Abstract
The plague-causing bacterium Yersinia pestis utilizes a contact-dependent (type III) secretion system (T3SS) to transport virulence factors from the bacterial cytosol directly into the interior of mammalian cells where they interfere with signal transduction pathways that mediate phagocytosis and the inflammatory response. The type III secretion apparatus is composed of 20–25 different Yersinia secretion (Ysc) proteins. We report here the structure of YscE, the smallest Ysc protein, which is a dimer in solution. The probable mode of oligomerization is discussed.
Keywords: Yersinia pestis, plague, type III secretion, YscE, crystal structure
Yersinia pestis, the causative agent of plague, utilizes a type III secretion system (T3SS) to inject effector proteins directly into the cytosol of mammalian cells. The concerted action of these effector Yops (Yersinia outer proteins) enables the bacterium to subvert the innate immune response of the infected organism and proliferate freely in its tissues (for review, see Navarro et al. 2005).
The effectors are delivered from the bacterium into the cytosol of mammalian cells through a large macro-molecular assembly termed the injectisome, which is composed of 20–25 different Yersinia secretion (Ysc) proteins (for review, see Ghosh 2004). This superstructure spans both bacterial membranes and the plasma membrane of the target cell. About half of the Ysc proteins have counterparts in other type III secretion systems and most of these also have structural homologs in the bacterial flagellum, suggesting that the two systems share a common evolutionary heritage (Blocker et al. 2003).
Although YscE is not broadly conserved among type III secretion systems, yscE mutants are unable to export the Yersinia effectors (Allaoui et al. 1995). Hence, YscE evidently plays a crucial role in either the assembly of the injectisome or the regulation of Yop secretion. Seeking to gain some insight into its function, we overproduced YscE in Escherichia coli, crystallized it, and solved its structure at a resolution of 1.8 Å.
Results and Discussion
Crystallization of YscE
Y. pestis YscE was initially produced as a C-terminally hexahistidine-tagged protein (YscE–His6). Although it was possible to grow large crystals of YscE–His6 with sharp edges, they diffracted X-rays very poorly (~7 Å). Therefore, a new expression vector was designed to fuse YscE to the C terminus of an N-terminally His-tagged form of the E. coli maltose-binding protein (MBP). Digestion of the His-MBP-YscE fusion protein with tobacco etch virus (TEV) protease yielded “untagged” YscE (which retained a single nonnative glycine residue on its N terminus). Crystals of untagged YscE grew under a variety of conditions, one of which was subsequently optimized to produce high-quality crystals that diffracted X-rays to 1.8 Å. These crystals, which were clearly different from the original crystals of YscE–His6, belonged to the orthorhombic space group P21212 with unit cell dimensions a = 82.97, b = 84.62, and c = 81.80 Å.
Description of the YscE monomer
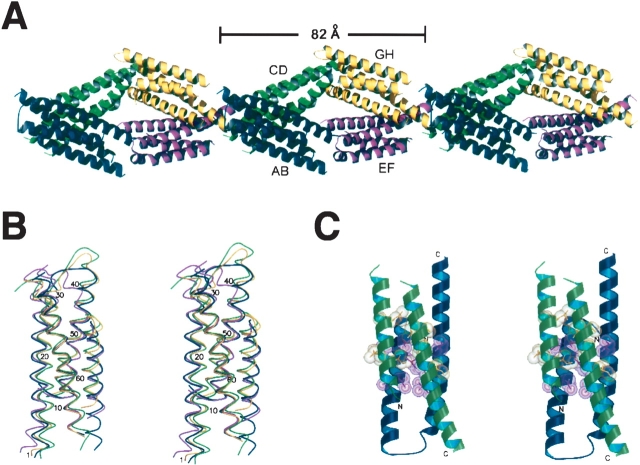
YscE crystallized as four dimers arranged in the form of a diamond-shaped pseudo-octamer in the asymmetric unit (Fig. 1A ▶). The YscE monomer is composed of two anti-parallel α-helices separated by a flexible loop. This helical hairpin fold is similar to structural elements found in many proteins. A structural alignment of the C-α traces from all of the subunits reveals differences in the helical axes, the separation between helices α1 and α2, and the loop region between them. Well-defined electron density was observed for the following residues in each subunit of the pseudo-octamer: A, 1–65; B, 1–33 and 41–66; C, 1–35 and 42–58; D, 1–66; E, 1–65; F, 1–32 and 47–66; G, 2–33 and 44–65; and H, 1–66. Hence, the loop regions in four of the monomers are disordered. Some loops are disordered because they do not contact neighboring molecules in the crystal lattice, while others adopt a rigid conformation either to make favorable contacts with neighboring molecules or to avoid steric clashes.
Figure 1.
(A) Architecture of the pseudo-octamer that defines the asymmetric unit of the YscE crystal lattice. (B) Structural alignment of the four YscE dimers that make up the asymmetric unit in the crystal lattice. C-α traces are shown in stereo view. Dimers AB–GH are rendered in blue, green, violet, and yellow, respectively. (C) Ribbon model of a YscE dimer in stereo view. Conserved residues at the interface between subunits (yellow and magenta) are depicted in stick and CPK format. Images were rendered by Molscript (Kraulis 1991) and Raster3D (Merritt and Murphy 1994).
Structure of the dimer
Although dynamic light scattering (DLS) suggested that YscE might be a tetramer, equilibrium ultracentrifugation and gel filtration experiments both indicated unambiguously that YscE is a dimer in solution (data not shown). The reason for this discrepancy is unclear, but it may be related to the elongated, rod-like structure of the YscE dimer. The crystal structure is also more consistent with that of a dimer than a tetramer because it is possible to construct eight structurally distinct tetramers from the crystal lattice but only one type of dimer.
The four copies of the YscE dimer in the asymmetric unit are superimposable with an RMSD value of 0.471 Å between dimers AB and EF, 0.474 Å between CD and GH, and 1.931 Å between AB and GH (Fig. 1B ▶). They differ in the relative orientation of their helical elements and all have different loop conformations. Dimers AB and EF form two sides of the diamond along the major axis, while CD and GH form the remaining sides in an end-to-end fashion (Fig. 1A ▶). The dimer interface is comprised of conserved residues (identical in orthologs of YscE from Y. enterocolitica, Photorabdus luninescens, and Pseudomonas aeruginosa) that create an interlocking network of hydrophobic interactions (Fig. 1C ▶); Ile17, Leu21, Ala24, Leu25, Ala50, Leu51, Ile55, Ile57, and Ile58 interact to form the hydrophobic core in the central region of the helical bundle. The buried surface areas of dimers AB, CD, EF, and GH are 2245, 2128, 2345, and 2206 Å2, respectively. The dimer interfaces represent ~30% of the total surface area of the interacting subunits. It is noteworthy that one subunit of each dimer has a disordered loop between its two α-helices. These loops do not make crystallographic contacts with neighboring molecules.
Unfortunately, the structure of YscE did not improve our understanding of its biological function. A DALI search (Holm and Sander 1993) of structures in the Protein Data Bank (PDB) (Berman et al. 2000) identified a number of anti-parallel four-helix bundles that exhibit some structural similarity to the YscE dimer, but not enough to suggest any functional relationship between them. Moreover, solvent-exposed residues that are conserved in orthologs of YscE from other organisms are relatively few in number and distributed evenly over its surface, providing few clues about the functionally important features of the structure. Nevertheless, the availability of the structure of YscE should facilitate future efforts to investigate its role in virulence.
Materials and methods
Protein expression and purification
The His6-MBP-YscE expression vector was constructed by Gateway recombinational cloning (Invitrogen). Briefly, the YscE ORF was amplified from Y. pestis genomic DNA using one primer to add a TEV protease cleavage site to the N terminus of YscE (5′-GAGAACCTGTACTTCCAGGGT ATGACACAATTAGAGGAGCAACTG-3′) and another to add an attB2 recombination site downstream of its C terminus (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTATTATTTTAGGTCTCCTGCTAC-3′). The resulting PCR amplicon was used as the template for a second round of PCR with the C-terminal primer (above) in conjunction with an N-terminal primer that placed an attB1 recombination site adjacent to the TEV protease recognition site (5′-GGGGAC AAGTTTGTACAAAAAAGCAGGCTCGGAGAACCTG TACTTCCAG-3′). The final PCR amplicon was inserted by recombinational cloning into the donor vector pDONR201 (Invitrogen) and its DNA sequence was confirmed. The yscE gene was then moved into destination vector pDEST-HisMBP to create the His6-MBP-YscE fusion vector for protein expression (pBA1493).
The His6-MBP-YscE fusion protein was overproduced in E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene). Single antibiotic-resistant colonies were used to inoculate 100 mL of Luria broth supplemented with 100 μg mL−1 ampicillin, 30 μg mL−1 chloramphenicol, and 0.2% (D+)-glucose monohydrate (Sigma-Aldrich). These cultures were grown by shaking (225 rev min−1) to saturation overnight at 37°C and then diluted 50-fold into several liters of fresh medium. When the cells reached early log phase (OD600nm = 0.3–0.5), the temperature was reduced to 30°C and isopropyl β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. Four hours later, the cells were recovered by centrifugation at 5000g for 10 min and stored at −80°C.
E. coli cell paste was suspended in ice-cold 50 mM MES (pH 6.5), 200 mM NaCl, 25 mM imidazole (buffer A) containing Complete EDTA-free protease-inhibitor cocktail (Roche Molecular Biochemicals). The cells were lysed with an APV Gaulin Model G1000 homogenizer at 69 MPa and centrifuged at 30,000g for 30 min at 4°C. The supernatant was filtered through a 0.45 μm polyethersulfone membrane and then loaded onto a 25 mL Ni-NTA Superflow affinity column (Qia-gen) equilibrated in buffer A. The column was washed with five column volumes of buffer A and then eluted with a linear gradient from 25 to 250 mM imidazole in buffer A. Fractions containing recombinant His6-MBP-YscE were pooled and concentrated sixfold using an Amicon YM30 membrane (Millipore). The concentrated sample was then digested overnight with 1 mg of His6-tagged TEV protease (Kapust et al. 2001) per 100 mg of fusion protein at 4°C, resulting in free YscE protein. The digest was diluted 1:6 with 50 mM MES (pH 6.5), 200 mM NaCl, to approximate an imidazole concentration of 25 mM and then applied to a 50 mL Ni-NTA column equilibrated with buffer A. The sample was eluted isocratically and the flow-through collected and concentrated to a volume of 6 mL. This sample was next applied to a 26/60 HiLoad Superdex 75 prep-grade column (Amersham Biosciences) equilibrated with 50 mM MES (pH 6.0), 200 mM NaCl. The peak fractions containing YscE were pooled and concentrated to 6–7 mg mL−1. Aliquots were flash-frozen with liquid nitrogen and stored at −80°C. The final product was judged to be >95% pure by SDS PAGE (data not shown). The molecular weight of YscE was confirmed by electrospray mass spectrometry.
Crystallization
Untagged YscE crystallized under a variety of conditions in the Hampton Research Crystal and Index screens. Two crystal forms were observed: hexagonal and orthorhombic. The former was grown from conditions with high ionic strength and the latter from polyethylene glycol screens with low salt concentration. The hexagonal crystals were much larger but did not diffract X-rays, whereas the smaller orthorhombic crystals (0.37 × 0.26 × 0.11 mm) scattered to a resolution of 1.8 Å. These diffraction-quality crystals took 10 d to grow under optimized conditions, which consisted of 26% PEG 3350, 0.1 M Na Acetate (pH 4.5), and 1% glycerol.
Data collection, structure solution, and refinement
Although the mother liquor was adequate for flash-cooling the YscE crystals for low-temperature data collection, 10% glycerol was required to effectively protect the crystals from ice formation. The structure was solved by the multiple isomorphous replacement with anomalous scattering (MIRAS) method. Preliminary screening of heavy-atom derivatives was done on a laboratory X-ray source yielding three candidates: Au, Pt, and Pb. The gold and lead derivatives were obtained by soaking the crystals in 10 mM K2(CN)2Au and 1 mM Pb(CH3COO)2 for 10 min, respectively; the platinum derivative was obtained by soaking the crystals in 1 mM K2Pt(CN)4 for 12 h. Before mounting, the derivatized crystals were back-soaked in cryoprotectant for 10 sec to remove excess heavy atoms that could interfere with the anomalous signal. The final native and derivative data employed in solving the structure, however, were collected at the SER-CAT insertion device beamline 22-ID (Advanced Photon Source, Argonne National Laboratory). The wavelengths were tuned to the absorption edge of each derivative after a fluorescence scan had been run to confirm heavy atom binding. The native and anomalous data were indexed, scaled, and merged using the HKL 2000 suite of programs (Otwinowski and Minor 1997). The processed data were combined into the MIRAS script to run in the automated phasing package SOLVE (Terwilliger and Berendzen 1999). The resulting phases were channeled into RESOLVE for density modification and automated model building. The software ARP/wARP 6.0 was used to place the remaining side chains (Morris et al. 2002). Manual correction of the model was accomplished with the graphics software O (Jones et al. 1991) and refinement was carried out with CNS (Brunger et al. 1998) and Refmac5 (Murshudov et al. 1997; Table 1). The atomic coordinates and structure factors for the YscE structure have been deposited in the PDB (Berman et al. 2000) with accession code 1ZW0.
Table 1.
Crystallographic data, phasing, and refinement statistics
| Data set | Native | K2(CN)2Au | K2Pt(CN)4 | Pb(CH3COO)2 |
| Wavelength (Å) | 0.9793 | 1.0396 | 1.0716 | 0.9495 |
| Resolution (Å) | 25–1.8 | 25–2.0 | 25–2.0 | 25–2.4 |
| Unique reflections | 54,088 | 39,909 | 40,494 | 21,729 |
| Completeness (%) | 99.8 | 99.7 | 99.3 | 94.4 |
| Redundancy | 7.3 | 14.2 | 14.5 | 3.9 |
| Rmerge (%)/last shell | 9.1/56.0 | 6.8/26.9 | 8.7/32.7 | 7.9/21.9 |
| No. of sites | 5 | 2 | 3 | |
| Phasing power | ||||
| Anomalous | 0.42 | 0.29 | 0.23 | |
| Isomorphous (centric/acentric) | 0.63/0.70 | 0.51/0.55 | 0.61/0.67 | |
| Figure of merit (20–2.2 Å) | ||||
| Phasing | 0.47 | |||
| After density modification | 0.70 | |||
| R-factor for FC vs. FP | 0.28 | |||
| Refinement resolution (Å) | 25–1.8 | |||
| Rcryst (%) | 22.5 | |||
| Rfree (%) | 25.9 | |||
| Ramachandran plot | ||||
| Most favorable (%) | 97.6 | |||
| Average B-factor (Å2) | 28.6 | |||
| B-factor from Wilson plot (Å2) | 33.3 | |||
| RMSD bonds (Å) | 0.025 | |||
| RMSD angles (°) | 1.99 | |||
| No. of molecules (AU) | ||||
| Polypeptide | 8 | |||
| Water | 335 | |||
Acknowledgments
We thank Florian Schubot, Joseph Tropea, Jerry Alexandratos, and Nicole Le-Ronde Le-Blanc for expert technical assistance and advice. Electrospray mass spectrometry experiments were conducted on the LC/ESMS instrument maintained by the Biophysics Resource in the Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute at Frederick. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31–109-Eng-38.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051706105.
References
- Allaoui, A., Schulte, R., and Cornelis, G.R. 1995. Mutational analysis of the Yersinia enterocolitica virC operon: Characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol. 18 343–355. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker, A., Komoriya, K., and Aizawa, S. 2003. Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc. Natl. Acad. Sci. 100 3027–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A., Adams, P., Clore, G., DeLano, W.,Gross, P., Grosse-Kunstleve, R., Jiang, J., Kuszewiski, J., Nilges, M., Pannu, N., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr.DBiol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster 3D version 2.0: A program for photorealistic molecular graphics. Acta Crystallogr. D 50 869–873. [DOI] [PubMed] [Google Scholar]
- Morris, R.J., Perrakis, A., and Lamzin, V.S. 2002. Arp/warp’s modelbuilding algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 58 968–975. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276A 307– 326. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]