Abstract
We present the results of the evaluation of the latest LiveBench-8 experiment. These results provide a snapshot view of the state of the art in automated protein structure prediction, just before the 2004 CAFASP-4/CASP-6 experiments begin. The last CAFASP/CASP experiments demonstrated that automated meta-predictors entail a significant advance in the field, already challenging most human expert predictors. LiveBench-8 corroborates the superior performance of meta-predictors, which are able to produce useful predictions for over one-half of the test targets. More importantly, LiveBench-8 identifies a handful of recently developed autonomous (nonmeta) servers that perform at the very top, suggesting that further progress in the individual methods has recently been obtained.
Keywords: protein structure prediction, LiveBench, CAFASP
Automatic protein structure prediction has become an important complement to the various sequencing and structural genomics projects (Fischer and Eisenberg 1997; Abbott 2001; Fischer et al. 2001). Dozens of automated structure prediction servers are currently available. Knowing which methods work best is thus important for the biologist users. Here we describe the results of the recently concluded LiveBench-8 (LB) experiment. As in previous even years (Fischer et al. 2000; Bujnicki et al. 2001; Fischer and Rychlewski 2003), this report is a snapshot view of the state-of-the-art in fold-recognition (FR) methods at the time that the CASP/CAFASP (Fischer and Rychlewski 2003; Lattman 2003) experiments are about to start.
LB continuously assesses the capabilities of automated servers using a relatively large number of prediction targets compiled every week from newly released protein structures, and provides an assessment of the servers’ capabilities approximately every half year. LB thus complements the CASP/CAFASP experiments (Fischer et al. 2003; Lattman 2003) which are held every two years using a significantly smaller number of prediction targets. Another large-scale evaluation project that focuses on other aspects of structure prediction is EVA (Rost and Eyrich 2001).
The last LB and CAFASP experiments (Fischer et al. 2003; Lattman 2003; Rychlewski et al. 2003) demonstrated that the so-called meta-servers—defined as servers that need as input the results of other participating servers—outperform all the individual, autonomous servers, and are already challenging most human expert predictors. Since then, new servers and meta-servers have been developed and evaluated in subsequent LB rounds. To obtain an updated snapshot of the predicting capabilities of current servers, and of their expected performance in future experiments, we report the main results from the recently completed LB-8.
LiveBench-8
LB-8 was carried out between June and December 2003, with the participation of 44 servers and meta-predictors, of which 12 correspond to newly developed servers. The set of targets used in LB is selected every week from the sequences of newly released protein structures that share no significant sequence similarity with previously determined structures, as determined by BLAST (Altschul et al. 1990) (i.e., newly released structures are included if a BLAST search against the sequences of previously determined structures results in no hits with e-values < 0.01). In LB-8, 172 targets were considered, which were divided into 73 “easy” and 99 “hard” targets. This division is based on whether a template of known structure can be identified by PSI-BLAST with an e-value < 0.001 (Altschul et al. 1997). Because only a handful of the targets correspond to novel folds, the majority of our 172 targets could, in principle, be modeled using previously determined templates. Structure comparison using DALI (Holm and Sander 1995) shows that models with MaxSub scores > 0.33 could be built for 130 targets; DALI could not find a template with MaxSub score > 0.20 for only 11 targets (the DALI scores are available at the LB Web site at http://bioinfo.pl/LiveBench after adding DALI in the server list window; further details regarding the LB’s experimental setup are also available at the LB Web site).
The LiveBench Web site is a comprehensive, interactive repository of LB results. Because LB considers a number of evaluation methods, the LB Web site allows the user to select the evaluation method to be used as well as the way the results are presented. In addition, the LB site includes data for server predictions that were not submitted immediately upon release of the targets. The LB Web site also allows the user to select the set of servers to be considered. Because of the versatility of the interactive system, there are many ways that one can interpret the LB results. Consequently, understanding the meaning of the LB results may not be straightforward for an outsider. To aid in the interpretation of the LB data, here we report the LB-8 results using the same simplified approach as the one used two years ago in our LB-4 report (Fischer and Rychlewski 2003). This approach is based on the measures of “overall sensitivity” and “overall specificity” (described below) as assessed by the evaluation method MaxSub (Siew et al. 2000). MaxSub is a program that measures the quality of a prediction, by assigning scores between 0.0 (an incorrect prediction) to 1.0 (a perfect prediction). A positive MaxSub score is considered to be a (partially) correct prediction. To further simplify the presentation of the LB results, we discuss the performance of the individual or autonomous servers separately from that of the meta-predictors, and consider only those servers that submitted predictions every week for all 172 targets. Consequently, some of the servers that submitted predictions at a later time, including those submitted after the preparation of this manuscript, are excluded from the results presented here. We refer to the servers’ names using their LB’s four-letter abbreviation and refer the reader to the LB-8 Web pages for their full names.
Sensitivity
Table 1 lists the overall sensitivities of the top performing autonomous servers. Overall sensitivity is defined as the percentage of targets for which a correct prediction is obtained (i.e., a positive MaxSub score; see above). The second column lists the overall sensitivity over all 172 targets, and the following two columns list the overall sensitivity when considering the easy and hard targets separately. In this and other tables, we highlight the three highest numbers in bold.
Table 1.
Overall sensitivity
| Server name | Overall sensitivity (total 172) | Correct easy (total 73) | Correct hard (total 99) |
| BASD | 65% | 93% | 44% |
| BASP | 65% | 93% | 43% |
| MBAS | 64% | 93% | 42% |
| SHGU | 62% | 92% | 40% |
| SFST | 62% | 95% | 37% |
| STMP | 62% | 95% | 38% |
| ORF2 | 62% | 92% | 40% |
| FFA3 | 61% | 92% | 38% |
| ORFs | 61% | 93% | 37% |
| PDBB | 45% | 82% | 12% |
The table shows that the top performing servers have very similar overall sensitivities, with up to 65% (112 correct predictions out of 179). While correct predictions were obtained for up to 95% of the easy targets, only up to 44% of the hard targets were correctly predicted. For comparison, the table shows the performance of the PDBB server, which is a local implementation of PSI-BLAST (Altschul et al. 1997). It is clear that FR servers outperform PDBB, both among the easy and the hard targets.
The sensitivities of the best performing autonomous servers in LB-4 were just above 50%, suggesting that there may be a slight improvement in LB-8 over the “old” LB-4 servers. However, because the sensitivities of some of the servers that participated both in LB-4 and in LB-8 are also higher in LB-8, such an improvement may be due in part to the differences in the test sets (LB-8 may have included more “easier” targets), to the growth of the sequence and structural databases, or both.
The autonomous servers with highest overall sensitivity were the recently developed series of “Meta-BASIC” servers (unpublished): BASD, BASP, and MBAS. BASD (Distal-BASIC) and BASP (Proximal-BASIC) are profile-comparison methods. BASD uses two versions of low stringency profiles generated after 5 PSI-BLAST iterations combined with RPS-BLAST searches, and BASP uses profiles generated after three iterations. MBAS (Meta-BASIC) is a local, autonomous meta-predictor, which uses six different versions of profile-alignment methods. We notice that five of the top ranks are now occupied by newly developed servers (BASD, BASP, MBAS, SFST, and STMP). The other ranks are occupied by servers that have also ranked among the top performers in previous experiments: SHGU (Fischer 2003), ORF2 (Rychlewski et al. 2000), FFA3 (Pawlowski et al. 2001), and ORF-s (Rychlewski et al. 2000). Because of the excellent performance of the new servers, other older servers that had ranked among the top performers in previous experiments; e.g., 3DPS (Kelley et al. 2000), INBG (Fischer 2000), MGTH (McGuffin and Jones 2003), ST99 (Karplus and Hu 2001), and the two versions of FUGUE (Shi et al. 2001), now occupy lower ranks. This suggests that there have been positive developments in the field, and that the new servers appear to entail an improvement over the older ones.
The sum of MaxSub scores is an additional sensitivity indicator, which assesses the quality (or completeness) of the generated models (Table 2). Using this measure, the most sensitive servers are SHGU (an old server from LB-4 that applies the 3D-SHOTGUN meta-prediction approach on locally generated data from INBGU) (Fischer 2003), BASD and MBAS. For comparison, PDBB scores 45% lower than the best servers. The sensitivities of the top servers among the easy targets are very similar, with the two recently developed commercial servers, SFST and STMP, being at the top following SHGU. SFST and STMP are two versions of a profile–profile alignment method developed by the same group, which uses specific gap penalties and composition-based statistics. Among the hard targets, the most sensitive servers are again BASD, BASP, and MBAS, the same servers scoring at the top on overall sensitivity. The difference in scores between the rank-1 and rank-9 servers is 9% and 19%, among the easy and hard targets, respectively. PDBB’s performance on the hard targets is significantly lower than that of the top servers, indicating the value of FR servers are for these cases.
Table 2.
MaxSub sensitivity scores on all 172, 73 “easy” and the 99 “hard” targets
| Server name | All (total 172) | Easy (total 73) | Hard (total 99) |
| SHGU | 4330 | 3198 | 1132 |
| BASD | 4191 | 2968 | 1223 |
| MBAS | 4166 | 2976 | 1190 |
| BASP | 4142 | 2987 | 1154 |
| SFST | 4118 | 3059 | 1059 |
| STMP | 4047 | 2989 | 1058 |
| FFA3 | 4001 | 2946 | 1055 |
| ORFs | 3979 | 2952 | 1026 |
| ORF2 | 3959 | 2929 | 1030 |
| PDBB | 2983 | 2737 | 246 |
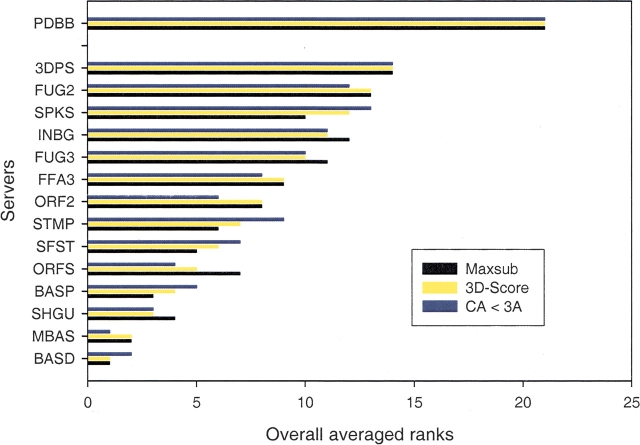
Because the differences among the top servers are in general not very large and because their relative ranking depends on the particular method of evaluation used, the exact rankings may not be very significant. To see whether the ranking is affected by the particular evaluation method applied, we also computed the sum of scores that each server obtains among the easy and hard targets, respectively, using, in addition to the total MaxSub score, the total scores obtained by two of the other evaluation methods applied in LB (Fig. 1 ▶). The results showed that the set of best performing servers identified above is almost the same regardless of the evaluation method used.
Figure 1.
Overall averaged ranks of the top autonomous servers in Live-Bench-8. The averages were computed separately using three of the LB evaluation methods (MaxSub, 3D-Score, and CA < 3A) by averaging the ranks obtained at each of the three evaluation categories (sensitivity on “easy” and “hard” targets and “specificity”). The overall averaged ranks vary little among the evaluation methods. The ranking was computed without considering meta-servers, which generally outperform the autonomous servers. Ranks 15–20 are not shown. It is clear that many fold-recognition servers outperform PDBB (a local implementation of PSI-BLAST), which received rank 21.
Specificity
Table 3 lists the overall specificities of the top LB-8 autonomous servers, computed as in LB-4: the average percentage of correct predictions with scores better than the first 10 false positives. The percentage is obtained by dividing the total specificity calculated in the LB-8 Web page, by the total number of targets (172), times 100. The table also lists for each server the score of its 8th incorrect prediction (eight wrong predictions amount to 5% of all targets). This can serve as a rough, optimistic estimation of the score below which the servers’ predictions become less reliable. For example, the FUG3 server had a specificity of 50% (total specificity score of 86.6, divided by 172 targets) and the score of its 8th wrong prediction is 4.93. This means that FUG3 approximately produced 87 correct predictions plus eight incorrect ones with scores higher than 4.93. Depending on the way each server scores its predictions, a more confident prediction can mean a larger or smaller score (shown with the “<” and “>” signs in the table). Probably the most valuable outcome of LB, rather than attempting to provide an exact ranking of the servers, may be the availability of these thresholds. The LB-8 Web pages include detailed information listing the scores of the first to 10th false positives of each server.
Table 3.
Overall specificity
| Server name | Overall specificity | 5% error rate score |
| BASD | 58% | >10.1 |
| MBAS | 57% | >11.2 |
| BASP | 55% | >9.23 |
| ORF2 | 54% | >22.1 |
| ORFs | 54% | >7.07 |
| SHGU | 54% | >20.1 |
| SFST | 53% | >36.0 |
| STMP | 51% | <0.001 |
| FUG3 | 50% | >4.93 |
| FUG2 | 50% | >4.93 |
| FFA3 | 49% | <−12.9 |
| PDBB | 37% | <.007 |
Table 3 shows that almost all of the servers that ranked at the top in sensitivity (see Tables 1, 2) are also the most specific (FFA3 scored 11th in specificity, and FUG3 scored ninth). The difference in specificity scores between rank-1 (BASD) and rank-9 (FFA3) is 18%. As expected, the servers’ specificities are lower than their sensitivities. For example, while FUG3’s overall sensitivity was 59% (correctly predicting 102 out of the 172 targets), its specificity was only 50%. That is, some of FUG3’s correct predictions had scores lower than that of its first incorrect prediction.
The overall specificities in LB-8 are also higher than those in LB-4 (the most specific servers in LB-4 had an overall specificity just below 50%), possibly suggesting that the new servers are slightly more specific, but also reflecting differences in the set of targets and/or the growth of the databases. As with the overall sensitivity results, the differences among the most specific servers are only slight.
Finally, to obtain an overall, single ranking we have computed the average rank that each server receives in each of the assessment categories: sensitivity on “easy,” sensitivity on “hard,” and specificity, using three different LB evaluation methods. Figure 1 ▶ depicts the average ranks of the top 14 autonomous servers plus that of PDBB, using each of the three evaluation methods. The figure confirms that the exact relative rankings can change slightly depending on the evaluation method used, but it demonstrates that the same top performing servers are identified regardless of how they are evaluated.
Meta-servers
Recent LB and CAFASP experiments have demonstrated that meta-servers clearly outperform the individual, autonomous servers. This is not surprising, since a well-designed meta-predictor should perform at least as good as the best of its input components. During LB-8, only three series of reliable, highly available meta-predictors were assessed: the PCONS/PMOD series (Lundstrom et al. 2001), the 3D-SHOTGUN series (Fischer 2003), and the newer 3D-JURY series (Ginalski et al. 2003). Each of these series includes a number of variants, totaling 15 different meta-servers. The top meta-predictors include representatives of each of the series, and have very similar performances, both in sensitivity and in specificity. Roughly, the best meta-predictors are about 7% more sensitive and more specific than the best of the individual servers. The difference in sensitivity is lower among the easy targets and more significant among the hard targets, of which, half are correctly predicted. The 3D-JURY series of meta-predictors, based on principles very similar to those of the PCONS/PMOD and 3D-SHOTGUN series, were developed during the last CAFASP experiment. While the PCONS/PMOD and 3D-SHOTGUN series use a small, fixed number of other autonomous servers as input (e.g., the 3DS3 3D-SHOTGUN meta-predictor uses as few as two external servers), the 3D-JURY servers are in fact meta-meta-predictors because they can use all the available information from other servers and other meta-servers, including those of the PCONS/PMOD and 3D-SHOTGUN series. Consequently, in LB-8, some of the meta-meta-predictors from the 3D-JURY series appear to be slightly superior to the others (see the LB-8 Web-pages for details). Despite the superior performance of meta-predictors, their utility is hampered by their dependence on external services and by their slow response time, sometimes requiring days before they can return a prediction. Local, autonomous, and fast servers such as SHGU and Meta-BASIC, that apply the meta-prediction principles on locally generated data, overcome some of these limitations because they thus provide the user with an increased performance both in correctness of the predictions and in response time.
Other servers
There were a number of other new, autonomous servers that participated in LB-8 for the first time, but did not rank at the top. Some of these were “unofficial” servers that entered LB-8 late, and that could not be properly evaluated because at the time LB-8 was closed they were not yet fully integrated into the LB communication protocol, or because only a small number of their results could be collected. There were also a number of servers that ranked at the top in previous LB or CAFASP experiments that did not participate in LB-8 or simply got lower ranks in LB-8.
Live Bench and CAFASP
Another additional large-scale assessment experiment initiated 2 years ago is PDB-CAFASP (Fischer et al. 2004). PDB-CAFASP is an extension of the LB and CAFASP experiments, with the only difference being the set of targets used. In PDB-CAFASP, only prerelease PDB entries are used as targets. Thus, the predictions in PDB-CAFASP are truly blind (the 3D structure is not known at the time of the prediction). The servers’ predictions are collected and stored until the 3D structures become available, at which point they can be evaluated and are reported in the PDB-CAFASP Web site. One attractive feature of PDB-CAFASP is that users can obtain in silico models for those proteins whose 3D structures have been solved but whose 3D coordinates are not yet publicly available. Unfortunately, not all the LB-8 servers participate in PDB-CAFASP, and an analysis of the performance of the various servers in PDB-CAFASP is out of the scope of this paper; we refer the readers to the corresponding Web sites at http://www.cs.bgu.ac.il/~dfischer/CAFASP3 and http://bioinfo.pl/PDB-Preview for details.
Based of the (slight) progress observed in LB-8, we expect that this year, the ongoing LB-9 and CAFASP-4 experiments will demonstrate further progress in automatic structure prediction. The past success of meta-predictors will probably result in the proliferation of new and better meta-predictors, which will continue to challenge the best human predictors. However, meta-predictors can only be as good as their components. As in LB-8, we expect that new, better, autonomous servers will continue to be developed. Probably one of the main lessons from LB-8 is that the newly developed, top-ranking autonomous servers BASD, BASP, MBAS, SFST, and STMP apply mainly sequence-based methods. This may suggest that most of the progress observed in LB-8 is focused on better recognition and modeling of relatively close family members. Future experiments may help identify whether any progress exists for the harder cases.
To extend the scope of CAFASP’s assessment, CAFASP-4 experiment this year introduced two new subcategories: assessment of domain prediction servers (DP), and model quality assessment programs (MQAPs). Identifying domains in a protein sequence is an essential component of the structure (and function) prediction process. The DP subcategory is aimed at evaluating the performance of current methods. The MQAP subcategory is aimed at evaluating the performance of methods that assign energies, pseudoenergies, or simply scores to a given model. Being able to identify near-native models is an important aspect of structure prediction, not only for ab initio methods, but also for refinement procedures. Past evaluations have demonstrated that MQAPs are not very good at this task, and this new subcategory will attempt to identify the strengths and limitations of current methods. Further information about the upcoming CAFASP-4 experiment can be obtained at http://www.cs.bgu.ac.il/~dfischer/CAFASP4.
Disclaimer
The authors are at the same time the organizers of CAFASP and LB and the developers of participating servers (including the autonomous servers SHGU, the BAS and ORF series, and the meta-servers of the 3D-SHOTGUN and 3D-JURY series). Thus, the results presented here cannot be considered as independent. However, it is important to emphasize that the results were obtained by fully automated evaluation methods, identical to those used in previous experiments.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04888805.
References
- Abbott, A. 2001. Computer modellers seek out “ten most wanted” proteins. Nature 409 4. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki, J.M., Elofsson, A., Fischer, D., and Rychlewski, L. 2001. LiveBench-1: Continuous benchmarking of protein structure prediction servers. Protein Sci. 10 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, D. 2000. Hybrid fold recognition: Combining sequence derived properties with evolutionary information. Pac Symp. Biocomput. 119–130. [PubMed]
- ———. 2003. 3D-SHOTGUN: A novel, cooperative, fold-recognition meta-predictor. Proteins 51 434–441. [DOI] [PubMed] [Google Scholar]
- Fischer, D. and Eisenberg, D. 1997. Assigning folds to the proteins encoded by the genome of Mycoplasma genitalium. Proc. Natl. Acad. Sci. 94 11929–11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, D. and Rychlewski, L. 2003. The 2002 Olympic Games of protein structure prediction. Protein Eng. 16 157–160. [DOI] [PubMed] [Google Scholar]
- Fischer, D., Elofsson, A., and Rychlewski, L. 2000. The 2000 Olympic Games of protein structure prediction; fully automated programs are being evaluated vis-a-vis human teams in the protein structure prediction experiment CAFASP2. Protein Eng. 13 667–670. [DOI] [PubMed] [Google Scholar]
- Fischer, D., Baker, D., and Moult, J. 2001. We need both computer models and experiments. Nature 409 558. [DOI] [PubMed] [Google Scholar]
- Fischer, D., Rychlewski, L., Dunbrack Jr., R.L., Ortiz, A.R., and Elofsson, A. 2003. CAFASP3: The third critical assessment of fully automated structure prediction methods. Proteins 53(Suppl 6): 503–516. [DOI] [PubMed] [Google Scholar]
- Fischer, D., Pas, J., and Rychlewski, L. 2004. The PDB-preview database: A repository of in-silico models of “on-hold” PDB entries. Bioinformatics 20 2482–2484. [DOI] [PubMed] [Google Scholar]
- Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. 2003. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19 1015–1018. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20 478–480. [DOI] [PubMed] [Google Scholar]
- Karplus, K. and Hu, B. 2001. Evaluation of protein multiple alignments by SAM-T99 using the BAliBASE multiple alignment test set. Bioinformatics 17 713–720. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299 499–520. [DOI] [PubMed] [Google Scholar]
- Lattman, E.E. 2003. Fifth Meeting on the Critical Assessment of Techniques for Protein Structure Prediction. Proteins 53(Suppl. 6): 33. [DOI] [PubMed] [Google Scholar]
- Lundstrom, J., Rychlewski, L., Bujnicki, J., and Elofsson, A. 2001. Pcons: A neural-network-based consensus predictor that improves fold recognition. Protein Sci. 10 2354–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L.J. and Jones, D.T. 2003. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics 19 874–881. [DOI] [PubMed] [Google Scholar]
- Pawlowski, K., Rychlewski, L., Zhang, B., and Godzik, A. 2001. Fold predictions for bacterial genomes. J. Struct. Biol. 134 219–231. [DOI] [PubMed] [Google Scholar]
- Rost, B. and Eyrich, V.A. 2001. EVA: Large-scale analysis of secondary structure prediction. Proteins Suppl 5 192–199. [DOI] [PubMed] [Google Scholar]
- Rychlewski, L., Jaroszewski, L., Li, W., and Godzik, A. 2000. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 9 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlewski, L., Fischer, D., and Elofsson, A. 2003. LiveBench-6: Large-scale automated evaluation of protein structure prediction servers. Proteins 53 (Suppl 6): 542–547. [DOI] [PubMed] [Google Scholar]
- Shi, J., Blundell, T.L., and Mizuguchi, K. 2001. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310 243–257. [DOI] [PubMed] [Google Scholar]
- Siew, N., Elofsson, A., Rychlewski, L., and Fischer, D. 2000. MaxSub: An automated measure for the assessment of protein structure prediction quality. Bioinformatics 16 776–785. [DOI] [PubMed] [Google Scholar]