Abstract
The TEL (ETV6)−AML1 (CBFA2) gene fusion is the most common reciprocal chromosomal rearrangement in childhood cancer occurring in ≈25% of the most predominant subtype of leukemia— common acute lymphoblastic leukemia. The TEL-AML1 genomic sequence has been characterized in a pair of monozygotic twins diagnosed at ages 3 years, 6 months and 4 years, 10 months with common acute lymphoblastic leukemia. The twin leukemic DNA shared the same unique (or clonotypic) but nonconstitutive TEL-AML1 fusion sequence. The most plausible explanation for this finding is a single cell origin of the TEL-AML fusion in one fetus in utero, probably as a leukemia-initiating mutation, followed by intraplacental metastasis of clonal progeny to the other twin. Clonal identity is further supported by the finding that the leukemic cells in the two twins shared an identical rearranged IGH allele. These data have implications for the etiology and natural history of childhood leukemia.
An extraordinary diversity of chromosomal molecular abnormalities has been identified in hematopoietic malignancies (1, 2). Among the most prominent are reciprocal chromosomal translocations that produce, via genetic recombination, in-frame fusion genes and hybrid proteins (3, 4). Although details of the mechanisms involved remain to be elucidated, many of these genes encode transcription factors; their novel products are thought to endow clonal advantage via the imposition of an altered pattern of gene regulation (3, 4). One of the most frequent gene fusions so far described is that between TEL (or ETV6) and AML1 (or CBFA2). This rearrangement, although cryptic at the level of chromosome karyotype, occurs in approximately 25% of the predominant subtype of pediatric cancer and leukemia—common acute lymphoblastic leukemia (cALL)—in children diagnosed between the ages of 2 and 10 years (5, 6). The translocation t(12;21)(p13;q22) in ALL consistently involves the fusion of the protein dimerization encoding 5′ region of the ETS-like gene TEL with almost the entire AML1 gene including its DNA binding region (with homology to Drosophila runt) and transactivation domain (reviewed in ref. 5). The chromosome 12 breakpoints cluster within a single intron of the TEL gene whereas AML1 breaks occur within the large and currently unsized first two introns of the AML1 gene on chromosome 21 (5–7). As with other fusion genes in leukemia, each patient’s intronic breakpoints and subsequent fusion sequence are unique, providing a stable genomic marker of the derivative clone of cells. In the context of the etiology and natural history of childhood ALL, a key issue is when and how the TEL-AML1 fusion gene is generated and whether this is an early or initiating event. We report here a molecular analysis of the genomic fusion region of TEL-AML1 in the unusual situation of concordant leukemia in monozygotic twins. This analysis provides unequivocal evidence that this genetic lesion can be acquired during fetal hematopoiesis in utero.
MATERIALS AND METHODS
Patients: Obstetric and Clinical History.
Twins nos. 3838 and 4518 (Dutch Childhood Leukemia Study Group) were diagnosed with ALL at ages 3 years, 6 months and 4 years, 10 months, respectively, at the Wilhelmina Children’s Hospital (The Hague, The Netherlands). Both had CD10+ B lineage ALL (cALL); in one case the leukemic blasts had cytoplasmic μ chains and on this basis was further subclassified as “pre-B” ALL. Cytogenetics were not very informative. It was performed only on case no. 4518 (bone marrow 59% blasts), and the karyotype was recorded as XX normal in six metaphases examined. The twins were identical in appearance and at birth there was a single or monochorionic placenta. Monozygosity has been confirmed with micro-satellite probes (A.M.F., data not shown). The pregnancy was uncomplicated and there was no exposure to diagnostic x-irradiation. The twins had no congenital malformations. Diagnostic samples—separated blood and bone marrow mononuclear cells—were stored as viable cells in liquid nitrogen and used in the current study as a source of DNA and mRNA. Percentage leukemic blasts in the diagnostic samples used for molecular analysis were: for twin 1 (no. 4518) bone marrow 59%, blood 2%; twin 2 (no. 3838) bone marrow 97%, blood 27%.
DNA Extraction.
High molecular weight DNA was extracted from diagnostic samples essentially as described (8).
Southern Blot Analysis.
Two micrograms of DNA was digested with BamHI, electrophoresed through 0.7% agarose transferred to Hybond N+ (Amersham), and hybridized to either a cDNA probe containing TEL exon 5 (9) or to a genomic fragment of the human IGH joining region (10). Similarly, confirmation of monozygosity was achieved by using the multilocus fingerprinting probe 33.15 (Cellmark Diagnostics, Abingdon, UK) on HinfI digests.
Reverse Transcription–PCR (RT-PCR) Analysis.
Total RNA was extracted from bone marrow or peripheral blood by acid guanidinium thiocyanate-phenol-chloroform extraction (11). cDNA was generated from 5 μg of total RNA by using random hexamers. The integrity of the cDNA was assessed by amplification of c-abl sequences by using primers ABLS1 5′-CAGCGGCCAGTAGCATCTGACTT-3′ and ABLAS1 5′-GCTTCACACTCCCCATT-3′. RT-PCR was performed with equivalent amounts of cDNA from control and twin samples in the presence of 1.8% formamide and in a final volume of 50 μl. Primers for the amplification of TEL-AML1 were TEL S841 5′-ATCATGCACCCTCTGATCCT-3′ and AML1 AS591 5′-ACGCCTCGCTCATCTTGCCT-3′. Cycle conditions were as follows: initial denaturing step at 94°C for 5 min followed by 30 cycles (94°C 1 min, 60°C 1 min, 72°C 2 min) and a final extension for 7 min at 72°C. PCR products (1/10th volume) were separated on 1.5% agarose gels.
Cloning of TEL Breakpoints.
Five micrograms of high molecular weight DNA from twin 1 was partially digested with BamHI and cloned into λ fix II by partial fill-in, packaged into Gigapack III Gold extracts and an XLI-blue MRA host (all Stratagene). The library was screened with the TEL exon 5 cDNA probe, and three positive clones were identified, one of which was further characterized by restriction mapping and sequencing. DNA was sequenced on an Applied Biosystems 373A automated DNA sequencer and further analyzed by Geneworks 2.1 (IntelliGenetics).
Breakpoint PCR analysis.
For the amplification of t(12;21) fusion sequences, PCR primers were designed from 500 bp of breakpoint specific sequence: forward: 5′-TGGGTGAT TCCTGTGTACAAG-3′, and reverse: 5′-CAAAC AGCCAGTAAAGGAACG-3′.
Patient DNA (200 ng) was mixed in a 50-μl reaction with 75 ng of each primer in 50 mM KCl/10 mM Tris⋅HCl, pH 8.3/1.5 mMMgCl2/0.01% gelatin/200 μM each dNTP/2½ units of Taq polymerase. After 1 min at 94°C, amplification was carried out for 1 min at 94°C, 1 min at 55°C, 1 min at 72°C for 35 cycles, followed by a 7-min incubation at 72°C. PCR products were separated on 1.5% agarose gels stained with ethidium bromide and visualized under UV light. Size markers were DNA marker VI (Boehringer Mannheim).
IGH gene rearrangements were examined by PCR, cloning, and sequencing essentially as described (12) except that 100 ng of DNA was used in 50-μl reactions. All sequences were identified by blast sequence searches through the National Center for Biotechnology Information (13) and by sequence comparison to published (14) IGH variable diversity and joining region sequences (15–17).
RESULTS
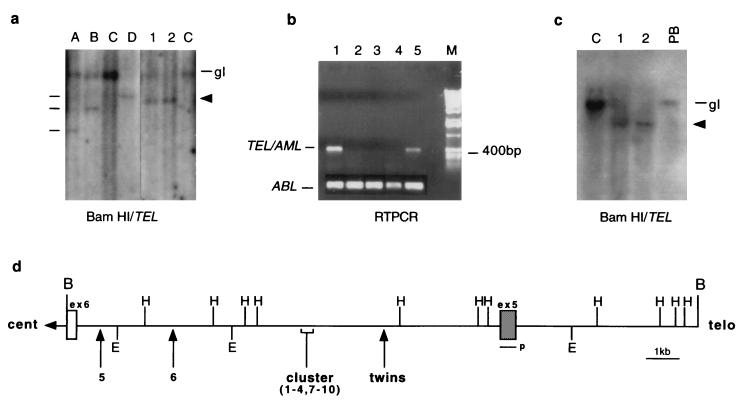
We first analyzed the TEL gene status in DNA isolated from leukemic bone marrow from the monozygotic twins at diagnosis with cALL. BamHI Southern blots hybridized to a cDNA probe containing TEL exon 5 (9) showed an apparently identical pattern of rearranged TEL restriction fragments, with additional loss of germ-line TEL sequences in twin 2 sample (Fig. 1a), suggesting deletion of the nonrearranged TEL allele as reported for other cases of ALL with TEL-AML1 fusion (18, 19). A germ-line (gl), nonrearranged TEL allele is present in the digest of the leukemic cell DNA from twin 1 (Fig. 1a, lane 1) but as the bone marrow sample from which this DNA was prepared had a mixture of leukemic and normal cells (59% blasts), we cannot say whether or not there was deletion of the nonrearranged TEL allele in the leukemic cells of twin 1. In contrast to the size concordance of rearranged TEL restriction fragments from twin DNA, diverse sizes of TEL rearrangement were seen in DNA from three other cases of ALL (Fig. 1a, lanes A, B, and D) known to express TEL-AML1 fusion transcripts by RT-PCR (C.M.P., data not shown). No TEL gene rearrangements were seen in the AML control (Fig. 1a, lanes C). To determine whether the TEL gene rearrangements detected by Southern blot involved fusion to AML1, we performed RT-PCR on RNA obtained from leukemic bone marrow cells from each twin. Fig. 1b shows that specific TEL-AML1 fusion transcripts (400 bp) were amplified from each twin cDNA.
Figure 1.
Molecular analysis of t(12;21) in identical twins. (a) BamHI Southern blot analysis of TEL rearrangement. The monozygotic twins, lanes 1 and 2, respectively, show a shared pattern of TEL rearrangement (indicated by arrow) and loss of heterozygosity. Lanes A, B, and D are t(12;21) controls whose different rearrangements are denoted by a line. Lanes C represent AML controls, and gl is germ line (20 kb). (b) RT-PCR analysis for TEL-AML fusion transcript. Lanes 2–4, childhood ALL cases negative for t(12;21). Lane 1, twin 1 (bone marrow at diagnosis 59% blasts), lane 5 twin 2 (bone marrow at diagnosis 97% blasts). (Lower) ABL RT-PCR. (c) The TEL rearrangement is nonconstitutive. BamHI Southern blot analysis showing a lack of TEL rearrangements in diagnostic peripheral blood of twin 1 (PB; 2% blasts). C is the AML control, 1 and 2 represent each twin, and gl is germ-line 20 kb. The rearranged TEL allele is indicated by an arrow. (d) Partial restriction map of the TEL breakpoint cluster region. The map shows only the relevant restriction sites and is adapted from ref. 14. B, BamHI; E, EcoRI; H, HindIII; p, probe. The location of the TEL breakpoint in the twins is indicated by an arrow in relation to 10 previously published breaks (9). Cent relates to centromere and telo to telomere.
We performed further Southern blots on twin 1 DNA prepared from a peripheral blood sample containing only 2% blast cells taken at diagnosis. The TEL rearrangement was not detectable in the peripheral blood sample, indicating that it was an acquired or nonconstitutive abnormality (Fig. 1c).
t(12;21) fusions involve the noncoding introns of TEL and AML1 and the breaks are scattered and diverse within the breakpoint cluster regions (5). As a consequence, each break and subsequent fusion junction is clonotypic and patient specific at the DNA level. The genomic fusion sequence therefore provides a unique marker of clonal identity and stable inprint of single cell origin. Sequencing of the TEL-AML1 fusion region in the twin pair therefore should provide unambiguous evidence for their clonal relationship as well as other possible clues to the recombination mechanisms. To our knowledge, no genomic fusion sequences of TEL-AML1 have been published to date. We cloned the TEL-AML1 fusion gene from twin 1 and sequence analysis confirmed identity to sequences within intron 5 of the TEL gene (Fig. 2). The twin sequence also revealed a small deletion (739 bp) compared with published sequence (14) before diverging into what we assume is AML1 intron sequence. (blasta sequence analysis has not yet identified any known homologies.) Several translocations that occur in B cell malignancies appear to result from illegitimate recognition and joining of sequences similar to those present in Ig gene rearrangements (20, 21). We noted that there is a sequence in the TEL gene, following the breakpoint in these twins, that has some features of V(D)J heptamer-spacer-nonamer recombination signal sequence: 5′-CACACAT-12-ACACATATA-3′. Further sequence analysis of the reciprocal AML-TEL gene fusion and fusion sequences from other patients will be required to determine the likelihood of V(D)J recombinase-mediated TEL-AML1 fusion. Some gene fusions in cancer cells may arise via homologous recombination between repeat families such as Alu (22, 23). Upstream of the TEL breakpoint in our twin pair is a 150-bp exact match for a human carcinoma cell-derived Alu RNA transcript (24) (Fig. 2) highlighting this as a candidate mechanism for recombination in TEL-AML1 fusion.
Figure 2.
Sequence comparison between TEL intron 5 and twin 1 breakpoint sequences. 5′ TEL refers to published intron 5 sequence from 31512 to 31600 (14). 3′ TEL refers to sequences 32339 to 32787. The twins sequence is shown underneath with a 739-bp deletion between the 5′ and 3′ TEL sequences. Twin sequence diverges from TEL at 32787 presumably into AML1 intron sequence. The sequence with ≈100% homology to a human carcinoma cell-derived Alu RNA transcript (NE36, ref. 24) is underlined. Arrows indicate PCR primers.
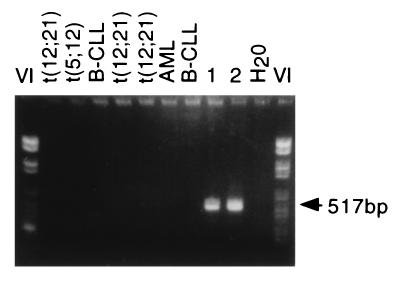
We next designed PCR primers to amplify across the breakpoint region of twin 1 DNA. To ensure that the small deletion was not a cloning artefact, the primers were designed to bridge this region in TEL as well as to span the breakpoint itself. Fig. 3 shows that only the expected fragment of 517 bp was present in twin 1 DNA and that an identically sized fragment was amplified from the DNA from twin 2. No PCR product was obtained from other patient material or cell lines with t(12;21) or from other types of acute leukemias. Sequencing of PCR products from both twins confirmed an identical breakpoint sequence (compare with Fig. 2). This finding suggests that the twin leukemias are derivatives of the same single cell or clone in which the unique TEL-AML1 fusion occurred.
Figure 3.
The TEL/AML1 genomic breakpoint is identical in each twin. Breakpoint-specific PCR primers were used to amplify the fusions from a number of patient DNAs with and without t(12;21). Only DNA from the twins gives the expected 517-bp product showing breakpoint sequence specificity.
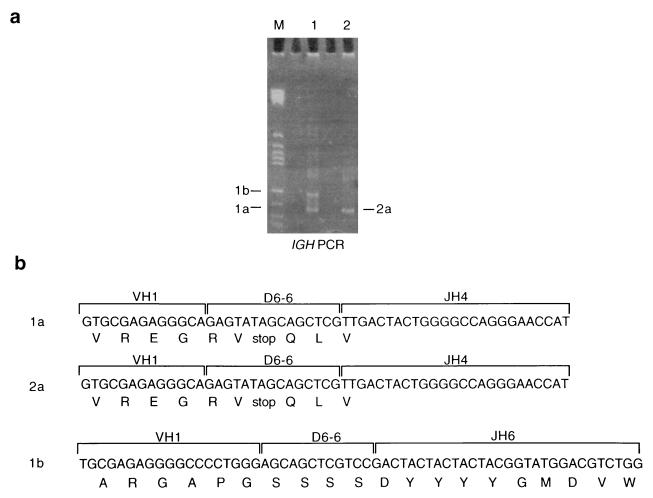
Because unique or clonotypic IGH and TCR gene rearrangements occur very early in B and T cell differentiation and are relatively stable, they also have provided markers of clonality in leukemia (reviewed in ref. 25). The finding of an identical T cell receptor (TCR) DNJHβ sequence in a pair of twins diagnosed with T cell malignancy at ages 9 and 10 years provided indirect, but compelling, evidence for a fetal origin coupled with protracted latency (12). PCR techniques have been used to detect IGH rearrangements by using a consensus joining (JH) primer in combination with a universal primer designed from within the conserved regions of the variable region gene families (VH) (26). We amplified the rearranged Ig genes from each twin DNA to evaluate the clonal relationship. Twin 1 DNA produced two PCR products of 104 bp and 124 bp (Fig. 4a, fragments 1a and 1b, respectively), whereas in twin 2 DNA only the 104-bp product (fragment 2a) was present. The identically migrating products from both twins were picked, cloned, and sequenced as previously described (12) and were found to have identical DNA sequences joining a VH1 family member to diversity region D6–6, joined to JH4 (Fig. 4b). There were no identifiable N or P region insertions present. The VDJH sequence, in both twin leukemic DNA samples, appears to encode a nonfunctional Ig gene product by formation of the same single stop codon in the diversity region (Fig. 4b). The presence of an identical clonal VDJ IGH gene rearrangement in twin leukemic DNA provides additional evidence for a common or single cell origin. The absence of N regions also accords with an origin of this clonal rearrangement during early fetal hemopoiesis (27). The larger rearranged fragment, distinct to twin 1, shows a VDJH join without N or P region insertions that joins another VH1 family member via D6–6 to JH6. It is likely that this represents a secondary allelic rearrangement associated with the presence of μ heavy chain protein in twin 1 leukemic cells and indicative of continuing recombinase activity after the TEL-AML1 fusion event.
Figure 4.
An IGH VDJ join is identical in each twin. (a) PAGE of PCR product from IGH gene rearrangements in the twin pairs showing two identically sized fragments and one additional band specific to twin 1. All bands were cut out, cloned, and sequenced. (b) Nucleotide and amino acid composition of the IGH products. VH, variable region; DH, diversity region; JH, joining region. V, D, and J motifs were identified from refs. 15–17.
DISCUSSION
In previous studies, very young (<1 year) identical twins with concordant ALL were found to share unique or clonotypic rearrangements of the MLL fusion gene (28, 29). Because this genetic abnormality was acquired rather than constitutive, it was argued that this finding could be explained only by a single cell origin in one fetus followed by spread of clonal progeny to the other twin via vascular anastomoses within a monochorionic placenta (30, 31) or even between separate placentas in the instance of a pair of infant twins that were dichorionic (29). The TEL-AML1 fusion gene in the older twins with concordant leukemia is therefore the same in terms of single cell origin and prenatal timing as the MLL gene fusion in infant ALL. It is also likely to have been the initiating clonal event occurring, in these cases, in a fetal B lineage progenitor cell already having acquired one nonfunctional VDJH rearrangement. The presence of an Alu repeat sequence and a possible heptamer-nonamer recombinase sequence close to the TEL gene breakpoint highlight potential mechanisms of recombination in the fetal cell that warrant further investigation.
Concordant leukemia in monozygotic twins with cALL differs from that in twin infants with MLL gene fusions in two respects. The twin pair reported here were aged 3 years, 6 months and 4 years, 10 months at diagnosis and therefore had a more protracted latency, following the proposed initiation in utero, than in infants with MLL gene fusions. Also, the concordance rate for leukemia differs substantially in twins with these two biologically and clinically distinct types of leukemia diagnosed at different ages. For twin infants, the concordance rate is very high. No accurate estimate is available but historically the figure has been estimated at 25% or 1 in 4 (32). Because only 60% of monozygotic twins have a monochorionic placenta (31), the concordance rate for those with the anatomical connections facilitating metastasis in utero could approach 100%. This result in turn would suggest that MLL gene fusions might be sufficient for leukemia and that secondary genetic changes, though often present (33), are nonessential. Alternatively, MLL gene fusions might promote rapid acquisition of necessary secondary genetic events. In contrast to the situation with infants, older twin children have a much lower concordance rate of cALL. Again, no accurate estimate of rate is available. We have collected nine such pairs via an international trawl, including three pairs from the United Kingdom since 1984. Based on expected United Kingdom incidence rates for ALL and for twinning, we calculate that concordance for cALL is no higher than 5% (34). Ninety-five percent discordance, as in other twin studies including Hodgkin’s disease (35), implies an important etiological role for postnatal exposures. The more protracted latency and lower concordance rate for cALL in older children compared with infant ALL accords with the notion, derived from epidemiological data, that an abnormal or delayed response to a common postnatal infection might be critically involved in promoting this type of leukemia (36). 12p− including loss of the normal TEL allele occurs in many cases of cALL with TEL-AML1 fusion (18, 19, 37) as in the twin pair reported here. Limited fluorescence in situ hybridization analysis suggests that this event is probably secondary to TEL-AML fusion (19) and therefore might represent the required secondary postnatal step in the genesis of cALL. A latency of several years for childhood leukemia is not unprecedented. We previously have described a pair of identical twins with T cell malignancy diagnosed at aged 9 and 11 years and in whom sharing of a clonotypic marker (T cell antigen receptor β sequence) suggested a fetal origin (12).
There is no reason to suppose that TEL-AML1 gene fusion and leukemia initiation will only occur in fetal hematopoiesis in the context of a monozygotic twin pregnancy. We therefore conclude that this same fetal event will have occurred at some frequency, and possibly commonly, in nontwinned children with cALL. Further support for this suggestion could come from the analysis of other, albeit rare, cases of concordant ALL in twins and from retrospective scrutiny by PCR of neonatal blood spots (Guthrie cards) for clonotypic TEL-AML1 fusion sequences as recently described for MLL-AF4 gene fusions in infant ALL (38).
Acknowledgments
We are grateful to Dr. O. Bernard for providing a TEL probe and Professor T. Rabbitts for a JH probe. We thank Dr. H. King for PCR primers and sequencing, Dr. E. Vandenberghe for preliminary molecular analysis of X chromosome polymorphisms, Drs. L. Wiedemann, J. Wiemels, and F. McBlane for helpful discussions, and Ms. B. Deverson for help in preparation of the manuscript. This work was supported by the Leukaemia Research Fund and the Kay Kendall Leukaemia Fund.
ABBREVIATIONS
- cALL
common acute lymphoblastic leukemia
- ALL
acute lymphoblastic leukemia
- RT-PCR
reverse transcription–PCR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession number AF044317).
References
- 1.Sawyers C L. Lancet. 1997;349:196–200. doi: 10.1016/S0140-6736(96)07535-6. [DOI] [PubMed] [Google Scholar]
- 2.Hagemeijer A, Grosveld G. In: Leukemia. Henderson E S, Lister T A, Greaves M F, editors. Philadelphia: Saunders; 1996. pp. 131–144. [Google Scholar]
- 3.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 4.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 5.Golub T R, Barker G F, Stegmaier K, Gilliland D G. Curr Top Microbiol Immunol. 1996;211:279–288. doi: 10.1007/978-3-642-85232-9_28. [DOI] [PubMed] [Google Scholar]
- 6.Bernard O A, Romana S P, Poirel H, Berger R. Leuk Lymphoma. 1996;23:459–465. doi: 10.3109/10428199609054854. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford A M, Molgaard H V, Greaves M F, Gould H J. EMBO J. 1983;2:997–1001. doi: 10.1002/j.1460-2075.1983.tb01533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romana S P, Poirel H, Leconiat M, Flexor M-A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 10.Rabbitts T H, Forster A, Milstein C P. Nucleic Acids Res. 1981;9:4509–4524. doi: 10.1093/nar/9.18.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Ford A M, Pombo-de-Oliveira M S, McCarthy K P, MacLean J M, Carrico K C, Vincent R F, Greaves M. Blood. 1997;89:281–285. [PubMed] [Google Scholar]
- 13.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Baens M, Peeters P, Gu O-C, Aerssens J, Marynen P. Genome Res. 1996;6:404–413. doi: 10.1101/gr.6.5.404. [DOI] [PubMed] [Google Scholar]
- 15.Ravetch J V, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Cell. 1981;27:583–591. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- 16.Corbett S J, Tomlinson I M, Sonnhammer E L L, Buck D, Winter G. J Mol Biol. 1997;270:587–597. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 17.Cook G P, Tomlinson I M, Walter G, Riethman H, Carter N P, Buluwela L, Winter G, Rabbitts T H. Nat Genet. 1994;7:162–168. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 18.Stegmaier K, Pendse S, Barker G F, Bray-Ward P, Ward D C, Montgomery K T, Krauter K S, Reynolds C, Sklar J, Donnelly M, et al. Blood. 1995;86:38–44. [PubMed] [Google Scholar]
- 19.Raynaud S, Cave H, Baens M, Bastard C, Cacheux V, Grosgeorge J, Guidal-Giroux C, Guo C, Vilmer E, Marynen P, Grandchamp B. Blood. 1996;87:2891–2899. [PubMed] [Google Scholar]
- 20.Lewis S M, Agard E, Suh S, Czyzyk L. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsden D A, McBlane J F, Van Gent D C, Gellert M. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 22.Chissoe S L, Bodenteich A, Wang Y-F, Wang Y-P, Burian D, Clifton S W, Crabtree J, Freeman A, Iyer K, Jian L, et al. Genomics. 1995;27:67–82. doi: 10.1006/geno.1995.1008. [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Alder H, Nakamura T, Schichman S A, Prasad R, Canaani O, Saito H, Croce C M, Canaani E. Cancer Res. 1994;54:2327–2330. [PubMed] [Google Scholar]
- 24.Sinnett D, Richer C, Deragon J-M, Labuda D. J Mol Biol. 1992;226:689–706. doi: 10.1016/0022-2836(92)90626-u. [DOI] [PubMed] [Google Scholar]
- 25.Waldmann T A. Adv Immunol. 1987;40:247–321. doi: 10.1016/s0065-2776(08)60241-2. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy K P, Sloane J P, Wiedemann L M. J Clin Pathol. 1990;43:429–432. doi: 10.1136/jcp.43.5.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasserman R, Galili N, Ito Y, Reichard B A, Shane S, Rovera G. J Exp Med. 1992;176:1577–1581. doi: 10.1084/jem.176.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford A M, Ridge S A, Cabrera M E, Mahmoud H, Steel C M, Chan L C, Greaves M F. Nature (London) 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 29.Gill Super H J, Rothberg P G, Kobayashi H, Freeman A I, Diaz M O, Rowley J D. Blood. 1994;83:641–644. [PubMed] [Google Scholar]
- 30.Clarkson B, Boyse E A. Lancet. 1971;i:699–701. doi: 10.1016/s0140-6736(71)92705-x. [DOI] [PubMed] [Google Scholar]
- 31.Strong S J, Corney G. The Placenta in Twin Pregnancy. Oxford: Pergamon; 1967. [Google Scholar]
- 32.Zuelzer W W, Cox D E. Semin Hematol. 1969;228:228–249. [PubMed] [Google Scholar]
- 33.Cimino G, Lanza C, Elia L, Lo Coco F, Gaidano G, Biondi A, Pastore C, Serra A, Canaani E, Croce C M, et al. Br J Haematol. 1997;96:308–313. doi: 10.1046/j.1365-2141.1997.d01-2044.x. [DOI] [PubMed] [Google Scholar]
- 34.Greaves M. Blood. 1993;82:1043–1051. [PubMed] [Google Scholar]
- 35.Mack T M, Cozen W, Shibata D K, Weiss L M, Nathwani B N, Hernandez A M, Taylor C R, Hamilton A S, Deapen D M, Rappaport E B. N Engl J Med. 1995;332:413–418. doi: 10.1056/NEJM199502163320701. [DOI] [PubMed] [Google Scholar]
- 36.Greaves M F. Lancet. 1997;349:344–349. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 37.Romana S P, Le Coniat M, Poirel H, Marynen P, Bernard O A, Berger R. Leukemia. 1996;10:167–170. [PubMed] [Google Scholar]
- 38.Gale K B, Ford A M, Repp R, Borkhardt A, Keller C, Eden O B, Greaves M F. Proc Natl Acad Sci USA. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]