Abstract
The three-dimensional structure of the rhodanese homology domain At4g01050(175–195) from Arabidopsis thaliana has been determined by solution nuclear magnetic resonance methods based on 3043 upper distance limits derived from NOE intensities measured in three-dimensional NOESY spectra. The structure shows a backbone root mean square deviation to the mean coordinates of 0.43 Å for the structured residues 7–125. The fold consists of a central parallel β-sheet with five strands in the order 1–5–4–2–3 and arranged in the conventional counterclockwise twist, and helices packing against each side of the β-sheet. Comparison with the sequences of other proteins with a rhodanese homology domain in Arabidopsis thaliana indicated residues that could play an important role in the scaffold of the rhodanese homology domain. Finally, a three-dimensional structure comparison of the present noncatalytic rhodanese homology domain with the noncatalytic rhodanese domains of sulfurtransferases from other organisms discloses differences in the length and conformation of loops that could throw light on the role of the noncatalytic rhodanese domain in sulfurtransferases.
Keywords: structural genomics, protein structure, rhodanese, At4g01050, high-throughput NMR, Arabidopsis thaliana
The RIKEN Structural Genomics/Proteomics Initiative (RSGI) is one of the major structural genomics projects in Japan. Its principal goals are to determine the three-dimensional structures of bacterial, mammalian, and plant proteins by X-ray crystallography and NMR spectroscopy, and to perform functional analyses with the target proteins. Arabidopsis thaliana was chosen by the RSGI within the framework of the National Project on Protein Structural and Functional Analyses as one of the target organisms for determining three-dimensional structures of representative members from all known protein families (Yokoyama et al. 2000). To select the target proteins, the collected cDNAs are clustered into families on the basis of their amino acid sequences, and families with no experimentally determined structures are selected. The most suitable protein or proteins within a selected family are produced by cell-free expression (Yokoyama 2003) and subjected to structure determination and functional studies. Domains within a protein are identified by a combination of computational and experimental methods, including limited proteolysis and NMR screening (Yokoyama 2003).
Here we report the solution structure of the 121-residue rhodanese homology domain protein comprising residues 175–295 from the gene At4g01050 on chromosome 4 from A. thaliana (Mayer et al. 1999), At4g01050(175–295), for which conventional sequence alignment did not reveal significant homology with proteins of known structure or function. Nevertheless, more sensitive algorithms for multiple sequence alignments such as Pfam (Bateman et al. 2002) provided tentative evidence that this sequence may have a rhodanese fold.
Structural modules of rhodanese domains have now been found in all the three major evolutionary phyla, even though the existence of rhodaneses in plants was highly controversial until different rhodanese isoforms were characterized in A. thaliana (Hatzfeld and Saito 2000; Nakamura et al. 2000; Papenbrock and Schmidt 2000). Rhodanese homology domains can be found as single-domain proteins, as tandemly repeated modules in which only the C-terminal domain bears the properly structured active site, or as members of multidomain proteins. Rhodanese homology domains are classified as catalytic when they contain a characteristic catalytic cysteine or as noncatalytic or inactive when another, often acidic or nonpolar, residue replaces the active-site cysteine.
In the last years, a considerable number of proteins with a rhodanese homology fold have been detected. The rhodanese domain fold was first observed in the crystal structure of bovine mitochondrial rhodanese, rhdbov (Ploegman et al. 1978), and later in the crystal structure of Azotobacter vinelandii rhodanese, rhdA (Bordo et al. 2000). These proteins are composed of two rhodanese domains with the same fold, of which the C-terminal domain shows the catalytic sulfur-transferase activity. However, the crystal structure of the GlpE protein from Escherichia coli, which displays cyanide sulfurtransferase activity in in-vitro assays (Ray et al. 2000), consists of a single catalytic rhodanese domain (Spallarossa et al. 2001), indicating that the non-catalytic N-terminal domain is not strictly necessary for the catalysis (Bordo and Bork 2002). A catalytic rhodanese domain was also observed in the solution structure of the periplasmic polysulfide-sulfurtransferase (Sud) protein (Lin et al. 2004), which has been proposed to serve as a polysulfide binding and transferase protein (Klimmek et al. 1998). Catalytic rhodanese domains have also been found in the crystal structures of the human cell cycle control phosphatases Cdc25A (Fauman et al. 1998) and Cdc25B (Reynolds et al. 1999). In spite of their very weak sequence similarity with the sulfurtransferases, sensitive profile analysis indicated that the two enzyme families share a common evolutionary origin (Hofmann et al. 1998). On the other hand, a noncatalytic rhodanese homology domain was detected in the crystal structure of the noncatalytic EB domain of the MAPK phosphatase MKP-3 (Farooq et al. 2001). In addition, the results of SMART resource (Letunic et al. 2004) provided indications that other, functionally distinct proteins such as dual-specific phosphatases, several ubiquitinating enzymes, and certain stress-response proteins may share the noncatalytic rhodanese homology domain (Fauman et al. 1998; Hofmann et al. 1998). It has been suggested that the frequent association of noncatalytic rhodanese homology domains with other protein domains is a consequence of its distinct regulatory role, possibly in connection with signaling (Bordo and Bork 2002).
In spite of the presence of catalytic or noncatalytic rhodanese homology domains in many living organisms, including bacteria, plants, and vertebrates, a possible general functional role and substrate specificity of the rhodanese homology domain has not yet been established unambiguously, and the role of the noncatalytic rhodanese homology domains, either found in sulfurtransferases or associated to other proteins, remains to be elucidated.
Results
Resonance assignment
The polypeptide backbone resonance assignments are complete except for the amide proton of Gly1 and the α protons of Gly134 (see Materials and Methods for the numbering of residues) in the nonnative expression tail regions (Pantoja-Uceda et al. 2004). The side-chain assignments of nonlabile protons are complete with the exception of Hζ of Phe33, Hδ of Lys58, Hɛ/Hζ of Phe83, and Hδ of Lys127. Among the labile side-chain protons, the amide groups of all asparagine and glutamine residues and the ɛ-proton resonance of Arg28 were assigned using intraresidual NOEs (Wüthrich 1986). The hydroxyl proton resonances of Thr75 and Ser87 could be observed and assigned. Overall, the assignments cover 98.4% of the chemical shifts of the nonlabile protons and backbone amide protons (Pantoja-Uceda et al. 2004). For all Xxx-Pro bonds, the trans conformation was confirmed independently by intense Xxx (Hα)-Pro (Hδ) sequential NOESY cross peaks (Wüthrich 1986) and by the 13Cβ and 13Cγ chemical shift differences (Schubert et al. 2002).
Structure determination
Statistics about the quality and precision of the 20 energy-minimized conformers that represent the solution structure of At4g01050(175–295) are summarized in Table 1. The structures are well defined and show excellent agreement with the experimental data. Of the 5411 cross peaks that had been identified in the three three-dimensional NOESY spectra, 98% could be assigned by the program CYANA (Herrmann et al. 2002; Güntert 2003). Almost 23 NOE distances constraints per residue, including 1112 long-range distance constraints between protons five or more residues apart in the sequence, were used in the final structure calculations with CYANA and in the subsequent restrained energy-refinement with the program OPALp (Koradi et al. 2000). No violations of NOE distance constraints larger than 0.16 Å were observed in the final 20 conformers, and no hydrogen bond constraints were used. The precision of the structure is characterized by RMSD values to the mean coordinates of 0.43 Å for the backbone and 0.84 Å for all heavy atoms excluding the unstructured regions at the chain termini of residues 1–7 and 126–134. Finally, the quality of the structures is also reflected by 79.2% of the (φ,ψ) backbone torsion angle pairs being found in the most favored regions and 20.4% being found within the additionally allowed regions of the Ramachandran plot according to the program PROCHECK-NMR (Laskowski et al. 1996).
Table 1.
Statistics for the NMR solution structure of At4g01050 (175–295)
| NOE distance constraints: | |
| Short-range, |i − j| ≤1 | 1392 |
| Medium-range, 1 < |i − j| < 5 | 539 |
| Long-range, |i −j| ≥5 | 1112 |
| Total | 3043 |
| Maximal violation | 0.14 Å |
| Number of violations > 0.2 Å | 0 |
| Final CYANA target function value | 0.74 Å2 |
| AMBER energy | −5021 kcal/mol |
| RMS deviations from ideal geometry: | |
| Bond lengths | 0.014 Å |
| Bond angles | 1.80° |
| RMSD to mean coordinates: | |
| Backbone N, Ca, C′of residues 7–125 | 0.43 Å |
| All heavy atoms of residues 7–125 | 0.84 Å |
| PROCHECK Ramachandran plot statistics: | |
| Most favorable regions | 78.6% |
| Additional allowed regions | 20.9% |
| Generously allowed regions | 0.4% |
| Disallowed regions | 0.1% |
Average values over the 20 final energy-minimized CYANA conformers. The final CYANA target function value was computed for the structure before energy minimization with OPALp.
Solution structure
At4g01050(175–295) is a small α /β domain with a central five-stranded parallel β-sheet in the order 1–5–4–2–3, (β1: Ser6-Ser8; β2: Ala22-Asp26; β3: Val49-Ser50; β4: Thr76-Leu80; β5: Ser101-Lys106), arranged in the conventional counterclockwise twist. This parallel β-sheet is surrounded by four α-helices (α1: Ala9-Thr18; α2: Thr30-Val36; α3: Gly55-Leu67; α4: Ser87-Asn97) and two 310-helices (α3′: Pro72-Asn74; α5: Leu116-Ser118). In addition, a small, two-stranded, antiparallel β-sheet is formed between β3′ (Ser38-Pro39) in the region connecting α2 and β3 and β6 (Trp122-Arg123), which may be involved in structural stabilization of the C-terminal segment of the protein.
The α-helices have most of the characteristic hydrogen bonds between the CO of residue i and the NH of residue i+4, whereas canonical hydrogen bonds linking the five parallel β-strands are also found in the calculated structures. The two-stranded, antiparallel β-sheet is stabilized by the hydrogen bonds between Ser38(HN) and Ile123(O) and Trp122(Hɛ1) and Val36(O) that are formed in 19 or more of the 20 conformers. Furthermore, hydrogen bonds involving side-chains have been observed in most of the conformers for Thr75(Hγ1) with Asp71(O) in helix α3′, Asn97(Hδ) with Leu93(O) in helix α4, and Trp115(Hɛ1) with Pro121(O) and Asn117(Hδ) with Gly111(O).
Discussion
Sequence analysis of expected rhodanese homology domains in A. thaliana
A PSI-BLAST (Altschul et al. 1997) search for sequence alignment revealed 13 sequences with an E value better than the standard threshold (0.005) in the whole database. These sequence homologs have unknown function and are found either in bacteria or eukaryotes. On the other hand, a more sensitive algorithm for multiple sequence alignments, Pfam (Bateman et al. 2002), provided tentative evidence that our sequence might have a rhodanese fold with an E value of 0.091. This evidence enabled an initial alignment of sequences of proteins from A. thaliana with an expected rhodanese homology domain. Nineteen different protein sequences containing 22 putative rhodanese homology domains could be identified in the A. thaliana genome. Three of these are grouped as tandem repeats, with the C-terminal domain hosting the active-site cysteine residue, whereas in the N-terminal domain, the same position is occupied by an aspartic acid. Twelve sequences are for single catalytic domains, and four domains are associated with other protein domains. Of these four domains, three are catalytic and one is a noncatalytic domain, again with the active-site cysteine replaced by an aspartic acid. Furthermore, 6 of 12 single catalytic domains are reported to be associated with the process of leaf senescence in A. thaliana (Oh et al. 1996), while the other six have unknown function. The proteins in which a catalytic rhodanese domain is associated with other domains have been described as peptidyl-prolyl cis-trans isomerases and molybdenum cofactors (Nieder et al. 1997), whereas the multidomain protein containing a noncatalytic rhodanese homology domain has been described as an extracellular calcium-sensing receptor (Han et al. 2003). Finally, the three tandem rhodanese domains are mercaptopyruvate sulfurtransferases (Hatzfeld and Saito 2000; Nakamura et al. 2000; Papenbrock and Schmidt 2000).
In spite of the low overall sequence identity, a few residues are conserved in the β-strands β2 and β4, which constitute the structural core of the protein. In β2, the sequence XDXR/S (X being hydrophobic residues) is conserved. In particular, Asp26, located in the C-terminal of β2, is strictly conserved in all the sequences. In β4, a set of four hydrophobic residues preceding the position occupied by cysteine or aspartic acid is conserved, except in the C-terminal domain of the sulfurtransferases, in which a serine residue instead of a hydrophobic residue precedes the catalytic cysteine. According to these data, we can conclude that these residues play an important role in the scaffold formation of the protein.
Rhodanese homology domains
A search for structural homologs using the DALI server (Holm and Sander 1996) revealed a high structural similarity between the present structure and the periplasmic poly-sulfide-sulfurtransferase (Sud) protein (Lin et al. 2004); catalytic and noncatalytic domains of the sulfur transfer proteins from Azotobacter vinelandii, rhdA (Bordo et al. 2000), and from Bos taurus, rhdbov (Gliubich et al. 1996); GlpE (Spallarossa et al. 2001); the catalytic domain of the Cdc25A phosphatase enzyme (Fauman et al. 1998); and the noncatalytic domain EB of the MAPK phosphatase MKP-3 (Farooq et al. 2001), all of which consist of rhodanese homology modules.
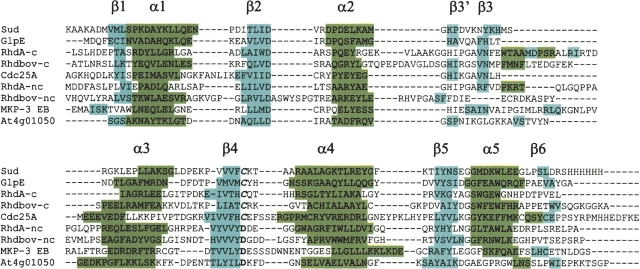
An alignment of the sequences of these proteins based on the optimal structural superposition with At4g01050(175–295) is shown in Figure 1 ▶. The extension and location of the regular secondary structure elements approximately coincide in all these proteins. However, some differences can be found. The catalytic and noncatalytic domains of rhdA and the catalytic domain of rhdbov have additional regular secondary structure elements inserted between β3 and α3. In addition, the helix α2 is shifted in the noncatalytic domain of rhdbov, while the β-strands β3′ and β3 are shifted in the noncatalytic domain EB of the MAPK phosphatase MKP-3. On the other hand, At4g01050(175–295) has a longer loop between β3 and β3′ and a shorter helix α5.
Figure 1.
Structure-based sequence alignment of the rhodanese and rhodanese homology domains with known structure according to the program DALI (Holm and Sander 1996). Domain names are as in Table 2. The catalytic position is shown in italics. Residues in helices or β-strands are highlighted in green and in cyan, respectively, and α-helices and β-strands are labeled.
The structure-based sequence alignment of these proteins allowed the identification of conserved amino acids that may define a signature of functional or structural importance. In spite of the dissimilar amino acid sequences of these proteins, several conserved residues at the ends of the β-strands β2 and β4 can be found. At the C-terminal end of β2, the sequence DXR is found in all of these proteins except the noncatalytic domains of rhdbov and rhdA, in which the arginine residue is replaced by a serine or a threonine, respectively. In the catalytic domain of the sulfur transfer proteins, the catalytic cysteine residue (Miller-Martini et al. 1994) is located in the loop between β4 and α4 (e.g., in the Cdc25 phosphatase, in GlpE, and in the Sud protein), whereas in At4g01050(175–295), in the EB domain of MKP-3 (Farooq et al. 2001), and in the noncatalytic domains of the sulfur transfer proteins, this sequence position is occupied by an aspartic acid. Although the MKP-3 EB domain is a noncatalytic domain of MKPs, it has recently been suggested that it is important for substrate specificity (Farooq et al. 2001), while the role of the noncatalytic rhodanese homology domain in sulfurtransferase proteins is not clear.
A summary of the structural alignment statistics is presented in Table 2. The best structural agreement is found for the monomer of the homodimeric periplasmic polysulfide-sulfurtransferase (Sud) protein (PDB entry 1QXN), where the only difference is the presence of an additional helix near the N terminus. The single catalytic rhodanese domain of the GlpE protein from E. coli (1GNO) displays a substantial shortening of the loop connecting β3′ with β3 and a different orientation of the helix α3. In the catalytic domain of the human cell cycle control phosphatase Cdc25A (1C25), the most significant differences are the orientation of the helices α1 and α3 and a shorter connecting loop between β3′ and β3.
Table 2.
Summary of structural alignment statistics
| PDB code | Z-score DALI | RMSD (Å) | Aligned Cα/total Cα | Sequence identity (%) | |
| Sud protein | 1QXN | 13.4 | 2.8 | 116/137 | 19 |
| GlpE | 1GN0 | 13.3 | 2.1 | 104/108 | 20 |
| RhdA-non catalytic domain | 1E0C | 10.3 | 2.7 | 104/132 | 14 |
| RhdA-catalytic domain | 1E0C | 10.6 | 2.6 | 108/126 | 15 |
| Rhdbov-non catalytic domain | 1RHS | 9.7 | 3.0 | 110/147 | 15 |
| Rhdbov-catalytic domain | 1RHS | 9.7 | 3.0 | 114/137 | 15 |
| Cdc25A | 1C25 | 9.3 | 2.5 | 108/161 | 13 |
| MKP-3 EB domain | 1HZM | 2.8 | 3.9 | 92/154 | 11 |
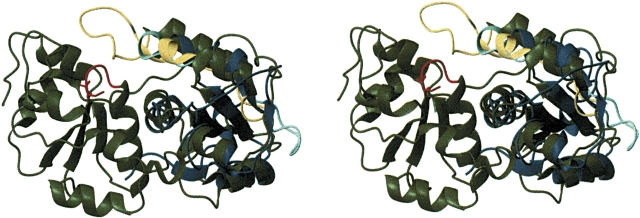
A superposition of the structure of At4g01050(175–295) with the noncatalytic domain of rhdA (1EOC) is shown in Figure 2 ▶. In the noncatalytic domains of rhdA and rhdbov, the loop connecting α2–(β3′)– β3 is shorter than in At4g01050(175–295), while the orientation of the helix α1, close to this loop, changes in the sulfurtransferases in order to maintain the compactness of the three-dimensional structure. Differences are also found in the length of the loops β2–α2 and β3–α3 located in the interdomain interface of rhdA and rhdbov, and thus close to the active-site cysteine, as well as in the orientation of the helix α3. At4g01050(175–295) exhibits the shortest loops. In rhdbov, both loops are longer than in At4g01050(175–295), whereas in the case of rhdA, only the loop β3–α3 is longer than in At4g01050(175–295). The different lengths of these loops could restrict the access to the active site pocket and induce a change in the shape of the protein surface in the proximity of the active site.
Figure 2.
Stereo view of At4g01050(175–295) superimposed on the non-catalytic domain of the sulfur transfer protein from Azotobacter vinelandii, rhdA (1EOC), displayed as ribbon diagrams (blue and green, respectively); the active-site loop in the catalytic domain of rhdA is drawn in red. The loops connecting α2–β3′, β3′– β3, and β3–α3 in At4g01050(175–295) are shown in cyan, and the loop connecting α2–β3 and β3–α3 in the noncatalytic domain of rhdA is shown in yellow.
Materials and methods
Protein sample
For the structure determination, a single 1.1-mM uniformly 13C-and 15N-labeled sample was prepared in 20 mM phosphate buffer (pH 6.0), 100 mM NaCl, 1 mM dithiothreitol, 0.02% NaN3 with the addition of D2O to 10% v/v, as described earlier (Pantoja-Uceda et al. 2004) using cell-free protein synthesis (Kigawa et al. 2004). The protein sample used for the NMR measurements comprised 134 amino acid residues, numbered 1–134 throughout this article. Residues 8–128 constitute the At4g01050(175–295) domain that is surrounded by nonnative flanking sequences of residues 1–7 and 129–134 that are related to the expression and purification system.
NMR spectroscopy
NMR experiments were performed at 25°C on Bruker DRX 600 or Bruker AV 800 spectrometers operating at 600.280 and at 800.350 MHz proton frequency, respectively. 1H, 15N, and 13C chemical shifts were referenced relative to the frequency of the 2H lock resonance of water.
Two two-dimensional and 11 three-dimensional NMR experiments were recorded with the uniformly 13C-/15N-labeled sample for the determination of the sequence-specific polypeptide backbone and side-chain chemical shift assignments (Pantoja-Uceda et al. 2004). For the collection of conformational constraints, a three-dimensional 15N-edited NOESY-HSQC and two 13C-edited NOESY-HSQC spectra for the aliphatic and aromatic regions (Muhandiram et al. 1993) were recorded with 80 ms mixing time. Water suppression was achieved either by selective presaturation or by including a WARTERGATE module (Piotto et al. 1992) in the original pulse sequences before the acquisition. For the three-dimensional 15N-edited NOESY-HSQC, the data matrix in the time domain was composed of 1024(HN) × 92(15N) × 256(1H) points. For the 13C-edited NOESY-HSQC spectra in the aliphatic and aromatic regions, the data matrices in the time domain were composed of 1024/1024(1H) × 68/64(13C) × 232/256 (1H) points. The time-domain data were multiplied by a squared sine-bell window function, with the corresponding shift optimized for every spectrum and zero-filled to at least twice the acquired number of data points before Fourier transformation. Baseline correction was applied in all dimensions. The raw NMR data were processed on a Linux computer, using the program NMRPipe (Delaglio et al. 1995). The program NMRView (Johnson and Blevins 1994) was used for interactive spectrum analysis.
Structure calculation
Peak lists for the NOESY spectra were generated by interactive peak picking, and peak volumes were determined by the automatic integration function of NMRView (Johnson and Blevins 1994). The three-dimensional structure was determined by combined automated NOESY cross-peak assignment (Herrmann et al. 2002) and structure calculation with torsion angle dynamics (Güntert et al. 1997) implemented in the program CYANA (Güntert 2003). The standard CYANA protocol of seven iterative cycles of NOE assignment and structure calculation, followed by a final structure calculation, was applied. Stereo-specific assignments for 61 isopropyl methyls and methylene groups were determined by the GLOMSA method (Güntert et al. 1991) before the final structure calculation by analyzing the structures obtained in the preceding, seventh NOE assignment/structure calculation cycle. Weak constraints on (φ,ψ) torsion angle pairs and on side-chain torsion angles between tetrahedral carbon atoms were used temporarily during the NOE assignment/structure calculation cycles in order to favor allowed regions of the Ramachandran plot and staggered rotamer positions, respectively. In each cycle, the structure calculation started from 100 randomized conformers and the standard CYANA-simulated annealing schedule (Güntert et al. 1997) was used with 10,000 torsion angle dynamics steps per conformer. The 20 conformers with the lowest final CYANA target function values were subjected to restrained energy minimization in a water shell with the program OPALp (Koradi et al. 2000), using the AMBER force field (Cornell et al. 1995) and only the NOE upper distance limits as constraints. PROCHECK-NMR (Laskowski et al. 1996) and MOLMOL (Koradi et al. 1996) were used to validate and to visualize the final structures, respectively.
Protein Data Bank accession number
The 20 energy-minimized conformers of the At4g01050(175–295) with the lowest CYANA target function values in the final structure calculation were deposited in the Protein Data Bank (PDB entry 1VEE).
Acknowledgments
We thank Yuki Kamewari and Hiroaki Hamana for technical support and Tomoko Nakayama for help with the preparation of the manuscript. D.P. was supported by a pre-doctoral grant from the Spanish Ministry of Science and Technology. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and by the Tatsuo Miyazawa Memorial Program of RIKEN Genomic Sciences Center.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041138705.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo, D. and Bork, P. 2002. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 3 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo, D., Deriu, D., Colnaghi, R., Carpen, A., Pagani, S., and Bolognesi, M. 2000. The crystal structure of a sulfurtransferase from Azotobacter vinelandii highlights the evolutionary relationship between the rhodanese and phosphatase enzyme families. J. Mol. Biol. 298 691–704. [DOI] [PubMed] [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117 5179–5197. [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Farooq, A., Chaturvedi, G., Mujtaba, S., Plotnikova, O., Zeng, L., Dhalluin, C., Ashton, R., and Zhou, M.M. 2001. Solution structure of ERK2 binding domain of MAPK phosphatase MKP-3: Structural insights into MKP-3 activation by ERK2. Mol. Cell 7 387–399. [DOI] [PubMed] [Google Scholar]
- Fauman, E.B., Cogswell, J.P., Lovejoy, B., Rocque, W.J., Holmes, W., Montana, V.G., Piwnica-Worms, H., Rink, M.J., and Saper, M.A. 1998. Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell 93 617–625. [DOI] [PubMed] [Google Scholar]
- Gliubich, F., Gazerro, M., Zanotti, G., Delbono, S., Bombieri, G., and Berni, R. 1996. Active site structural features for chemically modified forms of rhodanese. J. Biol. Chem. 271 21054–21061. [DOI] [PubMed] [Google Scholar]
- Güntert, P. 2003. Automated NMR protein structure calculation. Prog. NMR Spectrosc. 43 105–125. [Google Scholar]
- Güntert, P., Braun, W., and Wüthrich, K. 1991. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217 517–530. [DOI] [PubMed] [Google Scholar]
- Güntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Han, S., Tang, R., Anderson, L.K., Woerner, T.E., and Pei, Z.M. 2003. A cell surface receptor mediates extracellular Ca(2+) sensing in guard cells. Nature 425 196–200. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, Y. and Saito, K. 2000. Evidence for the existence of rhodanese (thio-sulfate:cyanide sulfurtransferase) in plants: Preliminary characterization of two rhodanese cDNAs from Arabidopsis thaliana. FEBS Lett. 470 147–150. [DOI] [PubMed] [Google Scholar]
- Herrmann, T., Güntert, P., and Wüthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319 209–227. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., Bucher, P., and Kajava, A.V. 1998. A model of Cdc25 phosphatase catalytic domain and Cdk-interaction surface based on the presence of a rhodanese homology domain. J. Mol. Biol. 282 195–208. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1996. Mapping the protein universe. Science 273 595–603. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Kigawa, T., Yabuki, T., Matsuda, N., Matsuda, T., Nakajima, R., Tanaka, A., and Yokoyama, S. 2004. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Genomics 5 63–68. [DOI] [PubMed] [Google Scholar]
- Klimmek, O., Kreis, V., Klein, C., Simon, J., Wittershagen, A., and Kroger, A. 1998. The function of the periplasmic Sud protein in polysulfide respiration of Wolinella succinogenes. Eur. J. Biochem. 253 263–269. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Güntert, P. 2000. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 124 139–147. [Google Scholar]
- Laskowski, R.A., Rullmann, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32 D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.J., Dancea, F., Löhr, F., Klimmek, O., Pfeiffer-Marek, S., Nilges, M., Wienk, H., Kroger, A., and Rüterjans, H. 2004. Solution structure of the 30 kDa polysulfide-sulfur transferase homodimer from Wolinella succinogenes. Biochemistry 43 1418–1424. [DOI] [PubMed] [Google Scholar]
- Mayer, K., Schuller, C., Wambutt, R., Murphy, G., Volckaert, G., Pohl, T., Dusterhoft, A., Stiekema, W., Entian, K.D., Terryn, N., et al. 1999. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402 769–777. [DOI] [PubMed] [Google Scholar]
- Miller-Martini, D.M., Hua, S., and Horowitz, P.M. 1994. Cysteine 254 can cooperate with active site cysteine 247 in reactivation of 5,5′-dithiobis(2-nitrobenzoic acid)-inactivated rhodanese as determined by site-directed mutagenesis. J. Biol. Chem. 269 12414–12418. [PubMed] [Google Scholar]
- Muhandiram, D.R., Farrow, N., Xu, G.-Y., Smallcombe, S.H., and Kay, L.E. 1993. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O. J. Magn. Reson. Ser. B 102 317–321. [Google Scholar]
- Nakamura, T., Yamaguchi, Y., and Sano, H. 2000. Plant mercaptopyruvate sulfurtransferases: Molecular cloning, subcellular localization and enzymatic activities. Eur. J. Biochem. 267 5621–5630. [DOI] [PubMed] [Google Scholar]
- Nieder, J., Stallmeyer, B., Brinkmann, H., and Mendel, R.R. 1997. Molybdenum cofactor biosynthesis: Identification of A. thaliana cDNAs homologous to the E. coli sulfotransferase MoeB. In Sulphur metabolism in higher plants (eds. W.J. Cram et al.), pp. 275–277. Backhuys Publishers, Leiden, The Netherlands.
- Oh, S.A., Lee, S.Y., Chung, I.K., Lee, C.H., and Nam, H.G. 1996. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant. Mol. Biol. 30 739–754. [DOI] [PubMed] [Google Scholar]
- Pantoja-Uceda, D., Lopez-Mendez, B., Koshiba, S., Kigawa, T., Shirouzu, M., Terada, T., Inoue, M., Yabuki, T., Aoki, M., Seki, E., et al. 2004. NMR assignment of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana. J. Biomol. NMR 29 207–208. [DOI] [PubMed] [Google Scholar]
- Papenbrock, J. and Schmidt, A. 2000. Characterization of a sulfurtransferase from Arabidopsis thaliana. Eur. J. Biochem. 267 145–154. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Ploegman, J.H., Drent, G., Kalk, K.H., and Hol, W.G. 1978. Structure of bovine liver rhodanese. I. Structure determination at 2.5 Å resolution and a comparison of the conformation and sequence of its two domains. J. Mol. Biol. 123 557–594. [PubMed] [Google Scholar]
- Ray, W.K., Zeng, G., Potters, M.B., Mansuri, A.M., and Larson, T.J. 2000. Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J. Bacteriol. 182 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, R.A., Yem, A.W., Wolfe, C.L., Deibel Jr., M.R., Chidester, C.G., and Watenpaugh, K.D. 1999. Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle. J. Mol. Biol. 293 559–568. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Labudde, D., Oschkinat, H., and Schmieder, P. 2002. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR 24 149–154. [DOI] [PubMed] [Google Scholar]
- Spallarossa, A., Donahue, J.L., Larson, T.J., Bolognesi, M., and Bordo, D. 2001. Escherichia coli GlpE is a prototype sulfurtransferase for the single-domain rhodanese homology superfamily. Structure 9 1117–1125. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Yokoyama, S. 2003. Protein expression systems for structural genomics and proteomics. Curr. Opin. Chem. Biol. 7 39–43. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S., Hirota, H., Kigawa, T., Yabuki, T., Shirouzu, M., Terada, T., Ito, Y., Matsuo, Y., Kuroda, Y., Nishimura, Y., et al. 2000. Structural genomics projects in Japan. Nat. Struct. Biol. 7 943–945. [DOI] [PubMed] [Google Scholar]