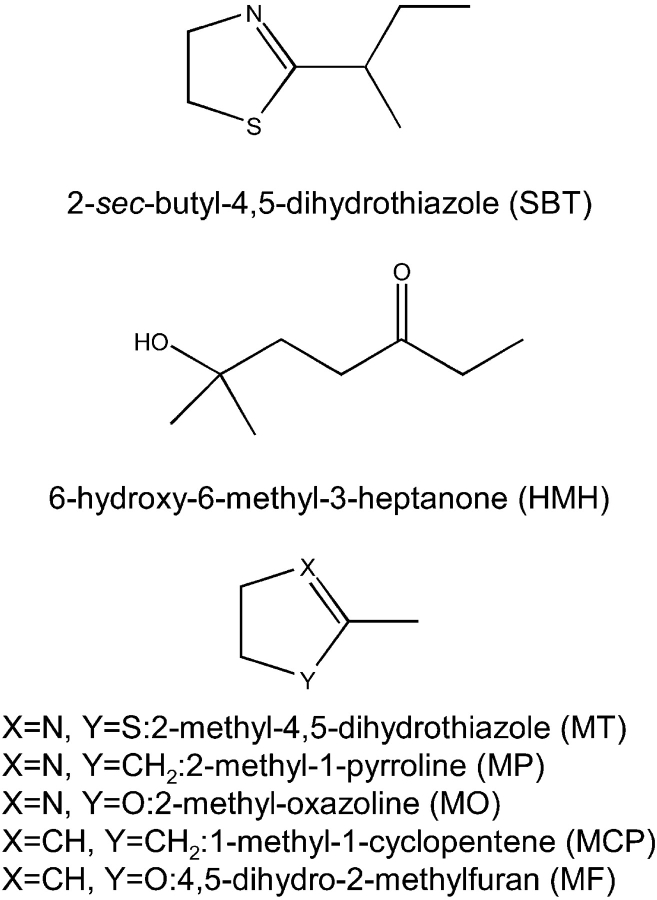
Abstract
The mouse pheromones (±)-2-sec-butyl-4,5-dihydrothiazole (SBT) and 6-hydroxy-6-methyl-3-heptanone (HMH) bind into an occluded hydrophobic cavity in the mouse major urinary protein (MUP-1). Although the ligands are structurally unrelated, in both cases binding is accompanied by formation of a similar buried, water-mediated hydrogen bond network between the ligand and several backbone and side chain groups on the protein. To investigate the energetic contribution of this hydrogen bond network to ligand binding, we have applied isothermal titration calorimetry to measure the binding thermodynamics using several MUP mutants and ligand analogs. Mutation of Tyr-120 to Phe, which disrupts a hydrogen bond from the phenolic hydroxyl group of Tyr-120 to one of the bound water molecules, results in a substantial loss of favorable binding enthalpy, which is partially compensated by a favorable change in binding entropy. A similar thermodynamic effect was observed when the hydrogen bonded nitrogen atom of the heterocyclic ligand was replaced by a methyne group. Several other modifications of the protein or ligand had smaller effects on the binding thermodynamics. The data provide supporting evidence for the role of the hydrogen bond network in stabilizing the complex.
Keywords: enthalpy, hydrogen bond, major urinary protein, pheromone, thermodynamics, water
There is widespread interest in the roles that water molecules play in the stabilization and function of macromolecules and their complexes. X-ray crystal structures of proteins typically reveal many localized water molecules on the protein surface, in crevices, and sometimes buried within the protein interior or in the interfaces between proteins and bound ligands (Janin 1999; Mattos 2002). In favorable cases, NMR experiments can reveal the residence times of these water molecules (Halle and Denisov 2001). However, the contributions made by water molecule interactions to the stability of a folded structure or a complex cannot be reliably deduced from structural and dynamic data. One approach to establishing the importance of buried water has been to compare the positions of these solvent molecules in multiple high-resolution structures, obtained for different members of the same protein family or for the same protein under different crystallization conditions (Sadasivan et al. 1998; Loris et al. 1999; Nakasako 1999). An alternative approach has been to disrupt interactions with the water molecules by mutation of the relevant protein groups; such experiments have indicated that some hydrogen bonds between protein groups and buried water molecules stabilize the structure (Takano et al. 1999a,b), whereas others appear to be energy-neutral (Langhorst et al. 2000; Xu et al. 2001). Thus, for any given system, detailed thermodynamic studies remain necessary to deduce the energetic roles of specific water molecules.
We report herein a thermodynamic study of a buried water-mediated hydrogen bond network in the complexes between mouse major urinary proteins (MUPs) and their pheromonal ligands (±)-2-sec-butyl-4,5-dihydrothiazole (SBT) and 6-hydroxy-6-methyl-3-heptanone (HMH) (Fig. 1 ▶). These pheromones are excreted in male mouse urine (Novotny et al. 1990); SBT induces estrus synchrony (Jemiolo et al. 1986) and puberty acceleration in females (Novotny et al. 1999a) and aggression in males (Novotny et al. 1985), whereas HMH has an accelerating effect on female puberty onset (Novotny et al. 1999b). MUPs (members of the lipocalin protein family) are a group of highly homologous soluble proteins that are either excreted in the urine or expressed in the nasal mucosa and lachrymal, parotid, sublingual, and submaxillary glands of mice (Finlayson and Braumann 1958; Sampsell and Held 1985; Shahan et al. 1987). They bind to pheromones, and are proposed to play roles in regulating the release of pheromones from urine (Bacchini et al. 1992; Robertson et al. 1993), the capture of pheromones in the nose, and/or the delivery of pheromones to their receptors (Pelosi 1994; Marie et al. 2000; Timm et al. 2001; Sharrow et al. 2002).
Figure 1.
Structures of the two pheromones (SBT and HMH) and the five pheromone analogs used in this study.
X-ray structures of MUP isoform-I (MUP-I) bound to SBT and HMH show that the ligand binds into an occluded hydrophobic cavity in the center of the MUP “β-clam” structure (Timm et al. 2001). In addition, there is a conserved hydrogen bond network, involving two buried water molecules that bridge the polar atoms of the ligand to backbone and/or side chain groups of the protein. Despite the two ligands being structurally unrelated, the hydrogen bond network is extremely similar in the two complexes (Fig. 2 ▶). The tyrosine residue that participates in the network (Tyr-120) is completely conserved among MUP isoforms, whereas the threonine residue (Thr-21) is retained in all the urinary isoforms and conservatively substituted by serine in the predominant nasal isoform (MUP-IV) (Sharrow et al. 2002).
Figure 2.
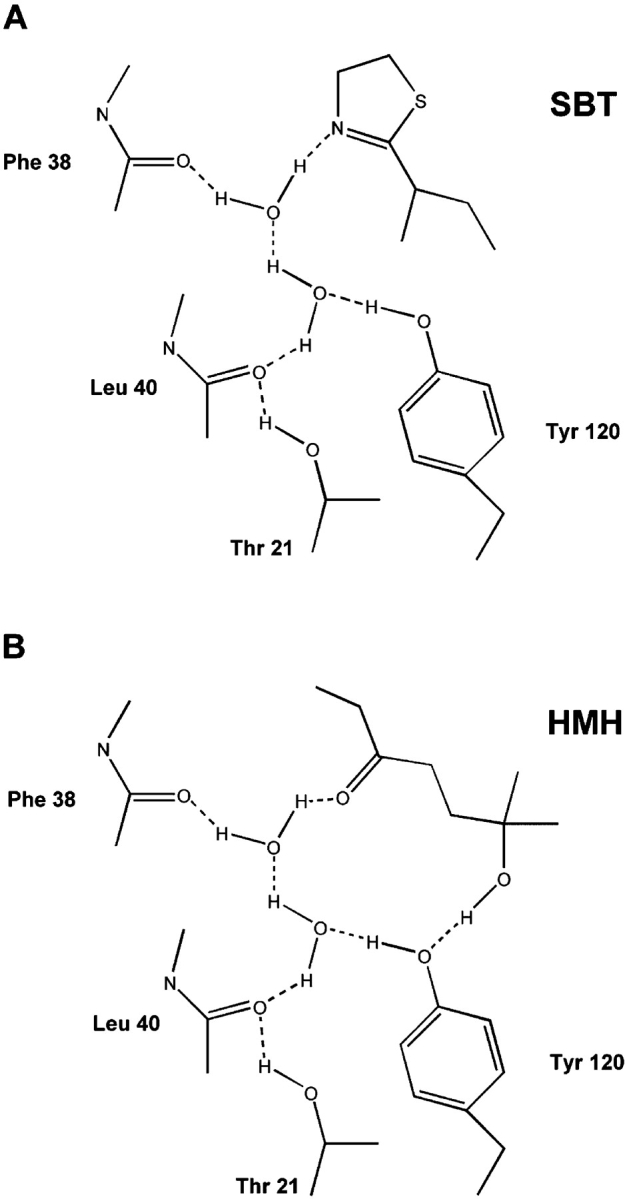
Water-mediated hydrogen-bonding network formed from MUP-I to (A) SBT and (B) as determined by X-ray crystallography (Marie et al. 2000). The hydrogen bonds are represented by dotted lines.
Although a large proportion of the buried intermolecular surface is hydrophobic, the binding of either MUP-I or MUP-IV to either SBT or HMH is characterized by a large favorable change in enthalpy (ΔH = −6 to −20 kcal/mol) and a smaller unfavorable change in entropy (−TΔS is in the range +3 to +8 kcal/mol at 30°C) (Sharrow et al. 2003). Furthermore, the binding enthalpy calculated from the MUP-I:SBT structure using the semiempirical approach of Freire and coworkers (Leavitt and Freire 2001; Velazquez-Campoy et al. 2001) is close to the observed value only when the buried water molecules are accounted for in the calculation, suggesting that interactions of the buried water molecules make a significant enthalpic contribution to stabilization of the complex.
To evaluate more directly the thermodynamic contributions of the water-mediated hydrogen bond network, we have now determined the thermodynamics of pheromone binding by MUP-I mutants in which the hydrogen-bonding residues Tyr-120 and/or Thr-21 have been mutated to Phe and/or Val, respectively. In addition, we have studied the binding of 2-methyl-4,5-dihydrothiazole (MT) and several commercially available analogs to MUP-IV. Together, these experiments demonstrate the effects of disrupting the hydrogen bond network by modification of either the protein or the ligand.
Results and Discussion
Pheromone binding by MUP-I mutants
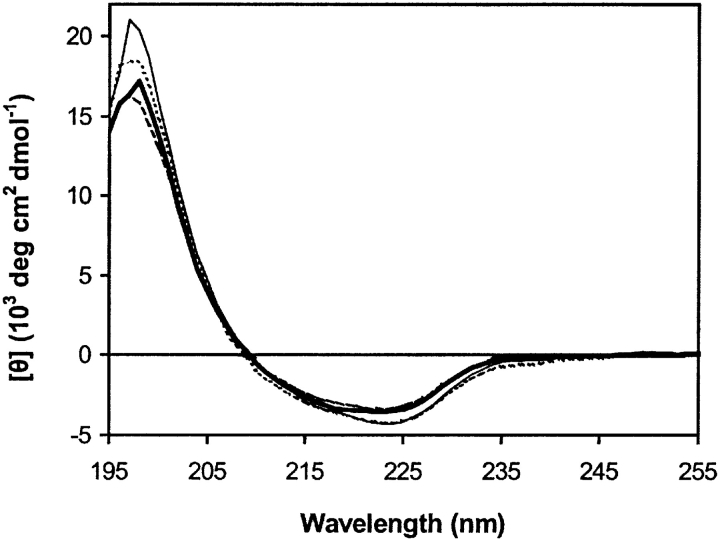
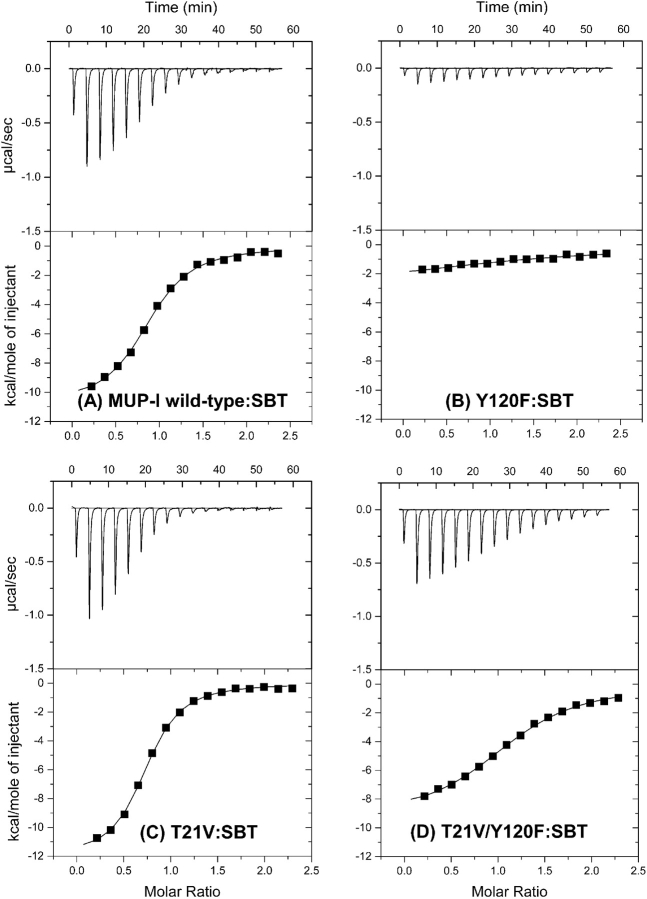
Wild-type MUP-I, the two single-mutants Y120F and T21V, and the double-mutant Y120F/T21V were expressed in Escherichia coli and purified as described previously (Zidek et al. 1999); the mutations were verified both by DNA-sequencing and protein mass spectrometry. The far-UV CD spectra of the three MUP-I mutants are very similar to that of wild type MUP-I (Fig. 3 ▶), indicating that the mutations do not induce any global conformational change of the protein, although minor changes in protein conformation cannot be excluded based on these data. ITC traces and fitted binding isotherms for the binding of wild type MUP-I and each mutant to SBT are shown in Figure 4 ▶ and the derived thermodynamic parameters for binding of SBT and HMH are listed in Table 1.
Figure 3.
Far-UV CD spectra of wild-type MUP-I (thick line), and the mutant MUP-I proteins: Y120F (thin solid line), T21V (dashed line); and T21V/Y120F (dotted line).
Figure 4.
ITC thermograms for binding of wild-type and mutant MUP-I proteins to SBT at 30°C. (A) Wild type, (B) Y120F, (C) T21V, and (D) T21V/Y120F.
Table 1.
Thermodynamic parameters for binding of MUP-I mutants to pheromones
| Protein | Ligand | Kd (μM) | Stoichiometry (n) | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) | Kd(mutant)/Kd(wild type) |
| Wild type | SBT | 1.1 ± 0.1 | 0.9 ± 0.1 | −10.9 ± 0.2 | +2.6 ± 0.2 | −8.3 ± 0.1 | 1 |
| T21V | SBT | 0.7 ± 0.1 | 0.7 ± 0.1 | −12.1 ± 0.1 | +3.6 ± 0.8 | −8.5 ± 0.1 | 0.64 |
| Y120F | SBT | 18 ± 10 | 1.4 ± 0.4 | −4 ± 2 | −3 ± 2 | −7 ± 3 | 16 |
| T21V/Y120F | SBT | 2.5 ± 0.1 | 1.1 ± 0.1 | −9.4 ± 0.1 | +1.6 ± 0.2 | −7.8 ± 0.1 | 2.3 |
| Wild type | HMH | 62 ± 4 | 0.8 ± 0.1 | −13 ± 1 | +7 ± 1 | −6 ± 1 | 1 |
| T21V | HMH | 51 ± 3 | 0.9 ± 0.1 | −12 ± 2 | +6.2 ± 0.2 | −6 ± 2 | 0.82 |
| Y120F | HMH | (no detectable signal) | |||||
| T21V/Y120F | HMH | (weakly exothermic signal) | |||||
All values were obtained by fitting ITC data at 30°C, as described in Materials and Methods.
Mutation of Tyr-120 to Phe dramatically influences the binding of both pheromones to MUP-I. For SBT, there is a very large reduction in the favorable binding enthalpy (ΔΔH = +6.9 kcal mol−1), which is partially compensated by an improvement in binding entropy (−TΔΔS = −5.6 kcal mol−1), resulting in an overall ~16-fold reduction in binding affinity (Δ ΔG = +1.3 kcal mol−1). Binding of MUP-I(Y120F) to HMH could not be observed by ITC, indicating that either the affinity had dropped more than ~10-fold or the binding enthalpy was close to zero (+11 < ΔΔH < +15 kcal mol−1) or both. Taken together, these data suggest that the hydrogen bond from the phenolic hydroxyl group of Tyr-120 to one of the bound water molecules (and possibly also to the hydroxyl group of HMH) provide essential enthalpic stabilization to the MUP-I:pheromone complexes, either directly or indirectly. It is also possible that the introduction of a Phe residue at position 120 gives rise to an unfavorable interaction between a bound water molecule and the Phe aromatic ring, which may be partially responsible for the reduced affinity and binding enthalpy of the mutant.
In contrast, the T21V mutation does not influence the affinity of SBT or HMH for MUP-I (the Kd values change by ≤1.5-fold), suggesting that the hydrogen bond between the Thr-21 side chain and the Leu-40 backbone CO group may not be a crucial component of the hydrogen bond network. It should be noted that the small free energy changes observed mask slightly larger (~1 kcal mol−1) but compensating changes in the binding enthalpy and entropy (Table 1). However, the latter are in opposite directions for the two ligands, complicating the physical interpretation of these observations.
Surprisingly, mutation of Thr-21 to Val within the context of the Y120F mutation (i.e., making the double mutant) results in recovery of much of the binding affinity that was lost upon mutating the Tyr to Phe. SBT binds to the double mutant ~sevenfold more tightly than to the Y120F single mutant (i.e., only ~2.3-fold more weakly than to wild-type MUP-I). The binding entropy and enthalpy are also similar for the double mutant and wild-type MUP-I. Again, the thermodynamics for HMH-binding are less easily quantified, although introducing the T21V mutation into the Y120F mutant clearly does increase the binding affinity and/or the favorable binding enthalpy. Thus, removing both of the protein side chain groups that participate in the water-mediated hydrogen-bond network yields a protein that is competent to bind both pheromonal ligands. Considering this result in isolation, one might be inclined to conclude that the backbone carbonyl groups of Phe-38 and Leu-40 (Fig. 2 ▶) are sufficient to maintain a stable hydrogen bond network. However, this would not account for the dramatic loss in affinity for the Y120F single mutant. An alternative explanation is that there is a change in the structure of the complex for the double mutant (e.g., a change in orientation of the ligand), but that the new mode of binding is not accessible for either single mutant. A further possibility is that the affinity difference between the single and double mutants results from differences in the free energy of the unbound rather than the bound states of these proteins. For example, the wild-type and single mutant proteins might all contain stabilizing interactions that are broken upon binding but the double mutant might not contain these same interactions. In this case, formation of the bound state would involve an additional energetic cost for the single mutants, resulting in lower affinity than the double mutant. Although binding of the wild-type protein would involve the same energetic sacrifice, this would be outweighed by additional favorable interactions in the wild-type bound state. More generally, it is important to note that any change in the structure, dynamics, or hydration of the protein (either in the free or bound state) can potentially influence the thermodynamics of ligand-binding. Detailed structural data on the free and pheromone-bound MUP-I variants would be required to address the influences of these factors.
MUP-IV binding by ligand analogs
As a complementary approach to mutation of MUP-I, we have also investigated the effect of modifying the ligand on the thermodynamics of MUP-binding. For convenience, these studies were performed using commercially available compounds with structural similarity to the SBT analog 2-methyl-4,5-dihydrothiazole (MT). We have shown previously that various 2-alkyl-4,5-dihydrothiazoles bind to MUP-I with a similar thermodynamic signature (large negative ΔH and small positive ΔS) to that observed for MUP-I:SBT binding, but that analogs with shorter alkyl chains have lower MUP-I affinity (Sharrow et al. 2003). The data support the proposal that the heterocyclic portions of all the 2-alkyl-4,5-dihydrothiazoles form similar interactions with MUP-I. We have also observed that the nasal isoform MUP-IV binds more than 100-fold more tightly than MUP-I to MT (S.D. Sharrow, M.V. Novotny, and M.U. Stone, un-publ.). Thus, the ligand analog studies were performed using MUP-IV, facilitating thermodynamic measurements with the weaker-binding analogs.
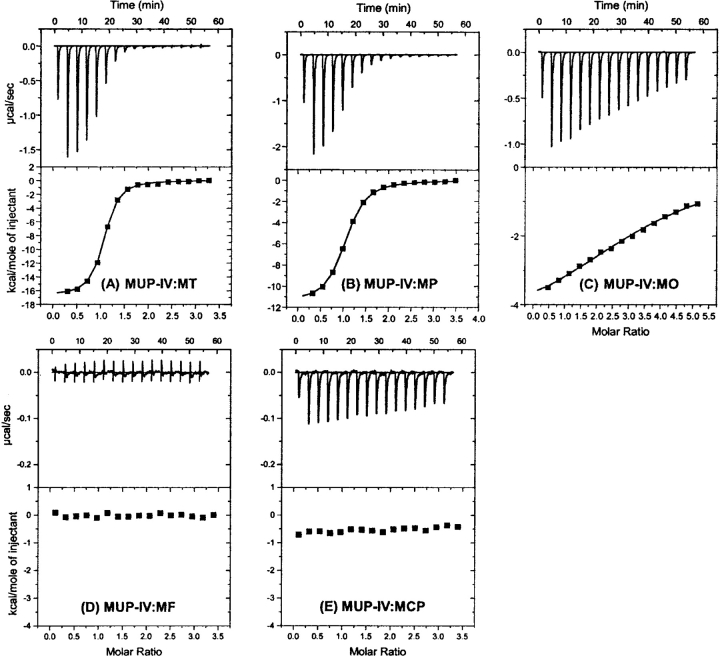
The structures of MT and the four MT analogs used in this study are shown in Figure 1 ▶. The four analogs each have one or both of the MT dihydrothiazole heteroatoms replaced by a methyne group, a methylene group, or an oxygen atom. Although the compound 2-methyl-2-thiophene (in which the nitrogen atom of MT is replaced by a methyne group) would have been an ideal analog to reveal the role of the ring nitrogen, this compound is not readily available (it has been reported only as a low-yield product from photocyclization of 5-thio-pent-1-yne (Dupuy et al. 1980) or as a minor volatile constituent of various biological samples) so it was not used in the current study. ITC data obtained for binding of MUP-IV to MT, and its analogs are shown in Figure 5 ▶ and the derived thermodynamic parameters are listed in Table 2.
Figure 5.
ITC thermograms for binding (at 30°C) of wild-type MUP-IV to the SBT analogs shown in Figure 1 ▶. (A) MT, (B) MP, (C) MO, (D) MF, and (E) MCP.
Table 2.
Thermodynamic parameters for binding of MUP-IV to pheromone analogs
| Ligand | Kd (μM) | Stoichiometry (n) | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) | Kd(analog)/Kd(MT) |
| MT | 0.2 ± 0.01 | 1.0 ± 0.1 | −16.7 ± 0.1 | +7.4 ± 0.1 | −9.3 ± 0.1 | 1 |
| MP | 1.0 ± 0.1 | 1.0 ± 0.1 | −11.6 ± 0.1 | +3.2 ± 0.1 | −8.4 ± 0.1 | 5.0 |
| MO | 25 ± 3 | 3.4 ± 0.1 | −5.1 ± 0.2 | −1.3 ± 0.2 | −6.4 ± 0.1 | 125 |
| MF | (no detectable signal) | |||||
| MCP | (weakly exothermic signal) | |||||
All values were obtained by fitting ITC data at 30°C, as described in Materials and Methods.
The ring sulfur atom of MT is expected to be buried in a hydrophobic environment in the MUP-IV complex but to be exposed to solvent in the free state. Replacement of this sulfur atom with a methylene group (in 2-methyl-1-pyrroline) reduces the affinity for MUP-IV ~4.8-fold and reduces the favorable binding enthalpy from −16.7 kcal/mol to −11.6 kcal mol−1. Similarly, replacement of the ring sulfur of MT with an oxygen atom (in 2-methyl-2-oxazoline) reduces the affinity for MUP-IV by more than two orders of magnitude and reduces the favorable binding enthalpy to −5.1 kcal mol−1. The more dramatic effect of introducing an oxygen atom rather than a methylene group may reflect the ability of oxygen to hydrogen bond with solvent in the free state, so that burial of oxygen in a hydrophobic environment in the complex is less favorable than burial of methylene. However, the fitted binding stoichiometry also changes (from 1.0 for 2-methyl-4,5-dihydrothiazole to 3.4 for 2-methyl-2-oxazoline), suggesting that 2-methyl-2-oxazoline may also interact with alternative binding sites on MUP-IV.
Upon subsequent replacement of the pyrroline or oxazoline ring nitrogen with a methyne group (in 1-methyl-1-cyclopentene or 4,5-dihydro-2-methylfuran, respectively), the binding enthalpy and/or affinity is dramatically reduced (Fig. 5 ▶; Table 2) to the point that ITC cannot yield a reliable estimate of the Kd. These observations are consistent with the nitrogen atom contributing to the highly favorable enthalpy of the hydrogen bond network. However, interpretation of the data for 4,5-dihydro-2-methylfuran is complicated by the possibility that the ligand could reorient in the binding site so as to place the oxygen atom in the location previously occupied by the ring nitrogen atom (of MT or 2-methyl-2-oxazoline). In this light, the observation that binding of 4,5-dihydro-2-methylfuran is so much weaker than binding of 2-methyl-2-oxazoline suggests that a nitrogen to water hydrogen bond may be preferred over an oxygen to water hydrogen bond, possibly because the orientations of the lone electron pairs on the oxygen are not optimal for hydrogen bonding to the structurally constrained water molecule.
Comparison to previous studies
It is instructive to compare the current results with several previous studies on the roles of hydrogen bonds to water in protein–ligand binding (Ladbury 1996). For example, in the binding of the protein FKBP-12 to its macrocyclic ligands (Connelly et al. 1994), residue Tyr-82 forms three hydrogen bonds to ordered water molecules in the free form of the protein, but they are replaced by a hydrogen bond to a ligand carbonyl group in the complex. Thermodynamic binding data for wild-type and mutant (Y82F) FKBP-12 collected in H2O and D2O indicated that the three water-to-tyrosine hydrogen bonds are (in combination) enthalpically more favorable but entropically destabilizing relative to the tyrosine-to-ligand hydrogen bond that replaces them. Although these enthalpy and entropy changes are in the opposite directions to those reported herein for the Y120F mutant of MUP-I, this presumably occurs because the water hydrogen bonds are important in the free state for FKBP-12 but the bound state for MUP-I. Thus, the conclusion that hydrogen bonds to water enthalpically stabilize the relevant state of the protein is consistent between the two studies.
A similar thermodynamic effect has been observed for zinc binding (coupled to folding) by the zinc-finger protein ZFY in which the hydrophobic core residue Phe-10 has been replaced by Leu (Lachenmann et al. 2002). Binding by the mutant is enthalpically more favorable and entropically less favorable than binding by the wild-type protein. The difference was proposed to reflect the thermodynamics of formation of a water network within a mixed polar/nonpolar cavity in the mutant protein. Lachenmann et al. (2002) commented that “if bound water molecules could be visualized in the crevice of an otherwise native-like structure, the Zn finger may be regarded as an explicit physical realization of solvent-mediated enthalpy–entropy compensation.” This is precisely the case for the MUP-I:pheromone complex, in which the water molecules have been visualized in the bound state (Timm et al. 2001), although there is no structure available for the free state.
Another well-characterized example of linkage between water structure and thermodynamics is the binding of hen egg white lysozyme (HEWL) to the Fv fragment of antibody D1.3, mediated through a network of bridging water molecules, including four that are completely buried in the binding interface (Bhat et al. 1994). The authors argued that a significant component of the favorable binding enthalpy and unfavorable binding entropy results from the formation of the water network, and this conclusion was supported by subsequent correlation of the thermodynamic changes with water activity (Goldbaum et al. 1996). Interestingly, however, water appears to play a different role in binding of HEWL to the antibody D44.1. In the latter case, both the enthalpy and entropy of binding are less negative at high water activity, indicating that interfacial water does not provide enthalpic stabilization and/or that removal of water from hydrophobic surfaces is a more important factor (Goldbaum et al. 1996).
Indeed, the variable role of water is a recurring theme in comparisons between structural and thermodynamic data. For example, the bacterial periplasmic binding protein OppA optimizes its interactions with 20 peptides containing the sequence Lys-X-Lys by using water molecules to pack the binding site matching the variable amino acid (“X”) (Sleigh et al. 1999). Depending on the nature of this side chain, the water molecules can form hydrogen bonds, shield charge–charge repulsions, or merely fill cavities between the protein and peptide. Although water-filled nonpolar cavities may initially appear to be unstable structures, it is important to consider not only the interactions of the water molecules in the binding interface, but also the degree of order of the interfacial water and the corresponding interactions and order of water at the binding surfaces of both binding partners in their free (hydrated) states. Ringe has noted that water molecules in nonpolar ligand binding sites can be displaced more readily than many surface water molecules by organic solvents (Ringe 1995), suggesting that formation of a nonpolar cavity containing water may be entropically advantageous. In addition, aromatic groups have the potential to accept hydrogen bonds from water, providing a possible source of enthalpic stabilization (Makhatadze and Privalov 1994a,b).
The role of water in broadening the ligand specificity of OppA contrasts with the role of water in the peptide binding interaction of the Src SH2 (phosphotyrosine-binding) domain (Henriques and Ladbury 2001). For the SH2 domain, water-mediated hydrogen bonds appear to be essential for maintaining the specificity for a glutamate residue in the second position after the phosphotyrosine residue. Indeed, it has been proposed (Henriques and Ladbury 2001) that the successful targeting of drugs to SH2 domains will require careful optimization of the water “mesh” at the binding interface. This may be generally true, especially for proteins in which water plays an important role in binding of natural ligands (Ladbury 1996; Henriques and Ladbury 2001).
Conclusion
The data obtained for the MUP-I mutants and the ligand analogs are consistent with the proposal that the water-mediated hydrogen bond network observed in the crystal structure has a significant stabilizing influence on MUP:ligand binding. Moreover, the calorimetric data indicate that the hydrogen bonds are both enthalpically favorable and entropically unfavorable, presumably because formation of optimal hydrogen bonding interactions requires immobilization of the ligand, the water molecules, and/or the relevant groups on the protein. The enthalpy–entropy compensation observed here is a general feature of many intermolecular interactions (Sharp 2001), including the interactions of water with proteins (vide supra).
Despite the qualitative conclusion that the hydrogen bond network stabilizes MUP:ligand complexes, reliable evaluation of the total energetic contribution of the hydrogen bond network is complicated by several factors. First, binding of MUP mutants and ligand analogs is weak and difficult to quantitate. Second, the hydrogen bonds are likely to be the major factors that specify the orientation of the ligand in the binding cavity, so ligand analogs do not necessarily bind in the same orientation as the natural ligands. Third, it is not possible to replace the water molecules themselves or the protein backbone groups with which they interact, so any disruption of the hydrogen bond network may be incomplete. Finally, the formation of the hydrogen bonding interactions may be cooperative not only with each other but also (positively or negatively) with other interactions such as van der Waals interactions between the nonpolar protein and ligand surfaces or hydrogen bonding interactions within the protein structure itself (including other hydration water molecules). Most of these difficulties are not specific to the current system, and are expected in the experimental analysis of any water-mediated hydrogen bond network.
Materials and methods
Materials
Genes encoding the MUP-I single mutants Y120F and T21V and the double-mutant Y120F/T21V were obtained using PCR mutagenesis (Papworth et al. 1996). Wild-type and mutant MUP-I and wild-type MUP-IV were expressed and purified following methods described previously (Zidek et al. 1999; Sharrow et al. 2002). The pheromones SBT and HMH and the SBT analogs 2-n-propyl-4,5-dihydrothiazole (n-PT) and MT were synthesized as described (Sharrow et al. 2002). 2-Methyl-1-pyrroline, 2-methyl-2-oxazoline, 4-5-dihydro-2-methyfuran, and 1-methyl-1-cyclopentane were purchased from the Aldrich Chemical Co.
Protein characterization
Protein purity was monitored by SDS-PAGE. Mass analysis was performed using a PerSeptive Biosystems Voyager-DE; RP MALDI-TOF mass spectrometer equipped with a nitrogen laser (337 nm). The spectra were collected in the linear mode, and were calibrated internally with both horse skeletal apomyoglobin and trypsin inhibitor. Both analyte and calibrants were mixed in 3,5-dimethyl-4-hydroxy-cinnamic acid. Signal averaged mass spectra for 64 laser pulses were collected, and data were smoothed with a Savitsky-Golay five-point smoothing algorithm using Grams/386 software. Eight to 10 independent measurements were taken for statistical analysis.
Circular dichroism
CD spectra were recorded at 30°C on 20-mM protein samples in 10 mM phosphate (pH 6.3) using 0.1-cm cuvettes and a Jasco J-715 spectropolarimeter. Each spectrum is the sum of 15 scans recorded at a scan rate of 200 nm/min and a resolution of 1 nm. Data for control scans, recorded using the same parameters, were subtracted from the scans of the protein samples to eliminate background signal resulting from the buffer.
Isothermal titration calorimetry
Isothermal titration calorimetry was carried out at 30°C using a Microcal MCS instrument located at the Keck Biophysics Facility, Northwestern University, Evanston, IL. Samples of MUP protein (2.4 mL of 13–500 μM protein in 10 mM phosphate, 0.02% NaN3 [pH 6.3]) were titrated with one 2-μL aliquot, then 15 4-μL aliquots of the pheromone or its analog (0.26–2.6 mM in the same buffer). The duration of each injection was ~20 sec, with a 210-sec recovery time between injections. The reference cell was filled with distilled water. To account for heats of dilution and mechanical mixing, control titrations were performed by injection of the ligand into buffer; binding data were corrected by subtraction of the dilution data prior to curve fitting. Corrected binding data were processed using the Origin ITC analysis software package supplied by Microcal. The data were fit well to the following equation (Holdgate 2001) describing the cumulative heat (Q) evolved from a series of injections when the ligand (L) binds to a set of identical, independent binding sites on the protein (P):
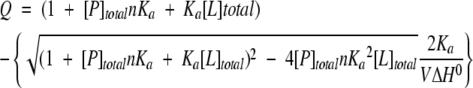 |
in which V represents the initial reaction volume. Nonlinear curve fitting of the first derivative of Q with respect to [L]total plotted against the molar ratio of ([L]total/[P]total) yielded the association equilibrium constant (Ka = 1/Kd), the binding stoichiometry (n = number of ligand binding sites per protein molecule), and the enthalpy of binding (ΔH0).
Electronic supplemental material
One figure (four panels) showing the ITC thermograms for binding of wild-type and mutant MUP-I proteins to HMH.
Acknowledgments
We acknowledge the use of instruments in the Keck Biophysics Facility at Northwestern University (http://x.biochem.nwu.edu/Keck/keckmain.html). This work was supported by grants awarded to M.J.S. from the NSF (MCB-0212746) and to M.V.N. from the National Institute of Deafness and Communication Disorders (DC 02418).
Abbreviations
HEWL, hen egg white lysozyme
HMH, 6-hydroxy-6-methyl-3-heptanone
ITC, isothermal titration calorimetry
MCP, 1-methyl-1-cyclopentene
MF, 4,5-dihydro-2-methylfuran
MO, 2-methyl-oxazoline
MP, 2-methyl-1-pyrroline
MT, 2-methyl-4,5-dihydrothiazole
MUP, major urinary protein
SBT, (±)-2-sec-butyl-4,5-dihydrothiazole
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04912605.
Supplemental material: see www.proteinscience.org
References
- Bacchini, A., Gaetani, E., and Cavaggioni, A. 1992. Pheromone binding-proteins of the mouse, Musmusculus. Experientia 48 419–421. [DOI] [PubMed] [Google Scholar]
- Bhat, T.N., Bentley, G.A., Boulot, G., Greene, M.I., Tello, D., Dall’Acqua, W., Souchon, H., Schwarz, F.P., Mariuzza, R.A., and Poljak, R.J. 1994. Bound water molecules and conformational stabilization help mediate an antigen–antibody association. Proc. Natl. Acad. Sci. 91 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, P.R., Aldape, R.A., Bruzzese, F.J., Chambers, S.P., Fitzgibbon, M.J., Fleming, M.A., Itoh, S., Livingston, D.J., Navia, M.A., Thomson, J.A., et al. 1994. Enthalpy of hydrogen bond formation in a protein–ligand binding reaction. Proc. Natl. Acad. Sci. 91 1964–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy, C., Crozet, M.P., and Surzur, J.M. 1980. Free radical heterocyclization of acetylenic thiols. Bull. Soc. Chim. France 7–8 II-361–II-373. [Google Scholar]
- Finlayson, J.S. and Braumann, C.A. 1958. Mouse proteinuria. Am. J. Physiol. 192 69–72. [DOI] [PubMed] [Google Scholar]
- Goldbaum, F.A., Schwartz, F.P., Eisenstein, E., Cauerhff, A., Mariuzza, R.A., and Poljak, R.J. 1996. The effect of water activity on the association constant and the enthalpy reaction between lysozyme and the specific antibodies D1.3 and D44.1. J. Mol. Recognit. 9 6–12. [DOI] [PubMed] [Google Scholar]
- Halle, B. and Denisov, V.P. 2001. Magnetic relaxation dispersion studies of biomolecular solutions. Methods Enzymol. 338 178–201. [DOI] [PubMed] [Google Scholar]
- Henriques, D.A. and Ladbury, J.E. 2001. Inhibitors to the Src SH2 domain: A lesson in structure–thermodynamic correlation in drug design. Arch. Biochem. Biophys. 390 158–168. [DOI] [PubMed] [Google Scholar]
- Holdgate, G.A. 2001. Making cool drugs hot: Isothermal titration calorimetry as a tool to study binding energetics. BioTechniques 31 164–166, 168, 170. [PubMed] [Google Scholar]
- Janin, J. 1999. Wet and dry interfaces: The role of solvent in protein–protein and protein–DNA recognition. Struct. Fold. Des. 7 R277–R279. [DOI] [PubMed] [Google Scholar]
- Jemiolo, B., Harvey, S., and Novotny, M. 1986. Promotion of the whitten effect in female mice by synthetic analogs of male urinary constituents. Proc. Natl. Acad. Sci. 83 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenmann, M.J., Ladbury, J.E., Phillips, N.B., Narayana, N., Qian, X., and Weiss, M.A. 2002. The hidden thermodynamics of a zinc finger. J. Mol. Biol. 316 969–989. [DOI] [PubMed] [Google Scholar]
- Ladbury, J.E. 1996. Just add water! The effect of water on the specificity of protein–ligand binding sites and its potential application to drug design. Chem. Biol. 3 973–980. [DOI] [PubMed] [Google Scholar]
- Langhorst, U., Backmann, J., Loris, R., and Steyaert, J. 2000. Analysis of a water mediated protein–protein interactions within RNase T1. Biochemistry 39 6586–6593. [DOI] [PubMed] [Google Scholar]
- Leavitt, S. and Freire, E. 2001. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 11 560–566. [DOI] [PubMed] [Google Scholar]
- Loris, R., Langhorst, U., Decanniere, K., De Vos, S., Bouckaert, J., Maes, D., Transue, T.R., and Steyaert, J. 1999. Conserved water molecules in a large family of microbial ribonucleases. Proteins 36 117–134. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I. and Privalov, P.L. 1994a. Energetics of interactions of aromatic hydrocarbons with water. Biophys. Chem. 50 285–291. [DOI] [PubMed] [Google Scholar]
- ———. 1994b. Hydration effects in protein unfolding. Biophys. Chem. 51 291–309. [DOI] [PubMed] [Google Scholar]
- Marie, A.D., Veggerby, C., Robertson, D., Gaskell, S.J., Hubbard, S.J., Martinsen, L., Hurst, J.L., and Beynon, R. 2000. Effect of polymorphisms on ligand binding by mouse major urinary proteins. Protein Sci. 10 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos, C. 2002. The dynamics of protein–water interactions are crucial for function in relation to protein structure, motion and adaptability to changes in the protein environment. Trends Biochem. Sci. 27 203–208. [DOI] [PubMed] [Google Scholar]
- Nakasako, M. 1999. Large-scale networks of hydration water molecules around bovine β-trypsin revealed by cryogenic X-ray crystal structure analysis. J. Mol. Biol. 289 547–564. [DOI] [PubMed] [Google Scholar]
- Novotny, M., Harvey, S., Jemiolo, B., and Alberts, J., 1985. Synthetic pheromones that promote inter-male aggression in mice. Proc. Natl. Acad. Sci. 82 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny, M., Jemiolo, B., and Harvey, S. 1990. Chemistry of rodent pheromones: Molecular insights into chemical signalling in mammals. In Chemical signals in vertebrates 5 (eds. D.W. MacDonald et al.), pp. 1–22. Oxford University Press, Oxford.
- Novotny, M.V., Ma, W., Wiesler, D., and Zidek, L. 1999a. Positive identification of the puberty-accelerating pheromone of the house mouse: The volatile ligands associating with the major urinary protein. Proc. R. Soc. Lond. B 266 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny, M.V., Ma, W., Jemiolo, S.B., Wiesler, D., Harvey, S., Xu, F., Xie, T.M., and Carmack, M.A. 1999b. A unique urinary constituent, 6-hydroxy-6-methyl-3-heptanone, is a pheromone that accelerates puberty in female mice. Chem. Biol. 6 377–383. [DOI] [PubMed] [Google Scholar]
- Papworth, C., Bauer, J., Braman, J., and Wright, D.A. 1996. Site-directed mutagenesis in one day with >80% efficiency. Strategies 9 3–4. [Google Scholar]
- Pelosi, P. 1994. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 29 199–228. [DOI] [PubMed] [Google Scholar]
- Ringe, D. 1995. What makes a binding site a binding site? Curr. Opin. Struct. Biol. 5 825–829. [DOI] [PubMed] [Google Scholar]
- Robertson, D., Beynon, R., and Evershed, R. 1993. Extraction, characterization, and binding analysis of two pheromonally active ligands associated with major urinary protein of house mouse (Mus musculus). J. Chem. Ecol. 19 1405–1416. [DOI] [PubMed] [Google Scholar]
- Sadasivan, C., Nagendra, H.G., and Vigayan, M. 1998. Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant Acta Crystallogr. D. Biol. Crystallogr. 54 1343–1352. [DOI] [PubMed] [Google Scholar]
- Sampsell, B.M. and Held, W.A. 1985. Variation in the major urinary protein multigene family in wild-derived mice. Genetics 109 549–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahan, K., Denaro, M., Gilmartin, M., Shi, Y., and Derman, E. 1987. Expression of six mouse major urinary protein genes in the mammary, parotid, sublingual, submaxillary, and lachrymal glands and in the liver. Mol. Cell. Biol. 7 1947–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh, S.H., Seavers, P.R., Wilkinson, A.J., Ladbury, J.E., and Tame, J.R.H. 1999. Crystallographic and calorimetric analysis of peptide binding to OppA protein. J. Mol. Biol. 291 393–415. [DOI] [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Funahashi, J., Hioki, Y., Kuramitsu, S., and Yutani, K. 1999a. Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme. Biochemistry 38 12698–12708. [DOI] [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Kubota, M., Funahashi, J., Fujii, S., and Yutani, K. 1999b. Contribution of hydrogen bonds to the conformational stability of human lysozyme: Calorimetry and X-ray analysis of Six Ser→Ala Mutants. Biochemistry 38 6623–6629. [DOI] [PubMed] [Google Scholar]
- Timm, D.E., Mueller, L., Zidek, L., and Novotny, M.V. 2001. Structural basis of pheromone binding to mouse major urinary protein (MUP-I). Protein Sci. 10 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Campoy, A., Luque, I., and Freire, E. 2001. The application of thermodynamic methods in drug design. Thermochim. Acta 380 217–227. [Google Scholar]
- Xu, J., Baase, W.A., Quillin, M.L., Baldwin, E.P., and Matthews, B.W. 2001. Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme. Protein Sci. 10 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek, L., Stone, M.J., Lato, S.M., Pagel, M.D., Miao, Z., Ellington, A.D., and Novotny, M.V. 1999. NMR mapping of the recombinant mouse major urinary protein I binding site occupied by the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry 38 9850–9861. [DOI] [PubMed] [Google Scholar]