Abstract
Many insect viruses survive for long periods by occlusion within robust crystalline polyhedra composed primarily of a single polyhedrin protein. We show that two different virus families form polyhedra which, despite lack of sequence similarity in the virally encoded polyhedrin protein, have identical cell constants and a body-centered cubic lattice. It is almost inconceivable that this could have arisen by chance, suggesting that the crystal lattice has been preserved because it is particularly well-suited to its function of packaging and protecting viruses.
Keywords: insects, viruses, polyhedra, powder diffraction, crystals
A wide range of insect viruses survive for long periods in harsh environments by occlusion within robust protein particles (http://www.agctr.lsu.edu/s265/young.htm). Some of these particles have a regular shape and are consequently known as “polyhedra.” They are composed primarily of a viral encoded protein (polyhedrin) and are 1–3 μm in size, and form surprisingly well ordered crystal lattices (Di et al. 1991). Virions become specifically embedded within the polyhedrin matrix, allowing polyhedra to transmit packages of infectious virus between hosts by oral-faecal routes. Ingested polyhedra dissolve in the high pH of the insect gut, releasing virus particles (Payne and Mertens 1983), which infect the gut epithelium, sometimes spreading to other cells. Progeny virus particles (and polyhedra) are subsequently released by excretion and/or disintegration of the host.
Polyhedra are produced by cytoplasmic polyhedrosis viruses (CPV) (icosahedral, ~70 nm diameter, segmented dsRNA viruses, family Reoviridae) and the unrelated nuclear polyhedrosis viruses (250–300 nm long/30–60 nm diameter, rod shaped, dsDNA viruses, family Baculoviridae). These viruses can infect the larvae of many insect species, including moths, butterflies, and flies. Indeed, baculoviruses in particular have been suggested as environmentally friendly and highly specific insecticides (Moscardi 1999). Here we show that although the polyhedrin proteins from these two families of viruses show no detectable amino acid similarity, analysis of powder diffraction data reveals that their crystals share essentially identical cell constants and a body-centered cubic lattice.
Results and Discussion
We have performed X-ray diffraction experiments on polyhedra from three viruses: the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and two cypoviruses (types 1 and 5), taken from infected Silkworm and Tussock Moth, respectively. AcMNPV polyhedra were purified from infected Spodoptera cell cultures and CPV polyhedra from infected insect hosts (see Materials and Methods). Growth in insect cells limits the size of polyhedra to a few micrometers, making single crystal analysis difficult. Powder diffraction experiments were therefore performed on slurries of polyhedra using synchrotron radiation (see Materials and Methods). The observed scattering pattern from an ideal (crystalline) powder is composed entirely of rings, produced by the overlapping patterns from a large number of randomly orientated crystallites. A very clear “powder pattern” was observed for all three polyhedra (Fig. 1A ▶), with sharp rings extending beyond 4 Å resolution, the technical limit in this experiment, suggesting that higher resolution could be achieved using a more specialized setup, and demonstrating that polyhedra from baculoviruses and cypoviruses possess well ordered crystalline lattices.
Figure 1.
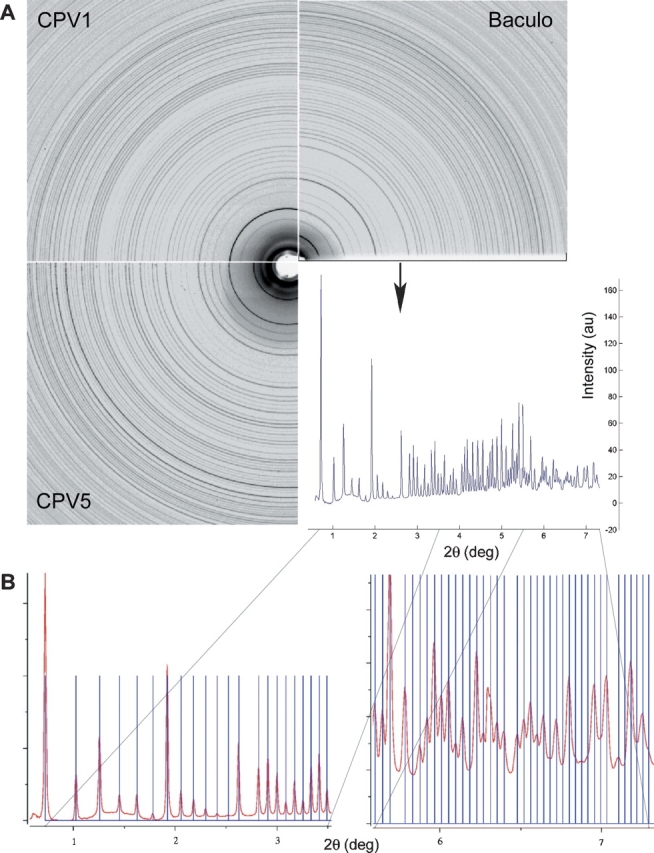
(A) Part of powder diffraction patterns for polyhedra of AcMNPV, CPV1, and CPV5. The nearly identical spacing of the intensity rings with a small opening angle shows the similarity of lattice constants of the different samples. Comparing the rings of CPV1 and CPV5, the small difference in the lattice constants is clearly visible, emphasising the precision of powder diffraction analysis. (B) The predicted peak positions for a cubic body-centered lattice of 103.7 ± 0.05 Å lattice constant, agree very well with the powder pattern measured for AcMNPV polyhedrin.
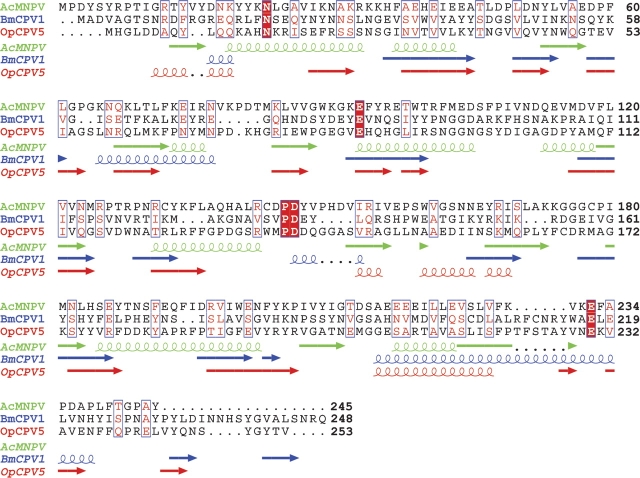
A systematic analysis was performed by summing the diffracted intensity along the circumference of individual rings and subtracting the background, allowing lattice constants to be determined accurately from peak positions. The diffraction patterns taken from the three viral polyhedra were indexed using CMPR (http://www.ncnr.nist.gov/programs/crystallography/software/cmpr/) generating an astonishing result. All three polyhedra have a cubic body-centered lattice. The lattice constants were found to be 103.7 Å for AcMNPV (245 residues), 103.8 Å for CPV1 (248 residues), and 102.9 Å for CPV5 (253 residues) polyhedrin; i.e., the lattice constants are nearly identical (within 1%). This lattice does not agree with that proposed earlier for CPV-1 polyhedra (Di et al. 1991), probably reflecting the higher quality and extent of the synchrotron data presented here (Fig. 1B ▶ shows the quality of fit to the data). Within the baculoviruses the polyhedrin genes show a high degree of conservation, and, similarly, within a CPV type the polyhedrin gene is highly conserved. However, although they are similar in size, the polyhedrins from different virus types show surprisingly little amino acid sequence similarity (the two CPVs have only 15% identity, while AcMNPV has no discernable similarity with either CPV [< 10% identity]; see Fig. 2 ▶). Unsurprisingly, secondary structure prediction programs such as PROFsec (Rost and Sander 1993) give good agreement for secondary structure prediction between CPV-1 and CPV-5 (nine strands of β-sheet are common between the two predictions, and two helices), but there is little agreement between AcMNPV (which is mainly helical) and CPV (Fig. 2 ▶). The intensities of the individual rings obtained from the different polyhedra differ even at low resolution (Fig. 1A ▶), confirming that their molecular structures are different.
Figure 2.
Alignment of the amino acid sequences for the polyhedrin gene of Autographa californica baculovirus (AcMNPV), Bombyx mori CPV type 1 (BmCPV1), and Orgyia pseudosugata CPV type 5 (OpCPV5) with predicted secondary structure assignments drawn underneath. Fully conserved residues are shown on a red background, and similar residues are highlighted red in a blue box (global similarity score as defined by Risler et al. [1988] > 0.7). The sequences were aligned with ClustalW and visualized using ESPript (Gouet et al. 1999), and the secondary structures were predicted by PROFsec (Rost and Sander 1993).
The polyhedra diffraction data are consistent with space groups I23 or I213 that together are found in only 0.6% of the > 29,000 entries in the Protein Data Bank, and no entry possesses cell dimensions within 1% of those of the polyhedra (Berman et al. 2002). It therefore seems almost inconceivable that the precise similarities observed between the crystals of the three different polyhedra could have arisen by chance, suggesting that the lattice has been selected because it is particularly well suited to its function of taking up and protecting the viruses. Since the viruses incorporated into the identical lattices are quite different from each other it seems plausible to suggest that the polyhedrin molecules could be engineered to design novel packaging activities. It is interesting to note that space group I23 possesses all of the icosahedral symmetry elements that are consistent with a crystal lattice (fivefold axes being prohibited) but no further elements. This would make it an ideal vehicle for selecting icosahedral viruses (or the hemispherical caps of the rod-like baculovirus) for inclusion into the developing polyhedron by specific polyhedrin–virion contacts (indeed inspection of the Protein Data Bank reveals that 13% of all crystal structures of viruses have been in this space group). It seems most likely that these generic packaging machines, which may be targets for engineering to design novel packaging activities, share a common ancestor; however, detailed three-dimensional structural analysis is required to resolve this issue.
Materials and methods
AcMNPV polyhedra were produced by infecting Spodoptera frugiperda cell cultures with wild-type baculovirus. CPV1 and CPV5 were obtained by infecting their respective insect hosts (Bombyx mori and Orgyia pseudosugata). Polyhedra were purified as described previously (Hill et al. 1999). Cypovirus type was confirmed by electropherotyping (Payne and Mertens 1983). In each experiment a slurry of polyhedra in a thin walled quartz capillary was placed in a monochromatic X-ray beam of wavelength 0.933 Å at beamline ID14-EH2 of the European Synchrotron Radiation Facility (Grenoble). The sample was kept at room temperature and rotated around 10° during a 10-sec exposure time. An ADSC CCD detector with 80 μm pixel size was used to record the images.
Acknowledgments
We thank the staff of ID14-EH2 at the ESRF, Grenoble, for support. The U.K. Medical Research Council and the Royal Society provided financial support.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051516405.
References
- Berman, H.M., Battistuz, T., Bhat, T.N., Bluhm, W.F., Bourne, P.E., Burkhardt, K., Feng, Z., Gilliland, G.L., Iype, L., Jain, S., et al. 2002. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 58: 899–907. [DOI] [PubMed] [Google Scholar]
- Di, X., Yu-Kun, S., McCrae, M.A., and Rossmann, M.G. 1991. X-ray powder pattern analysis of cytoplasmic polyhedrosis virus inclusion bodies. Virology 180: 153–158. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Hill, C.L., Booth, T.F., Stuart, D.I., and Mertens, P.P.C. 1999. Lipofectin increases the specific activity of cypovirus particles for cultured insect cells. J. Virol. Methods 78: 177–189. [DOI] [PubMed] [Google Scholar]
- Moscardi, F. 1999. Assessment of the application of baculoviruses for control of Lepidoptera. Annu. Rev. Entomol. 44: 257–289. [DOI] [PubMed] [Google Scholar]
- Payne, C.C. and Mertens, P.P.C. 1983. The cytoplasmic polyhedrosis viruses. In The reoviridae (ed. W.K. Joklik). Plenum Press, New York.
- Risler, J.L., Delorme, M.O., Delacroix, H., and Henaut, A. 1988. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol. 204: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Rost, B. and Sander, C. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232: 584–599. [DOI] [PubMed] [Google Scholar]