Abstract
Mammalian telomeres consist of long tandem arrays of double-stranded telomeric TTAGGG repeats packaged by the telomeric DNA-binding proteins TRF1 and TRF2. Both contain a similar C-terminal Myb domain that mediates sequence-specific binding to telomeric DNA. In a DNA complex of TRF1, only the single Myb-like domain consisting of three helices can bind specifically to double-stranded telomeric DNA. TRF2 also binds to double-stranded telomeric DNA. Although the DNA binding mode of TRF2 is likely identical to that of TRF1, TRF2 plays an important role in the t-loop formation that protects the ends of telomeres. Here, to clarify the details of the double-stranded telomeric DNA-binding modes of TRF1 and TRF2, we determined the solution structure of the DNA-binding domain of human TRF2 bound to telomeric DNA; it consists of three helices, and like TRF1, the third helix recognizes TAGGG sequence in the major groove of DNA with the N-terminal arm locating in the minor groove. However, small but significant differences are observed; in contrast to the minor groove recognition of TRF1, in which an arginine residue recognizes the TT sequence, a lysine residue of TRF2 interacts with the TT part. We examined the telomeric DNA-binding activities of both DNA-binding domains of TRF1 and TRF2 and found that TRF1 binds more strongly than TRF2. Based on the structural differences of both domains, we created several mutants of the DNA-binding domain of TRF2 with stronger binding activities compared to the wild-type TRF2.
Keywords: telomeres, tertiary structures, TRF2, TRF1, protein/DNA interactions, NMR, Myb domain
Telomeres are protein-DNA complexes that distinguish natural chromosome ends from damaged DNA. Mammalian telomeric DNA is composed of long tandem arrays of the double-stranded telomeric repeats of TTAGGG followed by a single-stranded DNA at the 3′ end. TRF1 and TRF2 are mammalian telomeric repeat-binding factors (Chong et al. 1995; Bilaud et al. 1997; Broccoli et al. 1997). TRF1 interacts with TIN2 (Kim et al. 1999), PINX1 (Zhou and Lu 2001), and tankyrase 1 and 2: poly (ADP-ribose) polymerases (Smith et al. 1998; Smith and de Lange 2000; Cook et al. 2002). In addition, Pot1 interacts with a complex of these proteins at the 3′ overhang consisting of single-stranded telomeric DNA (Loayaza and de Lange 2003). In contrast, TRF2 interacts with Rap1 (Li et al. 2000) and the Mre11 complex composed of Mre11, Rad50, and Nbs1 (Zhu et al. 2000).
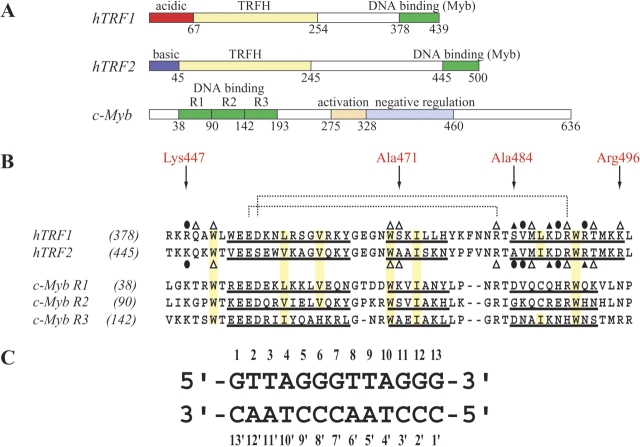
Overexpression of wild-type TRF1 induces telomere shortening in the presence of telomerase, and expression of a dominant-negative allele of TRF1 results in elongation of telomeres (van Steensel and de Lange 1997; Smogorzewska et al. 2000). In contrast, the expression of a dominant negative allele of TRF2 results in destabilization of chromosomes, chromosome end-to-end fusions, the activation of ATM/p53 or p16/RB pathways, and the induction of senescence or apoptosis (van Steensel et al. 1998; Karlseder et al. 1999, 2002; Smogorzewska and de Lange 2002). In addition, TRF2 can form t-loops by itself (Griffith et al. 1999; Stansel et al. 2001). Although the cellular functions of TRF1 and TRF2 are different from each other, they each contain similar functional domains, a central TRF-homology (TRFH) domain and a C-terminal DNA-binding domain (Chong et al. 1995; Bianchi et al. 1997; Smith and de Lange 1997). Their N-terminal domains differ: an acidic domain in TRF1, and a basic domain in TRF2 (Fig. 1A ▶; Broccoli et al. 1997). Both TRFH domains can form ho-modimer but not heterodimer (Bianchi et al. 1997; Broccoli et al. 1997; Smith and de Lange 1997; Fairall et al. 2001). Figure 1B ▶ shows that both DNA-binding domains of human TRF1 and TRF2 contain an amino acid sequence similar to each sequence of the three repeats in the c-Myb DNA-binding domain (Gonda et al. 1985; Klempnauer and Sippel 1987).
Figure 1.
Domain structures and amino acid sequences of the TRF2 and TRF1 DNA-binding domains with base sequence of the telomeric DNA used. (A) Comparison of domain structures of hTRF1, hTRF2, and c-Myb. basic, basic domain; acidic, acidic domain; TRFH, TRF homology domain; Myb, Myb domain; R1, R2, and R3, the first, second, and third repeats of the c-Myb DNA-binding domain, respectively; activation, transcription activation domain; negative regulation, transcription negative regulation domain. (B) Sequence alignment of DNA-binding domains of hTRF1, hTRF2, and three repeats of the DNA-binding domains of mouse c-Myb. Well conserved residues involved in the hydrophobic core are boxed in yellow. Two pairs of polar residues that form an intramolecular salt bridge are connected with broken lines. Three helical regions are underlined. Amino acids that interact with DNA bases are indicated by closed circles; amino acids that interact with the sugar-phosphate backbone are indicated by open triangles, and amino acids that interact with both of them are indicated by closed triangles. (C) Sequence of the DNA duplex. The numbering scheme used in this paper is shown.
The c-Myb protein is a transcriptional activator that binds to a consensus sequence of TAACNG and regulates the proliferation of hematopoietic cells (Biedenkapp et al. 1988; Ness et al. 1989; Weston and Bishop 1989; Nakagoshi et al. 1990; Tanikawa et al. 1993). Figure 1 ▶ shows that the DNA-binding domain of c-Myb consists of three imperfect tandem repeats, R1, R2, and R3, each consisting of 52 amino acids. The c-Myb repeats have very similar tertiary structures, containing three helices (Ogata et al. 1992, 1994, 1995), and the second and third helices form a helix-turn-helix (HTH) variation motif (Brennan and Matthews 1989; Harrison and Aggarwal 1990; Pabo and Sauer 1992). In a DNA complex of the minimal DNA-binding domain of c-Myb, R2R3, both R2 andR3 are closely packed in the major groove of DNA, recognizing a specific base sequence cooperatively (Ogata et al. 1994).
However, in a DNA complex of the DNA-binding domain of hTRF1, only the single Myb-like domain consisting of three helices can bind specifically to a double-stranded telomeric DNA with the sequence GTTAGGGTTAGGG (Fig. 1C ▶); the third helix recognizes the middle AGGG sequence in the major groove of DNA, and the N-terminal flexible arm interacts with the following TT sequence in the minor groove (Nishikawa et al. 2001). The homodimer of TRF1 binds not only to adjacent telomeric repeats but also to binding sites spaced far apart in an entirely independent manner (Bianchi et al. 1999), and to two different DNA molecules (Griffith et al. 1998). TRF2 also binds to double-stranded DNA. The DNA binding domain of hTRF2 (Thr445-Leu497) shows an ~59% sequence identity and 70% similarity to the corresponding region of hTRF1. Both hTRF1 and hTRF2, exhibiting different cellular functions, seem to hold a closely related DNA-binding mode. We previously reported the tertiary structures of the hTRF1 DNA-binding domain bound and unbound to a telomeric double-stranded DNA (Nishikawa et al. 2001). Here, we determined the tertiary structures of the hTRF2 DNA-binding domain bound and unbound to a telomeric double-stranded DNA with the same sequence of GTTAGGGT-TAGGG (Fig. 1C ▶), and we compared these structures with the corresponding structures of hTRF1. We observed small but significant structural differences between the DNA complexes of hTRF1 and hTRF2. In addition, we examined telomeric DNA-binding activities of both DNA-binding domains and found that hTRF1 binds more strongly than hTRF2. Based on the structural differences in the domains, the different DNA-binding abilities could be well explained. In addition, we created several mutants of the hTRF2 DNA-binding domain with stronger binding activities compared to the wild-type hTRF2.
Results
Structure determination
We determined the solution structures of the DNA binding domain of hTRF2 in its bound and unbound forms to the 13mer DNA duplex comprising the sequence GTTAGGGT-TAGGG (Fig. 1C ▶), with a conventional multidimensional heteronuclear NMR method using 15N- or 15N/13C-labeled protein. The DNA-binding domain, consisting of amino acids 438–500 of hTRF2, with a methionine residue at its N terminus was expressed in Escherichia coli BL21 (DE3), and the DNA duplex was prepared as reported previously (Nishikawa et al. 2001). For the DNA bound and unbound forms, the final sets of 20 and 25 structures were generated by simulated annealing, based on 1450 and 952 NMR-derived restraints, respectively, as summarized in Table 1.
Table 1.
Structural statistics for the 20 NMR structures of the DNA-bound form of the hTRF2 DNA binding domain and the 25 NMR structures of the DNA-free form
| Structural statistics for 20 structures of hTRF2 complex and 25 structures of hTRF2 | ||
| hTRF2 complex | HTRF2 | |
| Protein | ||
| Distance restraints | ||
| Intraresidue (i − j = 0) | 218 | 196 |
| Medium range (|i − j| < 5) | 573 | 511 |
| Long-range (|i − j| ≥ 5) | 231 | 203 |
| Total | 1022 | 910 |
| Dihedral angle restraints | 38 | 41 |
| DNA | ||
| Distance restraints | ||
| Intraresidue | 131 | |
| Sequential | 162 | |
| Interstrand | 4 | |
| Total | 297 | |
| Protein–DNA | 93 | |
| TOTAL | 1450 | |
| Statistic for structure calculations | <SA> | |
| R.m.s. deviations from experimental restraints | ||
| NOE(Å) | (4.70 ± 0.70) × 10−3 | (2.16 ± 0.04) × 10−3 |
| Dihedrals(deg.) | (1.93 ± 1.18) × 10−2 | (9.83 ± 3.46) × 10−2 |
| R.m.s. deviations from ideal restraints | ||
| Bonds(Å) | (1.10 ± 0.05) × 10−3 | (1.26 ± 0.03) × 10−3 |
| Angles(deg.) | (2.85 ± 0.04) × 10−1 | (4.58 ± 0.01) × 10−1 |
| Impropers(deg.) | (1.60 ± 0.07) × 10−1 | (3.35 ± 0.03) × 10−1 |
| Complex:residue 447–496 for protein, base-DNA 1–11(3′–13′) for DNA. | ||
| Free:residue 450–496 for protein. | ||
| R.m.s. deviations of atomic coordinates (Å) | backbone/all heavy atoms | |
| Protein | 0.43 ± 0.09/0.81 ± 0.08 | 0.34 ± 0.05/0.82 ± 0.06 |
| DNA | 0.43 ± 0.14/0.38 ± 0.13 | |
| Protein–DNA | 0.51 ± 0.11/0.68 ± 0.09 | |
| PROCHEK Ramachandran plot statistics (%) | ||
| Residues in most favored regions | 83.5 | 84.5 |
| Residues in additional allowed regions | 15.5 | 13.9 |
| Residues in generously allowed regions | 0.9 | 1.6 |
| Residues in disallowed regions | 0.0 | 0.0 |
Structure of the hTRF2 DNA-binding domain
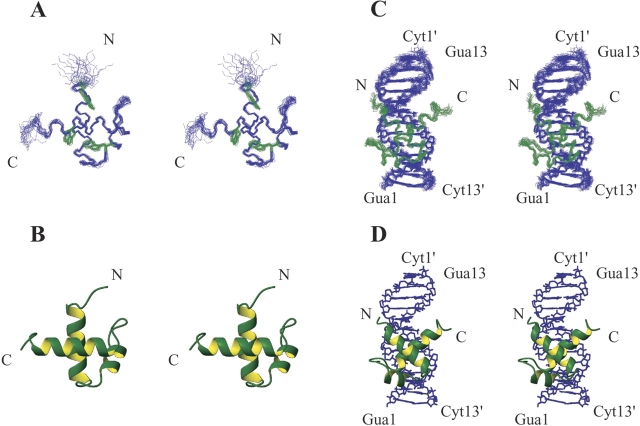
Figure 2A ▶ shows a stereo view of a superposition of the final 25 simulated annealing structures of the DNA unbound form, and Figure 2B ▶ shows a stereo view of the lowest-energy structure in the 25 structures. The overall root-mean-square deviation (rmsd) values between the 25 individual structures and an averaged structure are 0.34 ± 0.05 Å for backbone atoms and 0.82 ± 0.06 Å for all heavy atoms of amino acid residues Trp450–Arg496. The structures contain three helices, helix 1 from Val452 to Tyr465, helix 2 from Trp470 to Asn476, and helix 3 from Ala484 to Arg496 (helix3). These three helices are maintained by a hydrophobic core formed by Trp450, Val458, Val462, Trp470, Ile473, Phe479, Ile487, Trp491, and Met494, and also by two salt bridges formed by Glu454–Arg482 and Ser455–Arg490. The architecture of the three helices is a typical Myb motif found in the three repeats of the c-Myb DNA-binding domain as well as in the hTRF1 DNA-binding domain. The rmsd value of the 25 simulated annealing structures of the hTRF2 DNA-binding domain for the backbone atoms of amino acids Trp450–Arg496 against an averaged structure of the corresponding hTRF1 DNA-binding domain is 0.93 ± 0.07 Å.
Figure 2.
Structures of the DNA-binding domain of hTRF2. (A) Stereo view of the superposition of 25 NMR structures of the DNA-binding domain of hTRF2 in its DNA-free state with three tryptophan residues that form a hydrophobic core. The backbone atoms (N, Cα, C′) of the DNA-binding domain, amino acids Lys447–Asn500, are shown in blue, and three tryptophan residues are shown in green. (B) Stereo view of the lowest-energy structure of the 25 structures of the DNA-free form. (C) Stereo view of the superposition of 20 NMR structures of the DNA-binding domain bound to the 13mer DNA. The backbone atoms (N, Cα, C′) of the DNA-binding domain, amino acids Lys447–Asn500, are shown in green, and all heavy atoms of DNA are shown in blue. (D) Stereo view of the lowest-energy structure of the 25 structures of the hTRF2 complex with DNA.
Figure 2C ▶ shows a stereo view of a superposition of the simulated annealing 20 structures of the DNA-binding domain bound to the 13mer DNA duplex, and Figure 2D ▶ shows a stereo view of the lowest-energy structure in the 20 structures. The overall rmsd values between the 20 individual structures and an averaged structure are 0.51 ± 0.11 Å for the backbone atoms and 0.68 ± 0.09 Å for all heavy atoms of amino acid residues Lys447–Arg496 and DNA bases G1–G11 as well as the counterpart, C3′–C13′. The architecture of the DNA-binding domain in the DNA bound form was almost identical to the architecture of the DNA free form for amino acids Trp450–Arg496. However, upon the binding to DNA, like hTRF1 the N-terminal arm takes a rather rigid conformation, as shown by 15N-{1H} hetero-nuclear NOE values (Supplementary Fig. 1).
Telomeric DNA recognition of hTRF2
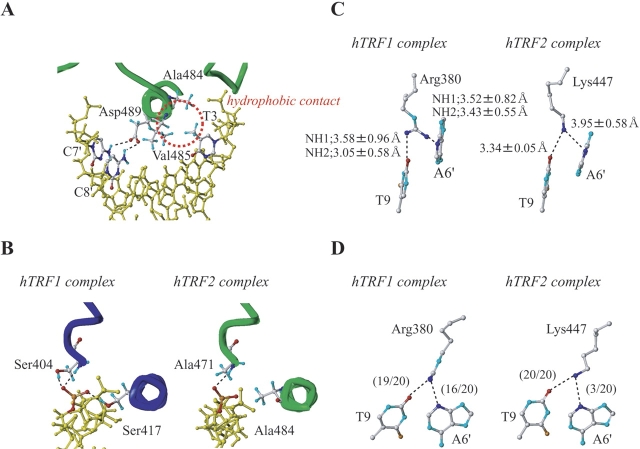
Figure 3A ▶ shows a summary of intermolecular contacts observed in the DNA complex of hTRF2. The third helix specifically recognizes the central T3A4G5G6G7 sequence in the major groove, and the N-terminal arm interacts with the following T8T9 region in the minor groove. Figure 4A ▶ shows that in the major groove, one of the methyl groups of Val485 and the methyl group of Ala484 make hydrophobic contacts with the methyl group of T3. G5 is recognized by Lys488. Asp489 probably interacts with both amino groups of C7′, the counterpart of G7, and C8′, the counterpart of G6 (Figs. 3B ▶, 4A ▶). The methyl groups of Met486, together with one of the methyl groups of Val485 and the methylene group of Asp489, forming a hydrophobic cluster, make contacts with bases of C7′ and C8′ and the backbone sugar of C7′. In the minor groove, Lys447 interacts with O2 of T9 (Fig. 4C ▶). In addition, the DNA-binding domain of hTRF2 makes a large number of nonspecific hydrophobic and electrostatic/hydrogen bonding interactions with backbone sugars and phosphate groups of DNA via Trp450, Trp470, Ala471, Lys488, Arg490, Arg492, and Thr493.
Figure 3.
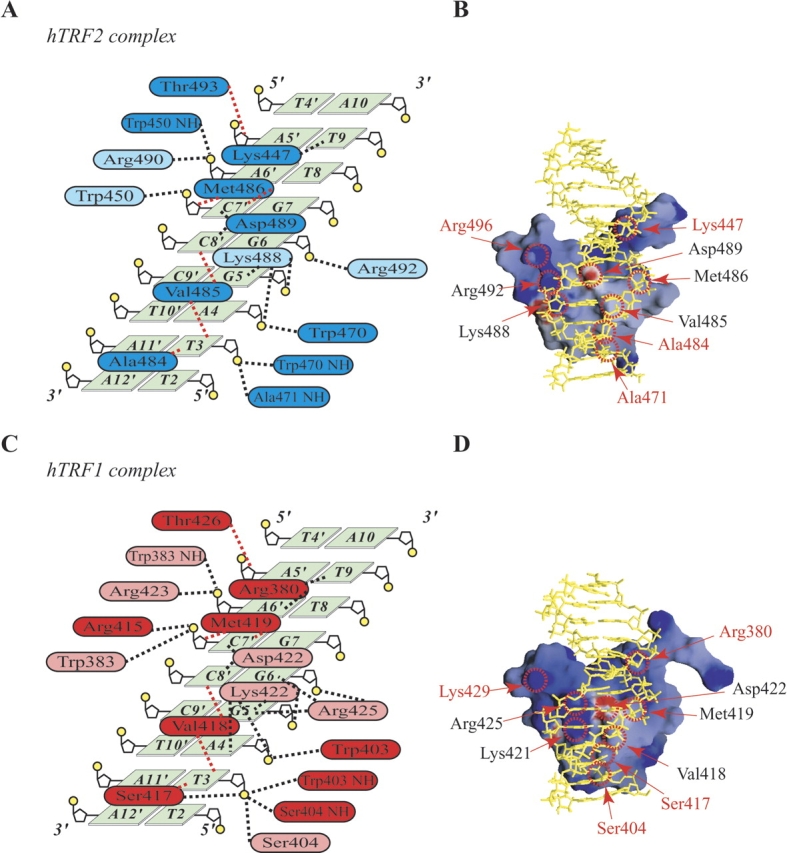
DNA recognition modes of DNA-binding domains of hTRF1 and hTRF2. (A) Summary of the intermolecular contacts observed in the hTRF2-DNA complex. Black broken lines indicate hydrophilic contacts, and red broken lines indicate hydrophobic contacts. For amino acids that interact with DNA, contact frequencies over 70% in the 20 NMR structures are shown in blue, and the frequencies between 20% and 70% are shown in light blue. The criteria of the hydrogen bond were set as N-H••D (O or N): N••D distance < 3.5 Å; N-H-D angle > 90°. (B) Electrostatics potential surface of the DNA-binding domain of hTRF2 bound to DNA. The amino acids colored in red differ between hTRF1 and hTRF2. (C) Summary of the intermolecular contacts observed in the hTRF1-DNA complex. Black broken lines indicate hydrophilic contacts, and red broken lines indicate hydrophobic contacts. For amino acids that interact with DNA, contact frequencies over 70% in the 20 NMR structures are shown in red, and the frequencies between 20% and 70% are shown in light red. The criteria of the hydrogen bond were set as N-H-D (O or N): N••D distance < 3.5 Å; N-H-D angle > 90°. (D) Electrostatics potential surface of the DNA-binding domain of hTRF1 bound to DNA. The amino acids colored in red differ between hTRF1 and hTRF2.
Figure 4.
Comparison of DNA recognition modes between hTRF1 and hTRF2. (A) DNA recognition of the DNA-binding domain of hTRF2. The red circle indicates hydrophobic contact containing methyl groups of Ala484, Val485, and T3. Black broken lines indicate hydrophilic contacts between Asp489 and C7′, C8′. (B) The interaction modes of Ser404/Ser417, Ala471/Ala484, and the phosphate group of T3. Black broken lines indicate hydrophilic contacts. (C) The interaction modes in the minor groove of DNA. Sidechains of Arg380 of hTRF1 and Lys447 of hTRF2, and DNA are shown. In hTRF1, the average distances over 20 structures between NH1 of Arg380 and O2 of T9, NH2 of Arg380 and O2 of T9, NH1 of Arg380 and N3 of A6′, and NH2 of Arg380 and N3 of A6′ are shown. In hTRF2, the average distances over 20 structures between NZ of Lys447 and O2 of T9, and NZ of Lys447 and N3 of A6′ are shown. (D) The number of hydrogen bonds in the determined structures of the hTRF1 complex and the hTRF2 complex, respectively. The criteria of the hydrogen bonds were set as N-H••D (O or N): N••D distance < 3.5 Å; N-H-D angle > 90°.
Structural comparison of the DNA complexes of hTRF2 and hTRF1
By comparing amino acid sequences of DNA-binding domains, consisting of 438–500 of hTRF2 and the corresponding sequence 371–433 of hTRF1, we found that 31 of 63 amino acids are identical; the two DNA-binding domains have very similar tertiary structures in their DNA bound and unbound forms. Among the identical 31 amino acids, the nine amino acids (Trp450, Trp470, Val485, Met486, Lys488 Asp489, Arg490, Arg492, and Thr493) of hTRF2 are responsible for the DNA binding of hTRF2; similarly in the DNA complex of the DNA-binding domain of hTRF1, the corresponding nine amino acids (Trp383, Trp403, Val418, Met419, Lys421, Asp422, Arg423, Arg425, and Thr426) participate in the interactions with DNA, as shown in Figure 3C ▶. Thus, the binding modes as well as the architectures of DNA complexes of the DNA-binding domains are very close to each other. The rmsd values between the averaged structures are 1.12 Å for backbone atoms of amino acids Lys447–Arg496 of hTRF2 and amino acids Arg380–Lys429 of hTRF1, 1.87 Å for heavy atoms of bases of G1–G11 and C3′–C13′ of DNA, and 1.62 Å for heavy atoms of both protein and DNA backbones.
By clarifying DNA interaction modes of both domains in detail, we found that four amino acids responsible for the DNA binding are different from each other: Lys447, Ala471, Ala484, and Arg496 in hTRF2 and the corresponding amino acids Arg380, Ser404, Ser417, and Lys429 in hTRF1. In the minor groove interaction by the flexible arm of hTRF2, Lys447 contacts only with T9; however, in the hTRF1 complex Arg380 likely interacts with N3 of A6′, the counterpart of T8, as well as O2 of T9. Figure 4C ▶ shows that in the hTRF2 complex the averaged distances over 20 calculated structures from the amide nitrogen of Lys447 to O2 of T9 and N3 of A6 are 3.34 ± 0.05 Å and 3.95 ± 0.58 Å, respectively. In the 20 calculated structures of the hTRF2 complex, for the distances between the atoms of Lys447 and N3 of A6′, only three structures out of 20 meet the hydrogen bonding criteria that we set as N••N distance < 3.5 Å, N-H-N angle > 90°. In contrast, Figure 4C ▶ shows that in the 20 calculated structures of the hTRF1 complex, the averaged distances between NH1 of Arg380 and O2 of T9, NH2 of Arg380 and O2 of T9, NH1 of Arg380 and N3 of A6′, and NH2 of the Arg380 and N3 of A6′ are 3.58 ± 0.96 Å, 3.05 ± 0.58 Å, 3.52 ± 0.82 Å and 3.43 ± 0.55 Å, respectively. Figure 4D ▶ shows that in most structures of the hTRF1 complex, Arg380 contacts both T9 and A6′ in the minor groove. However, in the hTRF2 complex, Lys447 interacts mainly with T9 and rarely with A6′. It is likely that hTRF1 interacts more strongly to the TT portion in the AGGGTT sequence than hTRF2.
In the hTRF2 complex, the amide hydrogen of Ala471 contacts with the phosphate group of T3, and the methyl group of Ala484, together with one of the methyl groups of Val485, makes hydrophobic contacts with the methyl group of T3 (Fig. 4A ▶, B). Similarly, in the hTRF1 complex, the amide hydrogen of Ser404 contacts the phosphate group of T3, and the methylene group of Ser417, together with one of the methyl groups of Val418, makes hydrophobic contacts with the methyl group of T3, at the same time, the hydroxyl group of Ser404 together with the hydroxyl group of Ser 417 contacts the phosphate group of T3, as shown in Figure 4B ▶. This suggests that for the recognition of the phosphate group of T3, hTRF1 has slightly stronger activity than hTRF2. Although in the hTRF1 complex Lys429 contacts with the phosphate group of T4′, the counterpart of A10, in the complex of hTRF2 the corresponding amino acid Arg496 is not likely to interact with the phosphate group of T4′. In this sense also hTRF1 seems to bind more strongly than hTRF2.
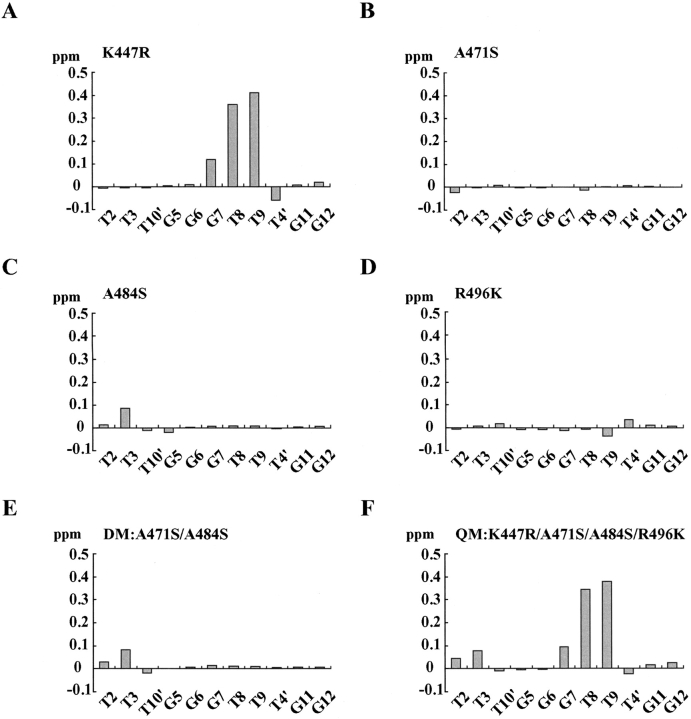
To check the contributions of the four amino acids described above for the DNA-binding activities of hTRF2 and hTRF1, we created six mutants of hTRF2 in which four critical amino acids (Lys447, Ala471, Ala484, and Arg496) are changed into the corresponding amino acids of hTRF1: four single mutants, K447R, A471S, A484S, and R496K, in which Lys447, Ala471, Ala484, and Arg496 are replaced by arginine, serine, serine, and lysine residues, respectively; one double mutant (DM), A471S/A484S, in which both Ala471 and Ala481 are substituted by serine residues; and one quadruple mutant, K447R/A471S/A484S/R496K, designated as QM, in which all four amino acids (Lys447, Ala471, Ala484, and Arg496) are replaced by the corresponding amino acids of hTRF1 simultaneously. One-dimensional (1D) NMR spectra of all of these mutants and the HSQC spectrum of QM suggest that the whole structures of all of the mutants are essentially identical to the wild-type structure of hTRF2 (Suppl. Figs. 2, 3).
The imino proton signal changes of the telomeric DNA bound to the hTRF2 DNA-binding domain and the mutants
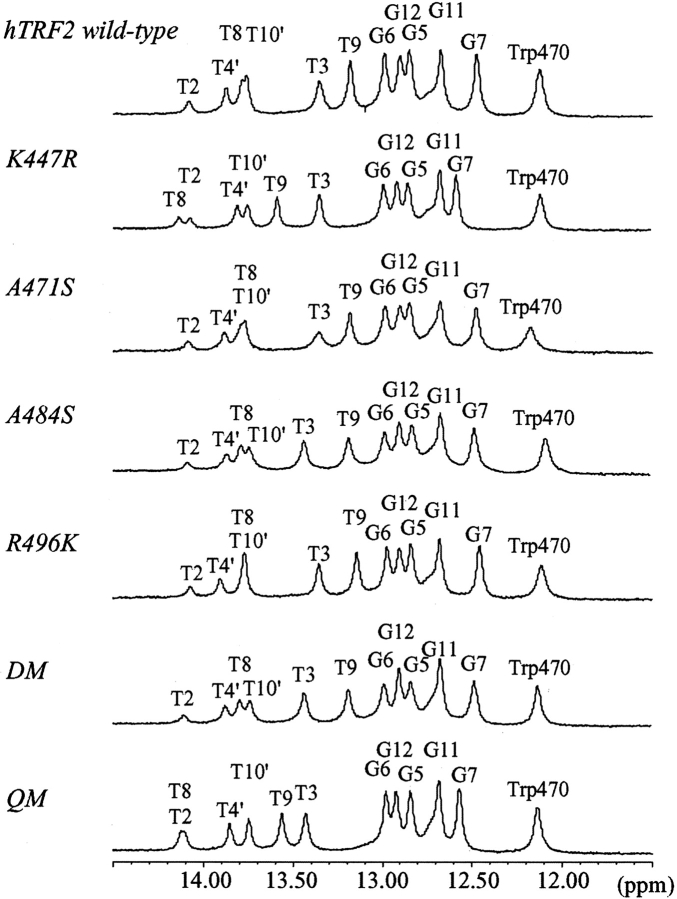
First we examined the DNA-binding modes of these mutants by 1D NMR. Figure 5 ▶ shows 1D NMR spectra of the imino proton signals of the 13mer DNA complexed with the wild type and the six mutants of hTRF2. The chemical shift changes of the imino proton signals of each mutant from the wild type are summarized in Figure 6 ▶. In K447R, the big chemical shift changes were observed for the imino protons of G7, T8, T9, and T4′, the counterpart of A10. This suggests that as found in the hTRF1 complex, the substituted arginine residue at 447 in the mutant of hTRF2 could interact with N3 of A6′ (the counterpart of T8) as well as O2 of T9 more strongly than the lysine residue of the wild-type hTRF2. In the spectra of A471S and A484S, small but significant chemical shift changes were observed for the imino protons of T2 and T3, respectively, and A471S/A484S shows both significant changes together with the imino proton shift change of T10′, the counterpart of A4. These observations might be related to the fact that the corresponding two serine residues in hTRF1 contact with the phosphate group of T3. In R496K, small chemical shift changes were observed in the signals of T9 and T4′ (the counterpart of A10), suggesting that the substituted lysine residue at 496 of hTRF2 contacts with the phosphate group of T4′ like hTRF1, while the arginine residue at 496 in the wild type is not likely to interact with the phosphate group of T4′. In the spectrum of QM, K447R/A471S/A484S/R496K, significant chemical shift changes of the imino protons were observed in T2, T3, G7, T8, and T9 in an additive manner of the chemical shift changes of each mutant, suggesting that the substituted four amino acids interact with DNA independently.
Figure 5.
NMR titration experiments of the wild-type, K447R, A471S, A484S, R496K, DM (A471S/A484S), and QM (K447R/A471S/A484S/R496K) to the telomeric double-stranded DNA with the sequence GT-TAGGGTTAGGG.
Figure 6.
The chemical shift changes of the imino proton signals of each mutant from the corresponding signals of the wild-type hTRF2. (A) K447R, (B) A471S, (C) A484S, (D) R496K, (E) DM (A471S/A484S), (F) QM (K447R/A471S/A484S/R496K).
The DNA-binding activities of hTRF1, hTRF2, and the mutants of hTRF2
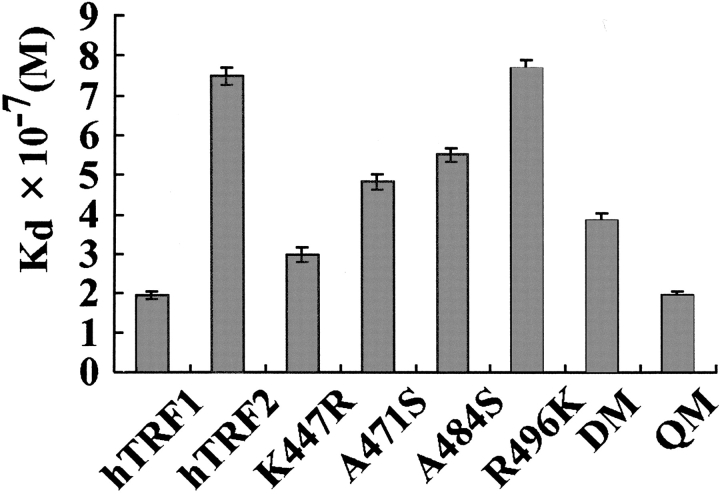
The binding activities of hTRF1, hTRF2, and the six mutants of hTRF2 with the 13mer telomeric double-stranded DNA 5′-GTTAGGGTTAGGG were examined using a surface plasmon resonance (SPR) apparatus, BIACORE. As expected, hTRF1 binds to DNA more strongly than hTRF2; in 10 mM HEPES-KOH, 3 mM EDTA, 180 mM KCl, and 0.003% Triton X-100 (v/v) (pH 6.8), the equilibrium dissociation constant Kd values are 2.0 × 10−7 M for the hTRF1 DNA-binding domain and 7.5 × 10−7 M for the hTRF2 DNA-binding domain; the binding ability of hTRF1 is about four times stronger than the ability of hTRF2 (Fig. 7 ▶). At the salt concentration of 150 mM KCl instead of 180 mM KCl in the otherwise same buffer, the equilibrium dissociation constant Kd values were measured for the wild type and the six mutants of hTRF2; for example, 1.8 × 10−7 M for the wild type and 2.4 × 10−8 M for QM (Suppl. Fig. 4). However, for the hTRF1 DNA-binding domain, a reliable Kd value could not be obtained, perhaps because of its stronger binding ability compared to QM in our present measuring system. Hereafter we therefore use the Kd values at the high salt concentration of 180 mM KCl, in which the Kd values were obtained with high qualities for all samples examined (Suppl. Fig. 5).
Figure 7.
The results of SPR analyses for equilibrium dissociation constant (Kd) values between DNA binding domains (TRF1, TRF2, K447R, A471S, A484S, R496K, DM, and QM) and the 13mer telomeric DNA in the buffer containing 10 mM HEPES-KOH, 3 mM EDTA, 180 mM KCl, and 0.003% Triton X-100 (v/v) (pH 6.8).
For the single mutants of hTRF2, Kd values were obtained as 3.0 × 10−7 M for K447R, 4.8 × 10−7 M for A471S, 5.5 × 10−7 M for A484S, and 7.6 × 10−7 M for R496K. Almost all mutants except R496K have stronger DNA-binding activities compared to the wild type; in particular, the mutant K447R binds to DNA 2.5 times stronger than the wild-type hTRF2. This suggests that the substituted arginine residue in the mutant could interact with N3 of A6′ (the counterpart of T8) as well as O2 of T9 in the minor groove, like hTRF1, while Lys447 of the wild type contacts T9 alone. This is well correlated to our observation of large chemical shift changes of the imino protons of T8 and T9 between the wild-type hTRF2 and K447R. As suggested by the small chemical shift changes of A471S and A484S from the wild type, both mutants bind slightly stronger than the wild type. The mutant DM (A471S/A484S) binds stronger to DNA than both single mutants in an additive manner: The Kd values are for DM, 3.9 × 10−7 M, 0.52 times smaller; for A471S, 4.8 × 10−7 M, 0.64 times smaller; and for A484S, 5.5 × 10−7 M, 0.73 times smaller than the wild-type value. Serine residues at 471 and 484 seem to bind to the backbone phosphate group independently, like hTRF1. Although R496K showed small chemical shift changes, its binding ability is very close to the wild-type ability; for amino acid 496 of hTRF2, the lysine and arginine residues seem to play a similar role in the interaction with DNA. As shown in Figure 3B,D ▶, just a positive charge at 496 of hTRF2 or at 429 of hTRF1 seems to be responsible for the interaction with phosphate backbone.
The mutant QM (K447R/A471S/A484S/R496K) binds to DNA more strongly than the wild-type hTRF2, with a binding ability similar to that of the hTRF1 DNA-binding domain; the Kd values are almost identical at 2.0 × 10−7 M. The Kd value of QM, 0.27 times smaller than the value of the wild-type hTRF2, is well explained by both Kd values: for K447R 3.0 × 10−7 M, 0.40 times smaller; for A471S/A484S, 3.9 × 10−7 M, 0.52 times smaller than the wild-type value. Arg447, Ser484, and Ser496 in QM likely interact with DNA independently. This may lead to the conclusion that only three amino acids, Lys447, Ala471, and Ala484 of hTRF2 and the corresponding amino acids, Arg380, Ser404, and Ser417 of hTRF1 are critical amino acids that clarify the DNA-binding activities of hTRF2 and hTRF1, because the substitution of Arg496 of hTRF2 to a lysine residue does not affect the DNA-binding ability of hTRF2.
Discussion
The DNA binding domains of hTRF1 and hTRF2 hold a very similar amino acid sequence closely related to the Myb domain found in the DNA-binding domain of c-Myb. We determined the solution structures of the DNA-binding domain of hTRF2 bound and unbound to DNA with the sequence GTTAGGGTTAGGG, and we compared their structures with the corresponding structures of hTRF1. We found that hTRF2 as well as hTRF1 recognize specifically the central AGGGTT sequence. The amino acid residues that are responsible for this recognition by hTRF2 are nearly conserved in hTRF1; however, by using a surface plasmon apparatus, we found that hTRF2 binds about four times more weakly to the telomeric DNA compared to hTRF1. To reveal the reason for the weaker binding activity of hTRF2, we examined the structures of both DNA-binding domains bound to the telomeric DNA in detail. In the DNA complex of hTRF2, Lys447 contacts T9 in the minor groove of DNA; however, in the DNA complex of hTRF1, the corresponding amino acid (Arg380) contacts T9 and also A6′ (the counterpart of T8) in the minor groove. By creating a mutant of hTRF2 in which Lys447 is substituted to an arginine residue, we have established that the difference between the lysine and arginine residues is the main contribution to the weaker binding activity of hTRF2 to the telomeric DNA compared to hTRF1. In addition, hTRF1 was found to have hydrogen bonds to a phosphate group by two serine residues at 404 and 417, which correspond to Ala471 and Ala484 of hTRF2. The double mutant in which both alanine residues at 471 and 484 of hTRF2 are replaced by serine residues has a stronger binding activity than the wild-type hTRF2. We conclude that the amino acids Arg380, Ser404, and Ser417 are critical players for the stronger binding activity of hTRF1 compared to hTRF2. Actually, a mutant in which Lys447, Ala471, and Ala484 of hTRF2 are replaced by arginine, serine, and serine residues has a binding activity similar to that of hTRF1.
In mammals, the looping structure called t-loop is formed in telomeres. T-loop is a large duplex telomeric loop that appears to be formed by the invasion of the 3′ overhang into double-stranded DNA. Electron microscopic studies showed that higher-order structure of telomeric DNA is effectively altered by TRF1 (Griffith et al. 1998; Bianchi et al. 1999). The alterations include pairing the two DNA duplex strands or looping linear strand by putting two distanced recognition half-sites close together. Since hTRF1 selects the two recognition sites with extreme spatial flexibility (Bianchi et al. 1999), it may greatly help to form and maintain the loop structure (Griffith et al. 1999). A model of the t-loop formation by TRF2 has been suggested in which the initiating step is the assembly of a TRF2 complex at the single-stranded (ss)/double-stranded (ds) telomeric junction either by direct binding or by sliding from an internal site. Next, the junction-bound TRF2 complex may fold backward to bind an internal site on the telomeric dsDNA to form a loop. Alternatively, TRF2 complex assembled at an internal site may interact with the end-bound complex to form a loop (Stansel et al. 2001). The present results show that hTRF2 binds to telomeric dsDNA via a similar binding mode but with a lower binding affinity compared to hTRF1. The different binding activities of hTRF1 and hTRF2 might correspond to their different roles in the formation of telomere structures. However, further studies are necessary to determine the detailed functional roles of hTRF1 and hTRF2 in the formation of telomeres.
In the telomeres of budding yeast, a telomeric protein, Rap1p, was found to recognize telomeric DNA (Shore 1994; Krauskopf and Blackburn 1996). The tertiary structure of the DNA-binding domain of scRap1p bound to a 13mer DNA with the sequence GGTGTGTGGGTGT was determined. It contains two subdomains, both closely related to the Myb domain (König et al. 1996). Each subdomain contacts a separated GGTGT sequence in a similar manner. The binding modes of scRap1p, hTRF1, and hTRF2 are very similar to each other; scRap1p holds two Myb-related subdomains, both of which recognize GGTGT sequence separately, and hTRF1 and hTRF2, both holding a single Myb domain that can recognize AGGGTT sequence, form a homodimer in telomeres. Although each subdomain of scRap1p alone has no specific DNA-binding ability at all, only the single Myb domain from hTRF2 as well as hTRF1 has a specific DNA-binding ability (Biaud et al. 1996). Thus the formation of homodimer is not necessary for specific bindings of hTRF1 and hTRF2, but at least for hTRF1 two Myb domains in the homodimer can recognize separate AGGGTT sequences in an entirely independent manner, forming a loop consisting of the sequence between the two recognition sites. This means that each Myb domain in the TRF1 homodimer connected by a flexible linker to the TRFH domain acts as an independent specific DNA-binding domain for the formation of tertiary structures of telomeres. Compared to the Myb domain of hTRF1, the hTRF2 Myb domain is found to have a weaker DNA-binding activity, which might be responsible for the flexible movement of hTRF2 upon the formation of tertiary structures of telomeres. Further studies are necessary to confirm the exact structural roles of TRF2 in the telomeres.
Materials and methods
Sample preparation
The human TRF2 DNA binding domain and its six mutants, consisting of amino acids 438–500 with a methionine residue at the N terminus were overexpressed in E. coli strain BL21(DE3) (Novagen) with the use of a pET23b vector. The cells were grown at 37°C, and when OD600 was reached to 0.5–0.6, 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) was added to induce the protein expression at 25°C. After an additional 9–12 h growth, the cells were harvested and resuspended in buffer (50 mM potassium phosphate buffer [pH 7.0], 5 mM EDTA, 100 mM NaCl). For isotope labeling, M9 minimal media containing 15NH4Cl (0.15%) and/or [13C]-glucose (0.2%) were used for culture. The cells were lysed by sonication on ice and then centrifuged (39,000g). The supernatant was subjected to the preceding purification. The DNA-binding domain was purified by ion exchange (P11; Whatman) and gel filtration (Superdex 75; Pharmacia) column chromatography. The identification and purity of the sample were assessed with MALDI-TOF mass spectroscopy and electrophoresis.
Each strand of a 13mer duplex DNA with the sequence 5′-GTTAGGGTTAGGG-3′ was purchased from Bex Co. Both strands were annealed by cooling slowly from 95°C in 50 mM potassium phosphate (pH 7.0), 1 mM EDTA, 150 mM KCl. The homogeneity of the sample was assessed by gel filtration.
The DNA complex of the DNA-binding domain was formed by gradual addition of the protein into the DNA solution until the equal molar ratio was reached. To avoid aggregation, we dissolved both protein and DNA in 50 mM potassium phosphate (pH 7.0) with 150 mM KCl, and the sample concentration was set lower than 1 mM before mixing. The solution was then concentrated, and the salt strength was diluted into 5 mM potassium phosphate using a Centricon (Amicon), with a 3-kDa cutoff membrane for the NMR experiments.
NMR spectroscopy
The DNA-free form of the DNA-binding domain was measured in 20 mM potassium phosphate buffer (pH 6.8), and the DNA bound form was in 5 mM potassium phosphate buffer (pH 6.9) in 10% (v/v) or 100% D2O. The protein concentrations of both samples were 1.0–1.5 mM. NMR experiments were carried out at 300 K for the DNA-free form and 303 K for the DNA-bound form on a Bruker DMX-600 and AVANCE-800 equipped with a triple-resonance gradient probe. Protein backbone resonance assignments were obtained from 3D HN(CO)CA, 3D HNCA, 3D HNCO (Grzesiek and Bax 1992a), 3D HN(CA)CO, 3D CBCANH, and 3D CBCA(CO)NH (Grzesiek et al. 1992b). Protein side chain resonance assignments were obtained from 3D HBHA(CO)NH (Grzesiek et al. 1992b), 3D HCCH-TOCSY (Kay et al. 1993), 3D HCCH-COSY, 3D 15N-edited NOESY, and 3D 15N-edite TOCSY experiments. 3JHNα coupling constants for backbone dihedral angle ϕ restraints were measured using 3D HNHA (Vuister and Bax 1993). DNA resonance assignments and intramolecular distance restraints were obtained from 2D NOESY, 2D TOCSY, and 2D DQF-COSY with a 13C or 13C/15N-filtered pulse scheme (Ogura et al. 1996). Intermolecular distance restraints were obtained in a 3D 13C-edited(F1), 13C-filtered(F3) NOESY experiment and a 3D 15N-edited(F2), 15N/13C-filtered(F3) NOESY experiment (Ogura et al. 1996). All NMR spectra were processed and analyzed using NMRPipe (Delaglio et al. 1995) and PIPP (Garret et al. 1991) software.
In the NMR titration experiments, the DNA-binding domain and six mutants were successively added into the 13mer telomeric DNA duplexes in increments of 0.25 molar equivalents. The concentrations of DNA were ~0.5 mM in 10 mM potassium phosphate buffer (pH 6.9) and 100 mM KCl. The chemical-shift changes of base-paired imino protons were monitored by 1D 1H NMR spectra at 293 K.
Structure calculations
Interproton distance constraints for the hTRF2 DNA-binding domain were derived from the cross-peak intensities of the NOESY spectra. NOEs were classified into four distance ranges; 1.8–3.0, 2.3–4.0, 2.3–5.0, and 2.3–6.0 Å, corresponding to strong, medium, weak, and very weak NOEs, respectively. In addition, torsion angle restraints were derived from the 3JHNα coupling constants. The restraint angle ranges were −90° < ϕ < −40° for 3JHNα < 5.5Hz and −160° < ϕ < −80° for 3JHNα > 8.5Hz. The intra-DNA NOEs were classified into four distance ranges, 1.8–3.0, 2.3–4.0, 2.3–5.0, and 2.3–6.0Å, corresponding to strong, medium, weak, and very weak NOEs, respectively. Pseudo-atom correction was applied to the upper limit. Hydrogen bond restraints within the DNA were used to maintain the base pairs. Watson-Crick base pairing was maintained in the DNA by the following hydrogen bond restraints: for GC base pair, rG(N1)–C(N3) = 2.95 ± 0.2 Å, rG(N2)–C(O2) = 2.86 ± 0.2 Å, and rG(O6)–C(N4) = 2.91 ± 0.2 Å and for TA base pair, rA(N6)–T(O4) = 2.95 ± 0.2 Å, and rA(N1)–T(N3) = 2.82 ± 0.2 Å. Loose torsion angle restraints for the DNA were used to alleviate problems associated with mirror images, covering both A- and B-form DNA conformers (α = −65° ± 50°, β = 180° ± 50°, γ = 60° ± 50°, ɛ = 180° ± 50°, and ζ = −85° ± 50°) (Omichinski et al. 1997). These hydrogen bond and torsion angle restraints for the DNA are justified, because the pattern of NOEs for the DNA is typical of B-DNA.
Initially, 200 structures of the only protein were calculated with simulated annealing protocols, using a Crystallography and NMR System (CNS; Yale University). Secondly, the structures of the hTRF2-DNA complex were calculated with simulated annealing protocols, starting from the 200 structures of the hTRF2 DNA-binding domain and B-form DNA. B-form DNA was placed 50 Å away from the protein in various orientations. In total, 200 structures of the hTRF2-DNA complex were calculated. Of these, 52 structures showed neither violation greater than 0.3 Å for the distance constraints nor 5° for the dihedral restraints. Finally, the 20 structures with the lowest energy were selected. All figures of molecular structures were drawn with SYBYL, MOLMOL, or GRASP.
Surface plasmon resonance analyses
DNA-binding activities of the DNA-binding domains of hTRF1 and hTRF2 as well as the six mutants were analyzed using a Biacore 3000 instrument. All of the experiments were performed at 293 K using a buffer containing 10 mM HEPES-KOH, 3 mM EDTA, 180 mM KCl, and 0.003% Triton X-100 (v/v) (pH 6.8). Flow cells of an SA streptavidin sensor chip were coated by bio-tinylated 13mer DNA with a sequence of GTTAGGGTTAGGG. The proteins were injected over flow cells for 3–5 min using a flow rate of 10μL/min until the reaction of protein with DNA had equilibrated. Bound proteins were removed with a 30-sec wash with 2M KCl. An equilibrium dissociate constant (Kd) was calculated from a Scatchard analysis of the RU values in the equilibrium region of the sensorgram at each analyte concentration. The affinity data were analyzed using BIAevaluation 3.2 software.
Protein Data Bank accession numbers
The PDB ID codes of the structures of the hTRF2 DNA-binding domain in DNA-free and bound states are 1VF9 and 1VFC, respectively.
Electronic supplemental material
Supplementary Figure 1 shows 15N-{1H} heteronuclear NOE values. Figure 2 ▶ shows HSQC spectra of the wild-type hTRF2 and QM. Figure 3 ▶ shows 1D NMR spectra of the wild-type hTRF2 and the six mutants. Figure 4 ▶ shows Kd values at 150 mM KCl for the wild-type hTRF2 and the six mutants. Figure 5 ▶ shows the error bars of Kd values at 180 mM KCl for hTRF1 and the wild-type and six mutants of hTRF2.
Acknowledgments
This work was supported by a Collaborative of Regional Entities for the Advancement of Technological Excellence (CREATE) in Yokohama from JST, and a Project of Protein 3000, Transcription and Translation, and Grants in Aid for Scientific Research from MEXT. The plasmid of the full length of hTRF2 was a kind gift from Dr. Titia de Lange at The Rockefeller University.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04983705.
Supplemental material: see www.proteinscience.org
References
- Bianchi, A., Smith, S., Chong, L., Elias, P., and de Lange, T. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, A.M., Stansel, R.M., Fairall, L.D., Griffith, J.D., Rhodes, D., and de Lange, T. 1999. TRF1binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biaud, T., Koerig, C.E., Binet-Brasselet, E., Ancelin, K., Pollice, A., Gasser, S.M., and Gilson, E. 1996. The telobox, a Myb-related telomereic DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 24 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkapp, H., Borgmeyer, U., Sippel, A.E., and Klempnauer, K.H. 1988. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335 835–837. [DOI] [PubMed] [Google Scholar]
- Bilaud, T., Brun, C., Ancelin, K., Koering, C.E., Laroche, T., and Gilson, E. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17 236–239. [DOI] [PubMed] [Google Scholar]
- Brennan, R.G. and Matthews, B.W. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264 1903–1906. [PubMed] [Google Scholar]
- Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17 231–235. [DOI] [PubMed] [Google Scholar]
- Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. 1995. A human telomeric protein. Science 270 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cook, D.B., Dynet, J.N., Chang, W., Shostak, G., and Smith, S. 2002. Role for the related polyADP-Ribose polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell Biol. 22 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Fairall, L., Chapman, L., Muss, H., de Lange, T., and Rhodes, D. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8 351–361. [DOI] [PubMed] [Google Scholar]
- Garret, D.S., Powers, R., Gronenborm, A.M., and Clore, G.M. 1991. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 99 214–220. [DOI] [PubMed] [Google Scholar]
- Gonda, T.J., Gough, N.M., Dunn, A.R., and de Blaquiere, J. 1985. Nucleotide sequence of cDNA clones of the murine myb proto-oncogene. EMBO J. 4 2003–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J., Bianchi, A, and de Lange, T. 1998. TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 278 79–88. [DOI] [PubMed] [Google Scholar]
- Griffith, J.D., Comenau, L., Rosenfield, S., Stansel, R.M., Bianch, A., Moss, H., and de Lange, T. 1999. Mammalian telomeres end in a large duplex loop. Cell 97 503–514. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax, A. 1992a. Improved three-dimensional triple resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson. 96 432–440. [Google Scholar]
- ———. 1992b. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114 6291–6293. [Google Scholar]
- Harrison, S.C. and Aggarwal, A.K. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59 933–969. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283 1321–1325. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Smogorzewska, A., and de Lange, T. 2002. Senesecence induced by altered telomere state, not telomere loss. Science 295 2446–2449. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Xu, G.-Y., Singer, A.U., Muhandiram, D.R., and Forman-Kay, J.D. 1993. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O sample of protein. J. Magn. Reson. B. 101 333–337. [Google Scholar]
- Kim, S.H., Kaminker, P., and Campisi, J. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer, K.H. and Sippel, A.E. 1987. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 6 2719–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König, P., Giraldo, R., Chapman, L., and Rhodes, D. 1996. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85 125–136. [DOI] [PubMed] [Google Scholar]
- Krauskopf, A. and Blackburn, E.H. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383 354–357. [DOI] [PubMed] [Google Scholar]
- Li, B., Oestreich, S., and de Lange, T. 2000. Identification of human Rap1: Implications for telomere evolution. Cell 101 471–483. [DOI] [PubMed] [Google Scholar]
- Loayaza, D. and de Lange, T. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 424 1013–1018. [DOI] [PubMed] [Google Scholar]
- Nakagoshi, H., Ngase, T., Kanei-Ishii, C., Ueno, Y., and Ishii, S. 1990. Binding of the c-myb proto-oncogene product to the simian virus 40 enhancer stimulates transcription. J. Biol. Chem. 265 3479–3483. [PubMed] [Google Scholar]
- Ness, S.A., Marknell, A., and Graf, T. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59 1115–1125. [DOI] [PubMed] [Google Scholar]
- Nishikawa, T., Okamura, H., Nagadoi, A., König, P., Rhodes, D., and Nishimura, Y. 2001. Solution structure of a telomere DNA complex of human TRF1. Structure 9 1237–1251. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Hojo, H., Aimoto, S., Naka, T., Nakamura, H., Sarai, A., Ishii, S. and Nishimura, Y. 1992. Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl Acad. Sci. 89 6428–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Srkikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S. and Nishimura, Y., et al. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79 639–648. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Hojo, H., Yoshimura, S., Zhang, R., Aimoto, S., Ametani, Y., Hirata, Z., and Nishimura, Y. 1995. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat. Struct. Biol. 2 309–320. [DOI] [PubMed] [Google Scholar]
- Ogura, K., Terasawa, H., and Inagaki, F. 1996. An improved double-tuned and isotope-filtered pulse scheme based on a pulse field gradient and a wideband inversion shaped pulse. J. Biomol. NMR 8 492–498. [DOI] [PubMed] [Google Scholar]
- Omichinski, J.G., Pedone, P.V., Felsenfeld, G., Gronenborn, A.M., and Clore, G.M. 1997. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat. Struct. Biol. 4 122–132. [DOI] [PubMed] [Google Scholar]
- Pabo, C.O, and Sauer, R.T. 1992. Transcription factors: Structural families and principles of DNA recognition. Annu. Rev. Biochem. 61 1053–1095. [DOI] [PubMed] [Google Scholar]
- Shore, D. 1994. RAP1: A protean regulator in yeast. Trends Genet. 10 408–412. [DOI] [PubMed] [Google Scholar]
- Smith, S. and de Lange, T. 1997. TRF1, a mammalian telomeric protein. Trends Genet. 13 21–26. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10 1299–1302. [DOI] [PubMed] [Google Scholar]
- Smith, S., Giriat, I., Schmitt, A., and de Lange, T. 1998. Tankyrase, a polyADP-ribose polymerase at human telomeres. Science 282 1484–1487. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A. and de Lange, T. 2002. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21 4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 20 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel, R.M., de Lange., T, and Griffith, J.D. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20 E5532–E5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa, J., Yasukawa, T., Enari, M., Ogata, K., Nishimura, Y., Ishii, S., and Sarai, A. 1993. Recognition of specific DNA sequence by the c-myb proto-oncogene product: Role of three repeat units in the DNA-binding domain. Proc. Natl Acad. Sci. 90 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B. and de Lange, T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Smogorzewska, A., and de Lange, T. 1998. TRF2 protects human telomeres from end-to-end fusion. Cell 92 401–413. [DOI] [PubMed] [Google Scholar]
- Vuister, G.W. and Bax, A. 1993. Quantitative J. correlation: A new approach for measuring homonuclear three bond J-HN-Hα coupling constants in 15N-enriched protein. J. Am. Chem. Soc. 115 7772–7777. [Google Scholar]
- Weston, K. and Bishop, J.M. 1989. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-Myb. Cell 58 85–93. [DOI] [PubMed] [Google Scholar]
- Zhou, X.Z. and Lu, K.P. 2001. The PIN2/TRF1-interacting protein PINX1 is a potent telomerase inhibitor. Cell 107 347–359. [DOI] [PubMed] [Google Scholar]
- Zhu, X.D., Kuster, B., Mann, M., Petrini, J.H., and de Lange, T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NSB1 with TRF2 and human telomeres. Nat. Genet. 25 347–352. [DOI] [PubMed] [Google Scholar]