BB) and antihuman brain creatine kinase antibody were from Fitzgerald Industries International. A kit for assaying creatine kinase activity was purchased from Diagnostics Chemical Ltd. The Amplified Alkaline Phosphatase Immun-blot kit was purchased from Bio-Rad and contained goat anti-rabbit secondary antibody; biotin; streptavidin; biotinylated alkaline phosphatase; and the substrates for alkaline phosphatase (AP color development components). The slot blot apparatus was purchased from Bio-Rad. The wash buffer was also purchased from Bio-Rad as a 10× stock solution (10× TBS). The pNPP substrate (Sigma 104 substrate) for the alkaline phosphatase ELISA reactions was obtained from Sigma-Aldrich. ELISA-compatible microtiter plates were from Costar and purchased through Fisher Scientific. Anti-mouse MT3 antibody was previously generated and characterized (Tokheim et al. 2005). These experiments established that the anti-serum was at least 100-fold more specific for MT3 than for MT1 and MT2.
Immobilized antibody preparation
Metallothionein-3 antibody (α-MT3) was immobilized on Affi-Gel Hz, Bio-Rad. Purified α-MT3 (20 mg/mL) was desalted with a 3 kDa spin filter and diluted into Affi-Gel Hz coupling buffer (Bio-Rad) at pH 5.5. The solution was oxidized for 1 h at room temperature using NaIO4. The NaIO4/α-MT3 mix was desalted using a 3 kDa spin filter to remove sodium salts. The Affil-Gel Hz gel was washed with coupling buffer. The oxidized, desalted α-MT3 (1 mL) was then mixed with the Affi-Gel Hz (2 mL) and incubated at room temperature for 24 h. After incubation the coupled antibody was washed with 0.5 M NaCl in 10 mM MOPS. The immunoaffinity matrix was stored in the cold room.
Immunoaffinity isolation
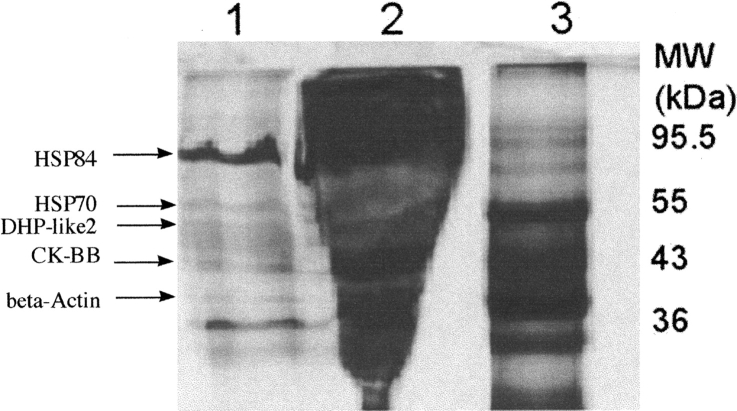
Immunoaffinity chromatography was done using α-MT3 antibody immobilized on Affigel-Hz (Bio-Rad Laboratories). Antibody had been first affinity purified using MT3 immobilized on Affigel-15 (Bio-Rad Laboratories). Brain extract was prepared from two mouse brains (Swiss Webster, Pel-Freez) with a combined wet weight of 0.78 g that were homogenized in 1.6 mL 0.01 M NaCl in 10 mM MOPS (pH 7.3). The homogenate was centrifuged (1 h at 3000 rpm) to pellet cell material, and the extract was pipetted off and filtered through a 0.45 micron syringe filter. The collected brain extract was applied to the immobilized antibody (2 mL) in 10 mM MOPS (pH 7.3) with 10 mM NaCl and containing PMSF and a cocktail of protease inhibitors (Sigma-Aldrich; catalog no. P8849). The slurry of immobilized antibody with brain extract was incubated overnight at 4°C with rocking. The beads were collected by centrifugation and washed three times for 20 min with 3 mL 10 mM MOPS (pH 7.3), with 10 mM NaCl with centrifugation after each wash. The resin was then washed with 10 mM MOPS (pH 7.3), with 150 mM NaCl until no protein was found identified by the Bradford method (Bradford 1976) in the recovered washes after centrifugation. Proteins bound to the immobilized antibody were then eluted with 1 mL 3 M NaSCN. The proteins in the eluant were recovered by centrifugation and shown to contain MT3 by slot blotting using α-MT3 antiserum. The eluant was concentrated using centrifugal concentrators with 3000 Da cutoff membrane and analyzed by SDS-polyacrylamide gel electrophoresis with silver staining (Fig. 1 ▶).
Figure 1.
Gel of immunoaffinity copurified proteins. Antibody against MT3 was immobilized on Affigel-Hz and used to isolated MT3 and associated proteins. Proteins were applied and eluted as described and analyzed by SDS-gel electrophoresis and mass spectrometry. Proteins identified by mass spectrometric analysis are labeled. Lane 1 is the recovered protein after elution with NaSCN. Lane 2 shows the proteins in the nonretained pool after incubation of whole-brain extract with the immobilized antibody. Lane 3 shows the positions of the prestained marker proteins with values of Mr shown.
SDS polyacrylamide gel electrophoresis/in gel digest
Protein separation by SDS-PAGE was done using 12% gels with detection by silver staining (Bio-Rad enhanced silver stain kit). Select bands were chosen for in-gel digestion. Digestion was done using the Investigator ProPrep instrument from Genomics Solutions (http://www.genomicsolutions.com) in the Proteomics facility of the University of Minnesota. Protein bands from the gel were excised and digested with trypsin (Sequence-Grade Modified Porcine) according to a published method with some modifications (Shevchenko et al. 1996). Silver-stained bands were destained with a 1:1 solution of potassium ferricyanate (30 mM) and sodium thiosulfate (100 mM) for 8 min and then washed four times with 1.0 mL of Nanopure water for 8 min. The gel slices were then covered with acetonitrile for 15 min, the solvent was decanted, and the slices were dried by speed vacuum. Dried gel slices were covered with a solution of ammonium bicarbonate (50 mM) containing CaCl2 (5 mM) and sequence grade modified porcine trypsin (final concentration 12 μg/mL, Promega). This mixture was kept on ice for 1 h and then at 37°C overnight once excess solution was discarded and 50 mM ammonium bicarbonate containing CaCl2 (5 mM) was added. The supernatants from the overnight incubations were removed and transferred to a different tube. The gel slices were washed once with 20 mM ammonium bicarbonate (20 min) and three times with 50% acetonitrile–5% formic acid. All supernatants were pooled, for each sample, and speed vacuumed to dryness.
Mass spectrometry analysis
Samples were spotted on a MALDI MS target and peptide fragments were recorded using a QSTAR XL instrument (Applied Biosystems, ABI). Full mass scans were collected in positive ion mode between m/z 500 and 3500 of the peptide mixtures for each sample. The software program Analyst QS (ABI) was then used to produce a peptide peak list for each mass spectrum. For the final searching, the list was trimmed to about 25 m/z peaks. Each peak list of measured peptide masses was then used to search the National Center for Biotechnology Information (NCBI) sequence database for protein identifications using Mascot (http://www.matrixscience.com). Searches were performed allowing for variable modifications that include methionine oxidation and carbamidomethyl addition, as well as allowing for up to one missed cleavage site on the peptide. Some peptides were further characterized by tandem mass spectrometry using the same instrument. Once selected, peptides are guided into the collision chamber whereupon they are fragmented by collision induced dissociation with argon. Data was analyzed using Analyst QS and Mascot.
Slot blot detection
Triplicate 200 μL samples of whole-brain homogenate were applied under gentle vacuum to Trans-Blot nitrocellulose membrane (Bio-Rad) using a slot blot apparatus (Bio-Rad). Each well was washed with 200 μL aliquots of TBS. After removing TBS with gentle suction, the membrane was removed from the apparatus and washed two times with TTBS (TBS with 0.05% Tween-20) and once with TBS. The membrane was divided into three identical pieces and with replicate samples incubated independently with different antibody solutions: α-creatine kinase BB (α-CK), α-heat-shock protein 84 (α-HSP84), and α-MT3. Incubations were allowed to proceed overnight at 4°C with rocking. After incubation, the antibody solutions were removed and the membranes rinsed twice with TTBS and once with TBS. Detection was done using the reagents of the Amplified Alkaline Phosphatase Immun-Blot Kit from Bio-Rad Laboratories. Briefly, the membrane was incubated for 1 h with the secondary antibody (biotinylated goat-antirabbit antibody in TBS); the membrane again washed as described, and the membrane incubated for 1 h with a conjugate of sterptavidin with biotinylated-alkaline phosphatase in TTBS. After a final washing procedure, the presence of antigen was detected by monitoring the color development after exposure to the substrate for the alkaline phosphatase-conjugated secondary antibody. For alkaline phosphatase, the substrate used was comprised of a mix of bromochloroindolyl chloride phosphate (BCIP) and nitroblue tetrazolium (NBT). BCIP is hydrolyzed by alkaline phosphatase and reacts with NBT to stain the membrane at the site of alkaline phosphatase. This reaction occurs only at sites of bound primary antibody. The reaction was stopped by the dilution of substrate solution with water.
Immunoprecipitation
Immunoprecipitation reactions were performed independently with antibodies directed against MT3 (α-MT3), HSP84 (α-HSP84), and the brain form of creatine kinase (α-CK). Antiserum (10 μL) was added to 50 μL brain extract and the solution was rocked overnight at 4°C. Following the incubation, 50 μL protein A sepharose, prewashed with 10 mM MOPS, was added and incubated for 1 h. Each sample was centrifuged and the supernatant was removed. Then the beads were washed three times with 10 mM MOPS and then SDS sample buffer was added to the pellet prior to boiling.
Creatine kinase assay
A creatine kinase assay kit was obtained from Diagnostics Chemical Ltd. and used to assess the amount of creatine kinase activity. The assay is based on the formation of NADPH, and can be monitored at 340 nm. Creatine kinase catalyzes the reaction of ADP with creatine phosphate to yield creatine and ATP. The reaction is coupled to the hexokinase-catalyzed formation of glucose-6-phosphate from glucose and ATP. Finally, glucose-6-phosphate is oxidized to glucono-6-lactone-6-phosphate by glucose-6-phosphate dehydrogenase. The oxidation is coupled to the formation of NADPH from NADP+.
Results
Immunoaffinity chromatography using α-MT3 antibody immobilized on Affigel-Hz (Bio-Rad Laboratories) was done to isolate specific MT3 binding proteins. Brain extract from Swiss-Webster mice was used as the source of proteins and the recovered samples from the immunoaffinity chromatography were separated by SDS-polyacrylamide gel electrophoresis with silver staining (Fig. 1 ▶). The sample in lane 2 represents the material from mouse brain extract not retained by the immunoaffinity column. It was noteworthy that only a limited amount of extract protein was retained by the column. Distinct bands observed in the recovered fraction were excised, washed, and digested with trypsin. Resulting peptides were recovered by extraction and analyzed by mass spectrometry. The peptide mass spectrum was collected for each band and compared to the MASCOT (Matrix Science) database for protein identification. Positive protein identification was confirmed by tandem mass spectrometry of the peptide samples to obtain sequence information that was compared to the MASCOT database. Up to five proteins (Table 1) have been identified co-precipitating with MT3: heat-shock protein 84 (HSP84; mouse homolog of human HSP90); heat-shock protein 70 (HSP70); dihydropyrimidinase-like protein 2 (DHP2); creatine kinase (CK, BB isoform); and β actin. The positions of these proteins are shown by the arrows in the figure. Unassigned protein bands shown were analyzed, but insufficient information was recovered for their identification by comparison to the MASCOT database.
Table 1.
Mass spectrometric identification of associated proteins
| Residues | Massa | Sequence | Identificationb |
| Heat-shock protein 84 | 22% sequence coverage | ||
| 73–82 | 1193.64 | IDILPNPQER | X |
| 83–95 | 1348.73 | TLTLVDTGIGMTK | |
| 96–107 | 1241.7 | ADLINNLGTIAK | |
| 149–168 | 2254.95 | HNDDEQYAWESSAGGSFTVR | |
| 187–196 | 1310.56 | EDQTEYLEER | X |
| 224–234 | 1292.53 | EISDDEAEEEK | |
| 276–284 | 1150.55 | YIDQEELNK | X |
| 292–306 | 1846.79 | NPDDITQEEYGEFYK | X |
| 307–319 | 1526.74 | SLTNDWEDHLAVK | |
| 379–392 | 1512.78 | GVVDSEDLPLNISR | X |
| 429–435 | 890.42 | FYEAFSK | X |
| 457–475 | 2175.94 | YHTSQSGDLPLNISR | |
| 482–491 | 1159.58 | SIYYITGESK | X |
| Heat-shock protein 70 | 26% sequence coverage | ||
| 37–49 | 1486.69 | TTPSYVAFTDTER | X |
| 57–71 | 1648.79 | NQVAMNPTNTVFDAK | |
| 113–126 | 1615.78 | SFYPEEVSSMVLTK | X |
| 138–155 | 1980.99 | TVTNAVVTVPAYFNDSQR | |
| 160–171 | 1198.67 | DAGTIAGLNVLR | X |
| 221–236 | 1690.72 | STAGDTHLGGEDFDNR | |
| 221–246 | 2907.32 | STAGDTHLGGEDFDNRMVNHFIAEFK | |
| 302–311 | 1252.61 | FEELNADLFR | X |
| 362–384 | 2259.14 | SINPDEAVAYGAAVQAAILSGD | |
| 513–524 | 1475.69 | EDIERMVQEAEK | |
| 540–550 | 1302.59 | NSLESYAFNMK | |
| 584–597 | 1744.8 | NQTAEKEEFEHQQK | |
| Dihydropyrimidinase-like protein 2 | 20% sequence coverage | ||
| 64–75 | 1293.69 | MVIPGGIDVHTR | X |
| 147–157 | 1245.63 | GIQEEMEALVK | |
| 346–361 | 1791.83 | DNFTLIPEGTNGTEER | X |
| 375–390 | 1724.8 | MDENQFVAVTSTNAAK | |
| 391–397 | 907.49 | VFNLYPR | X |
| 401–418 | 1914.96 | ISVGSDADLVIWDPDSVK | X |
| 452–467 | 1681.86 | IVLEDGTLHVTEGSGR | |
| 533–552 | 2168.06 | NLHQSGFSLSGAQIDDNIPR | |
| Creatine kinase | 33% sequence coverage | ||
| 33–43 | 1302.72 | VLTPELYAELR | X |
| 87–96 | 1245.62 | DLFDPIIEER | X |
| 108–130 | 2517.16 | TDLNPDNLQGGDDLDPNYVLSSR | |
| 139–148c | 1229.54 | GFCLPPHCSR | |
| 157–172 | 1601.83 | LAVEALSSLDGDLSGR | X |
| 253–265c | 1556.79 | FCTGLTQIETLFK | |
| 321–341 | 1963.92 | GTGGVDTAAVGGVFDVSNADR | X |
| 359–366 | 1030.55 | LLIEMEQR | X |
| 367–381 | 1655.82 | LEQGQAIDDLMPAQK | |
| Actin (β) | 32% sequence coverage | ||
| 3–11 | 944.54 | AVFPSIVGR | |
| 3–13 | 1187.68 | AVFPSIVGRSR | |
| 70–87 | 1953.06 | VAPEEHPVLLTEAPLNPK | |
| 171–180 | 1131.52 | GYSFTTTAER | X |
| 213–228 | 1789.88 | SYELPDGQVITIGNER | X |
| 266–286 | 2214.06 | DLYANTVLSGGTTMYPGIADR | |
| 310–333 | 2729.43 | KYSVWIGGSILASLSTFQQMWISK | |
| 334–346 | 1515.7 | QEYDESGPSIVHR | X |
a Peak identification by a QSTAR pulsar quadrupole MALDI-TOF-MS instrument. Theoretical m/z values are monoisotopic.
b Peptide analyzed by tandem mass spectrometry.
c Signifies carbaminomethyl modification on the peptide.
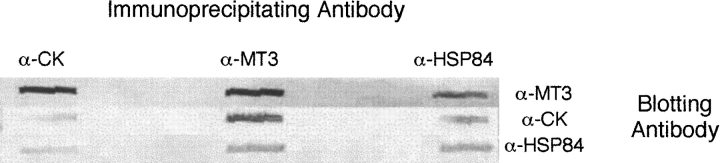
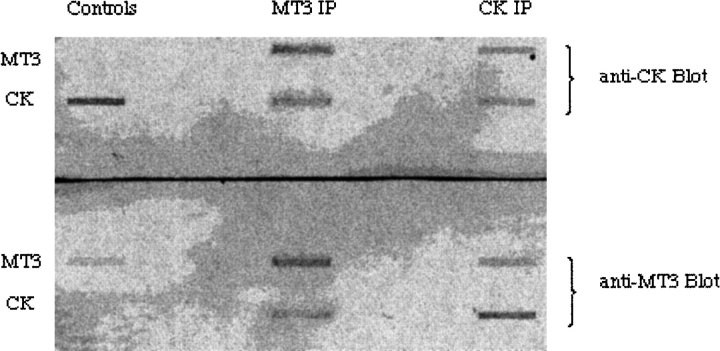
Two of these proteins have been further investigated: the brain isoform of creatine kinase (CK), and heat-shock protein 84 (HSP84). Antibodies to each of these proteins were purchased and confirmed the presence of these proteins in the recovered eluant by slot blotting. These antibodies and the anti-MT3 antibody were used in immunoprecipitation of brain extracts followed by slot blotting. Independent experiments showed the presence of all three proteins regardless of the precipitating antibody (Fig. 2 ▶). Together, these are convincing data that metallothionein-3, creatine kinase, and heat-shock protein 84 are in a multiprotein complex in normal mouse brains. Mixing of purified MT3 and CK followed by immunoprecipitation with both target antibodies showed that these two proteins interacted directly (Fig. 3 ▶). Within the larger macromolecular complex, it was reasonable to conclude that these two proteins are in contact. The proteins must exist minimally as a complex of these three proteins: MT3, CK-BB, and HSP84, because all three proteins are coprecipitated by each of the three antisera. HSP84 is hypothesized to be the central protein of a multiprotein complex with both MT3 and CK-BB associated. This is consistent with the chaperonin function assigned to this family of heat-shock proteins.
Figure 2.
Slot blot detection of immunoprecipitated complexes from mouse brain. Three samples of brain extract were treated independently with three different antibodies: α-MT3, α-HSP84, and α-CK. After incubation overnight at 4°C, Protein-A agarose was added and the mixture incubated for 1 h at 4°C. The beads were collected by centrifugation and washed with Mops-buffered saline. Proteins were eluted from the beads by heat treatment, and triplicate samples loaded into wells of a slot blot apparatus. Each well was probed with the three different antibodies to monitor for the presence of the three antigens.
Figure 3.
Slot blot detection of immunoprecipitated proteins. Duplicate samples of MT3 were mixed with creatine kinase in Mops-buffered saline. To replicate samples was added either α-MT3 (MT3 IP) or α-CK (CK IP). After incubation overnight at 4°C, Protein-A agarose was added and the mixture incubated for 1 h at 4°C. The beads were collected by centrifugation and washed with Mops-buffered saline. Proteins were eluted from the beads by heat treatment, and triplicate samples loaded into wells of a slot blot apparatus. Each well was probed with α-MT3 or α-CK to monitor for the presence of the antigens. Only the appropriate antigen was detected in the control samples (Controls) by their respective antibodies. The results showed that MT3 and CK directly associated under the conditions of the experiment, 4°C for 16 h.
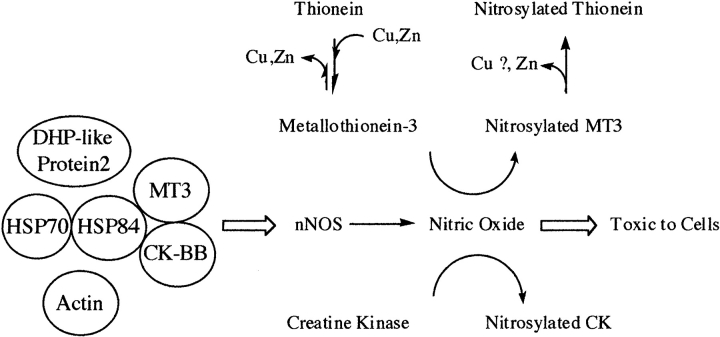
A hypothetical working model (Fig. 4 ▶) of this interaction has been developed based on these data and information published in other reports. MT3, HSP84, HSP70, DHP-2, and CK were all coprecipitated using anti-MT3 antiserum. Using specific anti-HSP84 and anti-CK antisera has demonstrated that MT3, HSP84, and CK are all immunoprecipitated in each case. HSP70 and HSP 84 coprecipitate (Gruppi and Wolgemuth 1993), and HSP90, the human variant of HSP84, can activate neuronal nitric oxide synthase (Song et al. 2001) with the resulting nitric oxide toxic to neurons. MT3 is neuroprotective against the toxic effects of nitric oxide (Montoliu et al. 2000). These same proteins are targets of modification with MT3, CK, and DHP-2 all nitro-sylated by nitric oxide in the brain (Jaffrey et al. 2001) and HSP84, CK, and DHP-like protein-2 are all targets of oxidation (Castegna et al. 2002a,b) in oxidative stress. MT3 and HSP70 (as well as nNOS) are induced in neural injury. Data such as these indicate the importance of these proteins in the health of neurons under changing oxygen environments. The action of nitric oxide on MT3 causes the release of zinc from Zn7-MT3 and may yield the apo-protein, thionein (Chen et al. 2002). The release of zinc may provide a feedback mechanism for regulating NOS activity as Zn2+ has been found to inhibit nNOS (Persecheni et al. 1995; Perry et al. 2000). Zinc in the native MT3 structure is in the α-domain, and evidence from earlier work indicates that Cd4- or Zn4-α-MT3 are very similar. The presence of zinc in MT3 may not be an absolute requirement inasmuch as CK did interact and coimmunoprecipitate with Cd7-MT3 (Fig. 3 ▶).
Figure 4.
Hypothesized model of interactions between MT3 and other brain proteins. Shown is a hypothetical association of proteins identified after immunoaffinity isolation. Shown as well is the possible involvement of nitric oxide synthase and nitric oxide, such as the nitrosylation of metallothionein-3 and creatine kinase. For metallothionein-3, nitrosylation has been shown to cause the loss of Zn2+ from the protein. Loss of Cu+ may also result from nitrosylation, but this has not been established.
Collectively, these observations indicate that these proteins are associated and targeted for similar changes in response to changes in the cellular environment. The hypothesized model adequately explains the interaction of MT3 and partner proteins with the conflicting interactions of nitric oxide and nitric oxide synthase. HSP84 is also critical to understanding the biological functions of nitric oxide synthase because HSP90 has been shown to activate human brain nNOS. The presence, however, of creatine kinase in the complex is intriguing, inasmuch as this protein has been found to be reduced in AD brains (Aksenov et al. 1997, 2001; David et al. 1998; Aksenova et al. 1999).
Functional consequences of MT3 interactions
Unfortunately, MT3 has no simple functional assay to assess changes in activity. One MT3 partner, creatine kinase, has a specific and important biological activity which can be assayed spectrophotometrically with monitoring of product formation at 340 nm. Using a creatine kinase assay kit from Diagnostics Chemical Ltd, no effect on creatine kinase activity was found with the addition of recombinant Cd7-MT3 to the assay mixture (0.57 μg/mL MT3 and 0.075 μg/mL CK). A MT3 knockout mouse strain has been reported in the literature (Erickson et al. 1997), and may prove valuable for insight about the relationship of MT3 with these partner proteins. Will the complexes remain in the same form in the absence of MT3? Immunocytochemical approaches will also be invaluable in establishing whether these same proteins are co-localized in the same regions of the brain.
Summary
MT3 was isolated using immunoaffinity chromatography. Select mouse brain proteins were copurified with the corollary conclusion that these proteins exist as cellular macromolecular complex. The identities of the interacting proteins was established using mass spectrometry an database comparisons. A working model for the functional consequences of these interactions is proposed, and will provide a basis for future directions. It is noteworthy that MT3 and all the identified partner proteins are induced by the same physiological processes, such as oxidative stress and neural injury. These links support the premise that these interactions will prove significant in understanding the roles of these proteins in the brain.
Acknowledgments
This research was supported by funds from the Minnesota Medical Foundation (B.L.M.) and from the Minnesota Medical Foundation and University of Minnesota Graduate School (I.M.A.). We fully appreciate the efforts of the staff of the Proteomics Analysis Core and the Mass Spectrometry Consortium for the Life Sciences of the University of Minnesota.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041113005.
References
- Aksenov, M.Y., Aksenova, M.V., Payne, R.M., Smith, C.D., Markesbery, W.R., and Carney, J.M. 1997. The expression of creatine kinase isoenzymes in neocortex of patients with neurodegenerative disorders: Alzheimer’s and Pick’s disease. Exp. Neurobiol. 146 458–465. [DOI] [PubMed] [Google Scholar]
- Aksenov, M.Y., Aksenova, M.V., Butterfield, D.A., Geddes, J.W., and Markesbery, W.R. 2001. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience 103 373–383. [DOI] [PubMed] [Google Scholar]
- Aksenova, M.V., Aksenov, M.Y., Payne, R.M., Trojanowski, J.Q., Schmidt, M.L., Carney, J.M., Butterfield, D.A., and Markesbery, W.R. 1999. Oxidation of cytosolic proteins and expression of creatine kinase BB in frontal lobe in different neurodegenerative disorders. Dement. Geriatr. Cogn. Disord. 10 158–165. [DOI] [PubMed] [Google Scholar]
- Amoureux, M.C., Van Gool, D., Herrero, M.T., Dom, R., Colpaert, F.C., and Pauwels, P.J. 1997. Regulation of metallothionein-III (GIF) mRNA in the brain of patients with Alzheimer disease is not impaired. Mol. Chem. Neuropathol. 32 101–121. [DOI] [PubMed] [Google Scholar]
- Boulanger, Y., Goodman, C.M., Forte, C.P., Fesik, S.W., and Armitage, I.M. 1983. Model for mammalian metallothionein structure. Proc. Natl. Acad. Sci. 80 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive for the quantitation of microgram quantitites of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Carrasco, J., Girault, M., Molinero, A., Penkowa, M., Moos, T., and Hidalgo, J. 1999. Metallothionein (MT)-III: Generation of polyclonal antibodies, comparison with MT-I + II in the freeze lesioned rat brain and in a bioassay with astrocytes, and analysis of Alzheimer’s disease brains. J. Neurotrauma 16 1115–1129. [DOI] [PubMed] [Google Scholar]
- Castegna, A., Aksenov, M., Aksenova, M., Thongboonkerd, V., Klein, J.B., Pierce, W.M., Booze, R., Markesbery, W.R., and Butterfield, D.A. 2002a. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part 1: Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 33 562–571. [DOI] [PubMed] [Google Scholar]
- Castegna, A., Aksenov, M., Thongboonkerd, V., Klein, J.B., Pierce, W.M., Booze, R., Markesbery, W.R., and Butterfield, D.A. 2002b. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II: Dihydropyrimidinase-related protein 2, g-enolase and heat shock cognate 71. J. Neurochem. 82 1524–1532. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Irie, Y., Keung, W.M., and Maret, W. 2002. S-Nitrosothiols react preferentially with zinc thiolate clusters of metallothionein-III through transnitrosation. Biochemistry 41 8360–8367. [DOI] [PubMed] [Google Scholar]
- David, S., Shoemaker, M., and Haley, B.E. 1998. Abnormal properties of creatine kinase in Alzheimer’s disease brain: Correlation of reduced enzyme activity and active site photolabeling with aberrant cytosol-membrane partitioning. Mol. Brain Res. 54 276–287. [DOI] [PubMed] [Google Scholar]
- Dutta, R., Sens, D.A., Somji, S., Sens, M.A., and Garrett, S.H. 2002. Metallothionein isoform 3 expression inhibits cell growth and increases drug resistance of PC-3 prostate cancer cells. Prostate 52 89–97. [DOI] [PubMed] [Google Scholar]
- Erickson, J.C., Sewell, A.K., Jensen, L.T., Winge, D.R., and Palmiter, R.D. 1994. Enhanced neurotrophic activity in Alzheimer’s disease cortex is not associated with down-regulation of metallothionein-III (GIF). Brain Res. 649 297–304. [DOI] [PubMed] [Google Scholar]
- Erickson, J.C., Masters, B.A., Kelly, E.J., Brinster, R.L., and Palmiter, R.D. 1995. Expression of human metallothionein-III in transgenic mice. Neurochem. Int. 27 35–41. [DOI] [PubMed] [Google Scholar]
- Erickson, J.C., Hollopeter, G., Thomas, S.A., Froelick, G.J., and Palmiter, R.D. 1997. Disruption of the metallothionein-III gene in mice: Analysis of brain zinc, behavior, and neuron vulnerability to metals, aging, and seizures. J. Neurosci. 17 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, S.H., Sens, M.A., Todd, J.H., Somji, S., and Sens, D.A. 1999a. Expression of MT-3 protein in the human kidney. Toxicol. Lett. 105 207–214. [DOI] [PubMed] [Google Scholar]
- Garrett, S.H., Sens, M.A., Shukla, D., Nestor, S., Somji, S., Todd, J.H., and Sens, D.A. 1999b. Metallothionein isoform 3 expression in the human prostate and cancer-derived cell lines. Prostate 41 196–202. [DOI] [PubMed] [Google Scholar]
- Gruppi, C.M. and Wolgemuth, D.J. 1993. HSP86 and HSP84 exhibit cellular specificity of expression and co-precipitate with an HSP70 family member in the murine testis. Dev. Genet. 14 119–126. [DOI] [PubMed] [Google Scholar]
- Jaffrey, S.R., Erdjument-Bromage, H., Ferris, C.D., Tempst, P., and Snyder, S.H. 2001. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3 193–197. [DOI] [PubMed] [Google Scholar]
- Kägi, J.H.R. 1993. Evolution, structure and chemical activity of class I metallothioneins: An overview. In Metallothionein III: Biological roles and medical implications (eds. K.T. Suzuki et al.), pp. 29–55. Birkhauser, Basel, Switzerland.
- Kägi, J.H.R. and Schaffer, A. 1988. Biochemistry of metallothionein. Biochemistry 27 8509–8515. [DOI] [PubMed] [Google Scholar]
- Klaassen, C.D. 1999. Metallothionein IV (ed. C.D. Klaassen). Birkhauser Verlaag, Basel, Switzerland.
- Masters, B.A., Quaife, C.J., Erickson, J.C., Kelly, E.J., Froelick, G.J., Zambrowicz, B.P., Brinster, R.L., and Palmiter, R.D. 1994. Metallothionein III is expressed in neurons that sequester zinc in synaptic vesicles. J. Neurosci. 14 5844–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu, C., Monfort, P., Carrasco, J., Palacios, O., Capdevilla, M., Hidalgo, J., and Felipo, V. 2000. Metallothioneien-III prevents glutamate and nitric oxide neurotoxicity in primary cultures of cerebellar neurons. J. Neurochem. 75 266–273. [DOI] [PubMed] [Google Scholar]
- Naruse, S., Igarashi, S., Furuya, T., Kobayashi, H., Miyatake, T., and Tsuji, S. 1994. Structures of the human and mouse growth inhibitory factor-encoding genes. Gene 144 283–287. [DOI] [PubMed] [Google Scholar]
- Otvos, J.D. and Armitage, I.M. 1980. Structure of the metal clusters in rabbit liver metallothionein. Proc. Natl. Acad. Sci. 77 7094–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R.D. 1995. Constitutive expression of metallothionein-III (MT-III), but not MT-I, inhibits growth when cells become zinc deficient. Toxicol. Appl. Pharmacol. 135 139–146. [DOI] [PubMed] [Google Scholar]
- Perry, J.M., Zhao, Y., and Marletta, M.A. 2000. Cu2+ and Zn2+ inhibit nitric-oxide synthase through an interaction with the reductase domain. J. Biol. Chem. 275 14070–14076. [DOI] [PubMed] [Google Scholar]
- Persechini, A., McMillan, K., and Masters, B.S. 1995. Inhibition of nitric oxide synthase activity by Zn2+ ion. Biochemistry 46 15091–15095. [DOI] [PubMed] [Google Scholar]
- Sens, M.A., Somji, S., Lamm, D.L., Garrett, S.H., Slovinsky, F., Todd, J.H., and Sens, D.A. 2000. Metallothionein isoform 3 as a potential biomarker for human bladder cancer. Environ. Health. Perspect. 108 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell, A.K., Jensen, L.T., Erickson, J.C., Palmiter, R.D. and Winge, D.R. 1995. Bioactivity of metallothionein-III correlates with its novel b-domain sequence rather than metal binding properties. Biochemistry 34 4740–4747. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylaminde gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Song, Y., Zweier, J.L., and Xia, Y.2001. Heat-shock protein 90 augments neuronal nitric oxide synthase activity by enhancing Ca2+/calmodulin binding. Biochem. J. 355 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokheim, A.M., Armitage, I.M., and Martin, B.L. 2005. Antiserum specific for the intact isoform 3 of metallothionein. J. Biochem. Biophys. Meth. (in press). [DOI] [PubMed]
- Tsuji, S., Kobayashi, H., Uchida, Y., Ihara, Y., and Miyatake, T. 1992. Molecular cloning of human growth inhibitory factor cDNA and its down-regulation in Alzheimer’s disease. EMBO J. 11 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, Y. and Ihara, Y. 1995. The N-terminal portion of growth inhibitory factor is sufficient for biological activity. J. Biol. Chem. 270 3365–3369. [DOI] [PubMed] [Google Scholar]
- Yu, W.H., Lukiw, W.J., Bergeron, C., Niznik, H.B., and Fraser, P.E. 2001. Metallothionein III is reduced in Alzheimer’s disease. Brain Res. 894 37–45. [DOI] [PubMed] [Google Scholar]