Abstract
Carbamoyl phosphate synthetase synchronizes the utilization of two ATP molecules at duplicated ATP-grasp folds to catalyze carbamoyl phosphate formation. To define the dedicated functional role played by each of the two ATP sites, we have carried out pulse/labeling studies using the synthetases from Aquifex aeolicus and Methanococcus jannaschii, hyperthermophilic organisms that encode the two ATP-grasp folds on separate subunits. These studies allowed us to differentially label each active site with [γ-32P]ATP and determine the fate of the labeled γ-phosphate in the synthetase reaction. Our results provide the first direct demonstration that enzyme-catalyzed transfer of phosphate from ATP to carbamate occurs on the more C-terminal of the two ATP-grasp folds. These findings rule out one mechanism proposed for carbamoyl phosphate synthetase, where one ATP acts as a molecular switch, and provide additional support for a sequential reaction mechanism where the γ-phosphate groups of both ATP molecules are transferred to reactants. CP synthesis by subunit C in our single turnover pulse/chase assays did not require subunit N, but subunit N was required for detectable CP synthesis in the traditional continuous assay. These findings suggest that cross-talk between domain N and C is required for product release from subunit C.
Keywords: carbamoyl phosphate synthetase, ATP, ATP-grasp, arginine, glutamine, pyrimidine
Synthesis of carbamoyl phosphate (CP) by carbamoyl phosphate synthetase (CPS) requires the coordinated utilization of two molecules of ATP per reaction cycle, as well as one molecule each of bicarbonate and ammonia (free or derived from glutamine through reaction on the glutamine amidotransferase domain of CPS) (Meister 1989). The ATP molecules react at two CPS domains (termed N for the one closest to the N terminus and C for the one closest to the C terminus) that share sequence identity and appear to have resulted from an ancestral gene duplication event (Lusty et al. 1983). The X-ray structure of Escherichia coli CPS (eCPS) demonstrated that both domains contain ATP-grasp folds and that the two folds are superimposable, with a root mean square deviation (RMSD) of 1.1 Å for 255 equivalent α carbons (Thoden et al. 1997, 1999b). Despite the strong structural similarity of the two ATP-utilizing domains, a wide variety of studies have shown that each of the two ATP molecules plays a dedicated and unique role in the synchronized synthesis of ATP (Meister 1989; Javid-Majd et al. 2000).
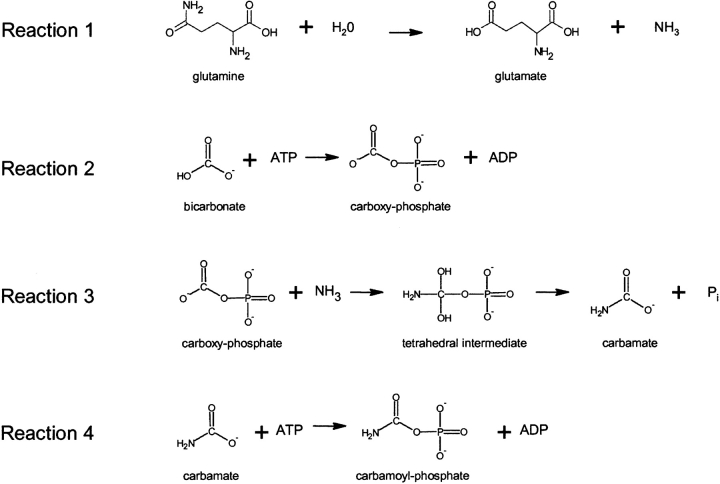
A multi-step CPS mechanism (Fig. 1 ▶) that was suggested by biochemical studies (Meister 1989) has been utilized to assign functions to the domains of the solved eCPS structure (Thoden et al. 1997, 1999b). The first reaction has been directly demonstrated and its localization determined: glutamine binding and cleavage occur on the glutamine amidotransferase domain (Thoden et al. 1999a). Reaction 2, carboxy phosphate formation from bicarbonate plus ATP has been established (Powers and Meister 1978; Raushel and Villafranca 1979; Wimmer et al. 1979) whereas it has not been possible to directly demonstrate carbamate formation (Reaction 3) or phosphorylation of carbamate (Reaction 4). Site-directed mutagenesis studies (Post et al. 1990) and oxidative inactivation data (Alonso et al. 1992), relying on two partial activities as probes, have suggested that carboxy phosphate formation (Reaction 2) is localized to domain N (the more N-terminal of the two ATP-grasp folds) and that carbamoyl phosphate formation (Reaction 4) is localized to domain C (the more C-terminal of the two ATP-grasp folds). However, domains N and C are potentially promiscuous in catalyzing these partial reactions. Bicarbonate-dependent ATPase activity was used as a probe for reaction 2, carboxy phosphate formation, yet bicarbonate might also serve as an analog for carbamate in reaction 4 (Rubio and Cervera 1995). ADP phosphorylation by CP was used as a probe of the reversal of reaction 4 yet might also reflect the reversal of reaction 2, with CP serving as an analog of carboxy phosphate (Rubio and Cervera 1995). In support of this latter possibility, the phosphorylation of ADP by CP is also catalyzed by several biotin-dependent enzymes, where carboxy phosphate is a known intermediate and where CP plays no known role (Knowles 1989). Additionally, phosphorylation of ADP by CP is catalyzed by glutamine synthetase (Tate et al. 1972) and formyltetrahydrofolate synthetase (Buttlaire et al. 1976), where presumably carboxy phosphate is acting as an analog of glutamyl-phosphate and formyl-phosphate, respectively.
Figure 1.
CPS reaction (sequential mechanism).
Inability to localize specific steps of the CPS reaction to domains N and C complicates the analysis of experiments investigating its mechanism. Thus, for instance, it has not been possible to test the critical distinguishing feature of a nucleotide switch mechanism that has been proposed as an alternative mechanism for CPS (Kothe et al. 1997). In both this mechanism and the sequential one of Figure 1 ▶, carboxy phosphate is formed at domain N from bicarbonate and ATP, and nucleophilic ammonia reacts with the activated carbonyl group to yield a tetrahedral intermediate. In the sequential mechanism (Fig. 1 ▶), the intermediate collapses to carbamate with phosphate as the leaving group, and, at domain C the carbamate reacts with ATP to yield ADP and CP. In the switch mechanism, the intermediate collapses directly to CP on domain N, with water as the leaving group, and with domain C acting as an ATP-driven molecular switch that allows the energetically unfavorable reaction to proceed on domain N.
Given that clear identification of the functional roles of domains N and C is required for elucidation of the CPS mechanism, many previous studies have been aimed at physically separating the two domains and analyzing their function. The majority of studies have utilized eCPS since its structure has been solved (Raushel et al. 1992; Guy and Evans 1996; Javid-Majd et al. 2000), but some have utilized eukaryotic CPSs (Guy and Evans 1996; Serre et al. 1999). A variety of CPS domain constructs, with varying boundary positions, have been expressed and analyzed. However, none of the domain N or domain C constructs has allowed definition of the distinct roles of the two ATP sites. The constructs have either been totally inactive (Raushel et al. 1992; Javid-Majd et al. 2000) or have failed to exclusively catalyze those reactions expected for domain-specific roles (Guy and Evans 1996; Serre et al. 1999). An alternative approach to defining the domain N and C functions, site-directed mutagenesis aimed at changing the substrate specificity of either of the two sites from ATP to GTP, is precluded by the architecture of the potassium-binding loop in CPS (Kothe and Powers-Lee 2004).
Whole-genome analysis of the extreme hyperthermophiles Aquifex aeolicus (Deckert et al. 1998) and Methanococcus jannaschii (Bult et al. 1996) has shown that the gene encoding the synthetase component of CPS is split, resulting in separately expressed N and C subunits. Interestingly, the boundary between A. aeolicus CPS (aCPS) subunits coincides almost exactly with that of eCPS domains whereas subunit N of M. jannaschii CPS (mCPS) is shortened by 74 amino acids (Fig. 2 ▶). Subunits N and C of aCPS have previously been expressed and characterized (Ahuja et al. 2001). These subunits were markedly promiscuous in their ability to catalyze the partial reactions. The two subunits had very similar bicarbonate-dependent ATPase activities, with values of 9.62 and 11.0 nmol/min/ mg for N and C subunits, respectively. Additionally, both subunits had significant levels of CP-dependent ATP synthesis activity, with values of 6.81 and 1.78 nmol/min/mg for N and C subunits, respectively. It is especially striking that, in contrast to the general expectation for these probes of specialized domain function, subunit N had more CP-dependent ATP synthesis activity than subunit C and subunit C had more bicarbonate-dependent ATPase activity than subunit N. The possibility that subunits N and C of aCPS have been incorrectly identified is eliminated by the strong sequence conservation with domains N and C of eCPS and the presence of the sequence motif RSSALASKA in aCPS N. This motif is present in every CPS identified so far and is only found in domain N.
Figure 2.
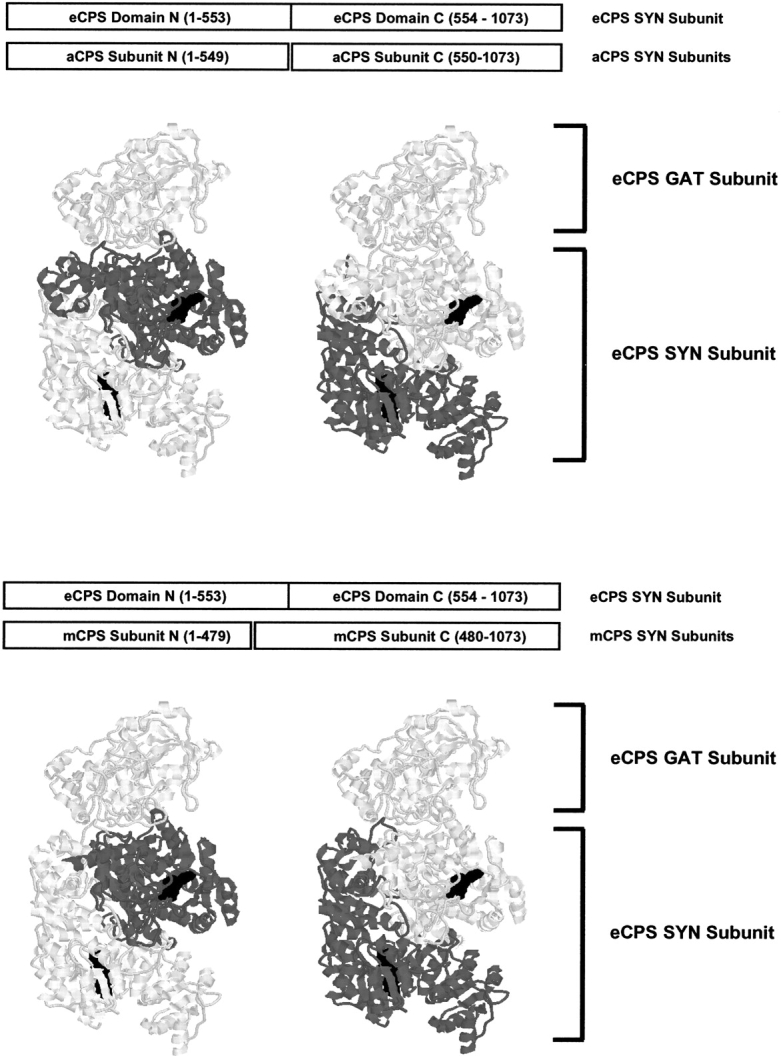
aCPS and mCPS synthetase component domain structures. In both aCPS and mCPS the synthetase component is expressed as two separate subunits. The block diagrams compare the extent of the N and C subunits to domains N and C in eCPS (numbering based on eCPS sequence). In the structure figures, residues in the eCPS structure (PDB file 1BXR) that correspond to subunits N or C of aCPS or mCPS are highlighted in dark gray. Bound nucleotide analogs are shown in black spacefill representation. Although all three CPSs contain a separate glutamine amidotransferase subunit (GAT), it is omitted for clarity with the exception of eCPS GAT in the structure figure, where it aids in orientation.
Although subunits N and C of aCPS do not possess unique partial activities that would allow definition of their functional roles in the coordinated synthesis of CP, they do constitute a unique mechanistic probe in that the isolated subunits are stably expressed and act independently. They also display another critical characteristic (Ahuja et al. 2001): neither subunit N or C can catalyze CP synthesis, yet a mixture of the two subunits displays activity, presumably by forming a heterodimer analogous to the one that occurs on a single polypeptide chain of eCPS (Fig. 2 ▶). We have taken advantage of these aCPS properties to apply a pulse/ chase procedure that differentially labels the ATP molecule bound to each subunit of aCPS and thereby allows definition of the dedicated functional role played by each subunit during synchronized synthesis of CP.
Results and Discussion
Pulse/chase labeling of A. aeolicus N and C subunits with [γ-32P]ATP
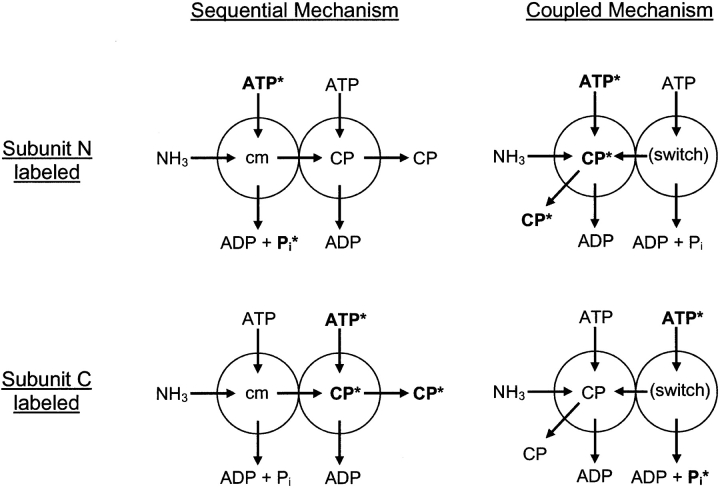
In these studies, a relatively large amount of subunit N or subunit C from aCPS was incubated briefly in a pulse solution containing [γ-32P]ATP, Mg2+ and K+ (to promote binding of the [γ-32P]ATP/Mg2+ complex) and lacking added sodium bicarbonate or ammonium chloride (to minimize ATP turnover). Next the protein was desalted so that free ATP was removed from the solution and the only labeled ATP was bound to the enzyme. The protein:[γ-32P]ATP complex was then added to a solution containing an excess of unlabeled ATP, ammonia, and sodium bicarbonate and either subunit N or C of aCPS. After brief incubation in the chase solution, the products ADP, Pi and CP were separated by paper chromatography and the relative amount of radioactivity associated with each product was determined. As outlined in Figure 3 ▶, the expected result if CP formation is localized to subunit N (nucleotide-switch mechanism) would be formation of [γ-32P]CP only when subunit N was labeled in the pulse solution and then interacted with subunit C in the chase solution to form a hetero-dimer that interacted with ammonia to carry out coordinated synthesis of CP. On the other hand, if CP formation is localized to subunit C (sequential mechanism), [γ-32P]CP would only be formed when subunit C is present in the pulse solution and subsequently formed an N/C heterodimer that interacted with ammonia to carry out coordinated synthesis of CP. As indicated in Figure 3 ▶, synchronized synthesis of CP by both subunits of the heterodimer would yield 32Pi from initial labeling of the subunit that is not dedicated to CP formation. Additional 32Pi would result from the bicarbonate-dependent ATPase activity intrinsic to both isolated subunits and the heterodimer, with this reaction occurring when water nonproductively interacts with the carboxy phosphate intermediate in the absence of an ammonia source. It should be noted that this reaction can occur to some extent in the absence of added bicarbonate, during the pulse labeling, since the estimated endogenous bicarbonate concentration is 0.4 mM (Anderson and Meister 1966) and the aCPS Km for bicarbonate is 7.9 mM (Ahuja et al. 2001).
Figure 3.
Schematic of pulse/chase study setup. Subunits N and C of aCPS or mCPS are represented by spheres (left sphere = subunit N, right sphere = subunit C) and the reactions that are proposed to occur on them in the sequential and coupled mechanism are indicated (cm = carbamate). For each mechanism, labeling of either subunit N or C with [γ-32P]ATP is represented. Labeled substrates and products are shown in bold, with the labeled phosphate indicated with an asterisk.
Localization of CP formation to subunit C of aCPS
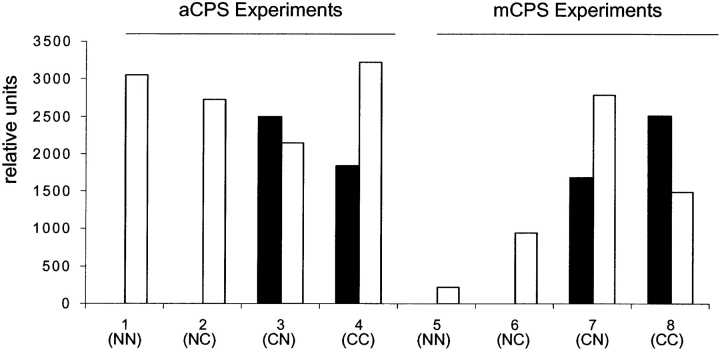
[32P]CP was formed when subunit C was pulse labeled (Experiments 3 and 4, Fig. 4 ▶) and was not formed when subunit N was pulse-labeled (Experiments 1 and 2, Fig. 4 ▶). These findings constitute the first direct demonstration of localization of CP formation on any CPS. Since the two ATP molecules play dedicated roles in the CPS reaction, these findings also establish that Pi is formed on domain N of CPSs. The fact that CP is derived from the C-terminal ATP fold of CPS is fully consistent with the sequential mechanism defined by reactions 1–4 (Fig. 1 ▶). However, the C-domain localization is not consistent with an essential feature of the switch mechanism and clearly rules out its functioning. It should be noted that earlier studies (Raushel et al. 1998; Rubio et al. 1998) have ruled out specific features of the switch mechanism as originally formulated but were not able to directly test this and other critical features. 32Pi formation was observed under all conditions utilized for the pulse/chase studies (Experiments 1–4, Fig. 4 ▶), as would be expected from the bicarbonate-dependent ATPase activity intrinsic to each isolated subunit and the heterodimer. Smaller amounts of 32Pi could also result from some break-down of the labile [32P]CP.
Figure 4.
aCPS and mCPS pulse/chase experiments. aCPS or mCPS subunits N or C were incubated with [γ-32P]ATP and then desalted into a solution containing either subunit N or C (e.g., NN: subunit N in the pulse solution, subunit N in the chase solution). The reaction products were separated by paper chromatography and amounts of labeled CP (black bars) and Pi (white bars) were determined by densitometric analysis.
Unexpectedly, [32P]-labeled CP also resulted when pulse-labeled subunit C was added to a solution containing subunit C (Experiment 4, Fig. 4 ▶) as well as when the chase solution contained subunit N (Experiment 3, Fig. 4 ▶). This finding was surprising since both the C and N subunits of aCPS are needed for detectable CP formation in the standard assay (Ahuja et al. 2001). The most straightforward explanation for the results of Experiment 4 (Fig. 4 ▶) is that subunit C of aCPS independently catalyzed the phosphorylation of carbamate by ATP (Reaction 4, Fig. 1 ▶). It is also probable that the [32P]-labeled CP of Experiment 3 (Fig. 4 ▶) was formed by subunit C independent of the presence of subunit N in the chase solution rather than by the coordinated functioning of the two subunits in reactions 1–4 (Fig. 1 ▶). It should be noted that, although carbamate per se was not added to the Fig. 4 ▶ experiments, it was present at appreciable concentrations since it exists in equilibrium with bicarbonate and ammonia (Rubio and Cervera 1995). The apparent ability of aCPS subunit C to catalyze phosphate transfer from ATP to carbamate, as documented is Figure 4 ▶, is contradictory to the demonstrated absence of this reaction under standard assay conditions (Ahuja et al. 2001). We confirmed that neither isolated subunit of aCPS demonstrated catalysis of CP formation from carbamate in a continuous assay by performing the standard CP formation assay in the absence and presence of 100 mM ammonium carbamate.
Taken together, these findings indicate that isolated subunit C of aCPS can carry out the single turnover production of CP that is detectable in the experiment of Figure 4 ▶, but cannot catalyze the multiple turnover synthesis necessary for detection in standard assays. A likely scenario is that, when isolated subunit C carries out the ATP-dependent phosphorylation of carbamate, there is no mechanism for efficient dissociation of the resultant CP from the enzyme surface. In the case of the heterodimeric enzyme carrying out multiple turnovers, there must be a cycle of conformational changes that communicate occupancy of the multiple active sites and that also promote dissociation of the products at appropriate times in the reaction cycle. Direct X-ray structural evidence for multiple conformations has not yet been obtained. However, the present findings suggest that cross-talk between domain N and C is required for product release from subunit C and thereby provide additional information upon which to base future studies aimed at defining the conformational steps of the reaction cycle.
Expression and characterization of N and C subunits from M. jannaschii (mCPS)
In order to confirm and possibly extend the aCPS findings, we carried out parallel studies with M. jannaschii CPS (mCPS), the other CPS known to have separate genes for domains N and C. Although these subunits had been sequenced and cloned as part of the whole genome analysis (Bult et al. 1996), they had not been previously expressed and analyzed. As described in Materials and Methods, we subcloned each of the subunits into the pET-28b vector, expressed the subunits in E. coli and purified them. SDS-PAGE confirmed that the subunits were the expected size (56 and 71 kDa, respectively, for subunit N and subunit C). Gel filtration analysis showed both monomeric and dimeric peaks for subunit C, with estimated molecular weights of 79,000 and 152,000 and some apparent self-association of subunit N, with a single peak of estimated molecular weight 73,000. When subunits N and C were combined, they catalyzed CP synthesis with a specific activity of 35 nmol/min/ mg. However, as with aCPS, neither subunit alone displayed CP synthesis that was detectable in our continuous assay.
Pulse/chase labeling of M. jannaschii N and C subunits with [γ-32P]ATP
As described above for the subunits from aCPS, a relatively large amount of subunit N or subunit C from mCPS was incubated briefly with [γ-32P]ATP, Mg2+ and K+, desalted so that the only labeled ATP was enzyme-bound, and then added to a solution containing an excess of ammonia, bicarbonate, and either subunit N or C of mCPS. The labeling pattern for the subunits of this CPS (Experiments 5–8, Fig. 4 ▶) parallels that for aCPS (Experiments 1–4, Fig. 4 ▶). 32Pi formation was observed under all conditions utilized for the pulse/chase studies (Experiments 5–8, Fig. 4 ▶) whereas [32P]CP was formed only when subunit C was pulse labeled (Experiments 7 and 8, Fig. 4 ▶) and was not formed when subunit N was pulse labeled (Experiments 5 and 6, Fig. 4 ▶). These mCPS findings confirm the localization of CP formation to subunit C. Additionally, they confirm that isolated subunit C of mCPS can carry out single turnover synthesis of CP.
Hyperthermophilic CPSs provide a critical mechanistic probe
The CPS N and C subunits from the extreme hyperthermophilic organisms have constituted a unique tool to analyze the mechanism of CPS and to link specific functional steps to specific structural locales. This occurrence of domains N and C of CPS as separate subunits is not unique relative to other hyperthermophilic proteins. In both A. aeolicus (Deckert et al. 1998) and M. jannaschii (Olsen and Woese 1996), many genes that are functionally grouped within operons in mesophilic organisms are dispersed throughout the genome and many proteins that occur as single polypeptides in mesophilic organisms are split into subunits in hyperthermophiles. However, the occurrence of functional CPS N and C subunits does appear to be unique to the hyperthermophilic enzymes. Increased structural rigidity in thermophilic enzymes has been proposed as the basis for the extremely low activity generally observed at mesophilic temperatures (Merz et al. 2000; Fitzpatrick et al. 2001). Such rigidity would be consistent with the low activity observed for aCPS and mCPS at 37°C and would also be consistent with the finding that these thermophilic N and C subunits are expressed as stable units that can associate to form a CP-synthesizing heterodimer. Furthermore, structural rigidity at the domain N/C interface is most likely essential to maintain “tunnel-ready” surfaces in the hyperthermophilic subunits as individually folded under physiological conditions. The solved structure of eCPS suggested two interior closed molecular tunnels (Thoden et al. 1997, 1999b). The existence of the first tunnel, allowing sequestered movement of uncharged ammonia from the amidotransferase domain to domain N, has been well documented in CPS and other amidotransferases (Raushel et al. 2003). Documented functioning of the second tunnel, proposed to allow sequestered movement of carbamate from domain N to domain C, has remained elusive (Kim and Raushel 2004), but the operation of the sequential reaction mechanism supported by the present data would be consistent with such an interior closed channel.
Use of structurally similar substrates (bicarbonate and carbamate) to yield structurally similar products (carboxy phosphate and carbamoyl phosphate) at similar ATP-grasp folds (RMSD of 1.1 Å for 255 equivalent α carbons) is integral to the CPS mechanism but has also yielded cognate substrate ambiguity and cognate partial reaction promiscuity that have confounded most direct approaches to defining the functions of the two ATP-grasp folds. Even more complexity in experimental design and analysis results from the interior tunnels and lids covering the active sites (Thoden et al. 1997, 1999b), which limit active site access to externally supplied substrates and intermediates in the intact mesophilic CPSs. The present demonstration of carbamate phosphorylation on subunit C of thermophilic CPSs therefore provides critical mechanistic information that has remained elusive in many previous studies with mesophilic CPSs in both intact and truncated forms.
Materials and methods
Strains, plasmids, and recombinant DNA methods
Molecular biology techniques were performed essentially as described by Sambrook et al. (1989). XL1-Blue and Top10 E. coli were used for DNA manipulations. Rosetta (DE3) E. coli (Novagen) encoding tRNAs for the rare codons AUA, AGG, AGA, CUA, CCC, and GGA were used for expression of mCPS constructs. Construction and expression of the aCPS constructs have been described previously (Ahuja et al. 2001). Clones of M. jannaschii genomic DNA encoding the mCPS N and C subunits were obtained from the ATCC (ATCC numbers 624,673 and 624,531, respectively). PCR was used to amplify the open reading frames and introduce convenient restriction sites for subcloning into pET-28b (Novagen). The mCPS N ORF was amplified with primers CACGCGCCATGGGACATATGGAGAGTATTAAAA AAGTAATGG (NdeI site underlined, ORF start codon bolded) and CGCGGATCCTTAAGCATTCAATTTCTTAATTTC (BamHI site underlined, stop codon bolded). The resulting product was digested with NdeI and BamHI and ligated into pET-28b, resulting in a construct with an N-terminal 6xHis-tag and thrombin cleavage site (sequence MGSSHHHHHHSSGLVPRGSH), followed by the entire mCPS N ORF. For mCPS C, primers CACGCGCCATGG GACATATGGATATGGAAAAATTAAAGG (NdeI site underlined, ORF start codon bolded) and CGTCGTCGACTTAGGAG CTCGCAAACCTATTTTTTATCCTTGCATC (SalI site underlined, stop codon bolded) were used and the NdeI/SalI-digested PCR product cloned into pET-28b. In addition to the N-terminal 6×His-tag and thrombin cleavage site identical to mCPS N, this construct has a three-residue C-terminal extension (sequence ASS), resulting from the introduction of a SacI site between the last codon of the ORF and the stop codon. All constructs were verified by sequencing.
Protein purification and activity determination
Purification of aCPS constructs has been described previously (Ahuja et al. 2001). For purification of mCPS N and C subunits, E. coli strain Rosetta (DE3) transformed with pET-28b encoding the N or C subunit was grown to stationary phase at 37°C in Terrific Broth containing 100 mg/L ampicillin, then diluted 100-fold into 1 L of the same medium and grown to an OD600 of about 0.6. The culture was induced for 4 h with 1 mM IPTG, harvested by centrifugation at 5,000g for 10 min and resuspended in 50 mL Buffer A (20 mM Tris, 500 mM NaCl, 5 mM imidazole, pH 7.6) with added 2 mM EDTA and 0.2 mM each phenylmethylsulfonyl fluoride, pepstatin, antipain, leupeptin, chymostatin, and aprotinin. The cells were disrupted by sonication (six 15-sec pulses, with cooling intervals between pulses) and cell debris was removed by centrifugation at 16,000g for 20 min. Cleared lysate was heated for 20 min to 70°C and centrifuged for 15 min at 16,000g to remove denatured E. coli proteins. Protein in the supernatant was precipitated by addition of ammonium sulfate to 95%, followed by a 20 min centrifugation at 16,000g and resuspension in Buffer A. Protein was applied to a 5 mL HiTrap column (Amersham-Pharmacia) charged with Ni2SO4 and equilibrated in Buffer A, and eluted with a linear gradient of 5–500 mM imidazole in Buffer A (ÄKTA FPLC, Amersham-Pharmacia). Bound protein eluted at ~100 mM (mCPS N) or 75 mM (mCPS C) imidazole. mCPS-containing fractions were pooled and concentrated by addition of solid ammonium sulfate to 95%. The protein was precipitated by centrifugation at 16,000g for 20 min, and the pellet resuspended in 100 mM potassium phosphate, 1 mM EDTA, pH 7.6 for application to a Hi-Load 16/60 Superdex 200 column. mCPS was eluted from this column in 100 mM potassium phosphate, 1 mM EDTA (pH 7.6), and stored at −80°C for further use.
CP synthesis was measured as citrulline formation by coupling CP formation to the reaction of ornithine transcarbamoylase in the presence of ornithine (Guthohrlein and Knappe 1968). Assay mixtures contained 50 mM HEPES, 100 mM KCl, 20 mM MgSO4, 40 mM NaHCO3, 1 mM DTT, 5 u/mL ornithine transcarbamoylase, 10 mM ornithine and 10 mM ATP, final pH 7.6. Assays aimed at determining the ability of aCPS and mCPS N or C to directly phosphorylate carbamate additionally contained 100 mM ammonium carbamate. Assays were performed at 37°C in order to prevent inactivation of the mesophilic ornithine transcarbamoylase or decay of the labile CP and carbamate. Protein concentration was determined by the Lowry assay (Lowry et al. 1951) or from mea-surements of A280, using molar extinction coefficients (57,760 M−1 cm−1 for aCPS N, 39,770 M−1 cm−1 for aCPS C, 25,400 M−1 cm−1 for mCPS N, and 43,730 M−1 cm−1 for mCPS C) calculated according to Pace et al(1995).
Pulse/chase analysis
For pulse/chase analysis, either the N or C subunit of aCPS or mCPS was incubated for ~2 min in the presence of [γ-32P]ATP. This, and all subsequent steps were carried out at room temperature. The pulse solution consisted of 50 mM HEPES, 100 mM KCl, 10 mM ornithine, 10 mM β-mercaptoethanol, 10 mM MgSO4, 0.1 or 1 mM [γ-32P]ATP and 1–1.8 nmol CPS N or C in a final volume of 50 μL. [γ-32P]ATP (specific activity 6000 Ci/ mmol, radiochemical concentration 10 mCi/mL) was from Perkin Elmer Life Sciences, Inc., and the specific radioactivity in the assay ranged from 0.2–4.5 μCi/nmol. The pulse solution was applied to a Quick Spin G-50 Sephadex column (Roche Applied Science) and the CPS:[μ-32P]ATP complex was separated from unbound [μ-32P]ATP by centrifugal desalting (2 min at 1100g) into 90 μl of chase solution. The chase solution consisted of (prior to dilution by ~60 μL of effluent from the Quick Spin column) 56 mM HEPES, 111 mM KCl, 11 mM ornithine, 11 mM β-mercaptoethanol, 88 mM MgSO4, 66 mM ATP, 333 mM NH4Cl, 33 mM NaHCO3 and 1–1.8 nmol CPS N or C. Immediately after the end of the 2 min centrifugation step, 4–10 μL of the reaction mixture was spotted 2 cm from the bottom of a piece of Whatman 3MM chromatography paper and developed for 1 h in the GW3 solvent system in order to separate ATP, CP, and inorganic phosphate as described previously (Wood 1961). The GW3 solvent system consists of 32 mL n-butanol, 16 mL n-propanol, 20 mL acetone, 20 mL 80% formic acid, 12 mL 30% trichloroacetic acid and 0.3 g EDTA (Wood 1961). The chromatograms were dried, exposed on Fuji medical X-ray film and subjected to densitometric analysis using the program ImageJ, developed by Wayne Rasband at the NIH (http://rsb.info.nih.gov/ij/).
Acknowledgments
This work was supported by National Institutes of Health Grants DK54423 (S.P.-L.) and GM/CA60371 (D.R.E).
Abbreviations
aCPS, Aquifex aeolicus carbamoyl phosphate synthetase
cm, carbamate
CP, carbamoyl phosphate
CPS, carbamoyl phosphate synthetase
CPSs, plural form for carbamoyl phosphate synthetase
eCPS, Escherichia coli carbamoyl phosphate synthetase
mCPS, Methanococcus jannaschii carbamoyl phosphate synthetase
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041041305.
References
- Ahuja, A., Purcarea, C., Guy, H.I., and Evans, D.R. 2001. A novel carbamoyl-phosphate synthetase from Aquifex aeolicus. J. Biol. Chem. 276 45694–45703. [DOI] [PubMed] [Google Scholar]
- Alonso, E., Cervera, J., Garcia-Espana, A., Bendala, E., and Rubio, V. 1992. Oxidative inactivation of carbamoyl phosphate synthetase (ammonia). Mechanism and sites of oxidation, degradation of the oxidized enzyme, and inactivation by glycerol, EDTA, and thiol protecting agents. J. Biol. Chem. 267 4524–4532. [PubMed] [Google Scholar]
- Anderson, P.M. and Meister, A. 1966. Bicarbonate-dependent cleavage of adenosine triphosphate and other reactions catalyzed by Escherichia coli carbamyl phosphate synthetase. Biochemistry 5 3157–3163. [DOI] [PubMed] [Google Scholar]
- Bult, C.J., White, O., Olsen, G.J., Zhou, L., Fleischmann, R.D., Sutton, G.G., Blake, J.A., FitzGerald, L.M., Clayton, R.A., Gocayne, J.D., et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273 1058–1073. [DOI] [PubMed] [Google Scholar]
- Buttlaire, D.H., Himes, R.H., and Reed, G.H. 1976. Formyltetrahydrofolate synthetase-catalyzed formation of ATP from carbamyl phosphate and ADP. Evidence for a formyl phosphate intermediate in the enzyme’s catalytic mechanism. J. Biol. Chem. 251 4159–4161. [PubMed] [Google Scholar]
- Deckert, G., Warren, P.V., Gaasterland, T., Young, W.G., Lenox, A.L., Graham, D.E., Overbeek, R., Snead, M.A., Keller, M., Aujay, M., et al. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392 353–358. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, T.B., Killer, P., Thomas, R.M., Jelesarov, I., Amrhein, N., and Macheroux, P. 2001. Chorismate synthase from the hyperthermophile Thermotoga maritima combines thermostability and increased rigidity with catalytic and spectral properties similar to mesophilic counterparts. J. Biol. Chem. 276 18052–18059. [DOI] [PubMed] [Google Scholar]
- Guthohrlein, G. and Knappe, J. 1968. Modified determination of citrulline. Anal. Biochem. 26 188–191. [DOI] [PubMed] [Google Scholar]
- Guy, H.I. and Evans, D.R. 1996. Function of the major synthetase subdomains of carbamyl-phosphate synthetase. J. Biol. Chem. 271 13762–13769. [DOI] [PubMed] [Google Scholar]
- Javid-Majd, F., Mullins, L.S., Raushel, F.M., and Stapleton, M.A. 2000. The differentially conserved residues of carbamoyl-phosphate synthetase. J. Biol. Chem. 275 5073–5080. [DOI] [PubMed] [Google Scholar]
- Kim, J. and Raushel, F.M. 2004. Access to the carbamate tunnel of carbamoyl phosphate synthetase. Arch. Biochem. Biophys. 425 33–41. [DOI] [PubMed] [Google Scholar]
- Knowles, J.R. 1989. The mechanism of biotin-dependent enzymes. Annu. Rev. Biochem. 58 195–221. [DOI] [PubMed] [Google Scholar]
- Kothe, M. and Powers-Lee, S.G. 2004. Nucleotide recognition in the ATP-grasp protein carbamoyl phosphate synthetase. Protein Sci. 13 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe, M., Eroglu, B., Mazza, H., Samudera, H., and Powers-Lee, S. 1997. Novel mechanism for carbamoyl-phosphate synthetase: A nucleotide switch for functionally equivalent domains. Proc. Natl. Acad. Sci. 94 12348–12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, O.H., Rosebrough, N.J., Farr, A.I., and Randall, R.J. 1951. J. Biol. Chem. 193 265–275. [PubMed] [Google Scholar]
- Lusty, C.J., Widgren, E.E., Broglie, K.E., and Nyunoya, H. 1983. Yeast carbamyl phosphate synthetase. Structure of the yeast gene and homology to Escherichia coli carbamyl phosphate synthetase. J. Biol. Chem. 258 14466–14477. [PubMed] [Google Scholar]
- Meister, A. 1989. Mechanism and regulation of the glutamine-dependent carbamyl phosphate synthetase of Escherichia coli. Adv. Enzymol. Relat. Areas Mol. Biol. 62 315–374. [DOI] [PubMed] [Google Scholar]
- Merz, A., Yee, M.C., Szadkowski, H., Pappenberger, G., Crameri, A., Stemmer, W.P., Yanofsky, C., and Kirschner, K. 2000. Improving the catalytic activity of a thermophilic enzyme at low temperatures. Biochemistry 39 880–889. [DOI] [PubMed] [Google Scholar]
- Olsen, G.J. and Woese, C.R. 1996. Lessons from an Archaeal genome: What are we learning from Methanococcus jannaschii? Trends Genet. 12 377–379. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Vajdos, F., Fee, L., Grimsley, G., and Gray, T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post, L.E., Post, D.J., and Raushel, F.M. 1990. Dissection of the functional domains of Escherichia coli carbamoyl phosphate synthetase by site-directed mutagenesis. J. Biol. Chem. 265 7742–7747. [PubMed] [Google Scholar]
- Powers, S.G. and Meister, A. 1978. Carbonic-phosphoric anhydride (carboxy phosphate). Significance in catalysis and regulation of glutamine-dependent carbamyl phosphate synthetase. J. Biol. Chem. 253 1258–1265. [PubMed] [Google Scholar]
- Raushel, F.M. and Villafranca, J.J. 1979. Determination of rate-limiting steps of Escherichia coli carbamoyl-phosphate synthase. Rapid quench and isotope partitioning experiments. Biochemistry 18 3424–3429. [DOI] [PubMed] [Google Scholar]
- Raushel, F.M., Miles, B.W., Post, L.E., and Post, D.J. 1992. Mutational analysis of two putative domains within the large subunit of carbamoyl phosphate synthetase from Escherichia coli. Bioorg. Med. Chem. Lett. 2 319–322. [Google Scholar]
- Raushel, F.M., Mullins, L.S., and Gibson, G.E. 1998. A stringent test for the nucleotide switch mechanism of carbamoyl phosphate synthetase. Biochemistry 37 10272–10278. [DOI] [PubMed] [Google Scholar]
- Raushel, F.M., Thoden, J.B., and Holden, H.M. 2003. Enzymes with molecular tunnels. Acc. Chem. Res. 36 539–548. [DOI] [PubMed] [Google Scholar]
- Rubio, V. and Cervera, J. 1995. The carbamoyl-phosphate synthase family and carbamate kinase: Structure-function studies. Biochem. Soc. Trans. 23 879–883. [DOI] [PubMed] [Google Scholar]
- Rubio, V., Llorente, P., and Britton, H.G. 1998. Mechanism of carbamoyl phosphate synthetase from Escherichia coli—binding of the ATP molecules used in the reaction and sequestration by the enzyme of the ATP molecule that yields carbamoyl phosphate. Eur. J. Biochem. 255 262–270. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Serre, V., Guy, H., Penverne, B., Lux, M., Rotgeri, A., Evans, D., and Herve, G. 1999. Half of Saccharomyces cerevisiae carbamoyl phosphate synthetase produces and channels carbamoyl phosphate to the fused aspartate transcarbamoylase domain. J. Biol. Chem. 274 23794–23801. [DOI] [PubMed] [Google Scholar]
- Tate, S.S., Leu, F.Y., and Meister, A. 1972. Rat liver glutamine synthetase. Preparation, properties, and mechanism of inhibition by carbamyl phosphate. J. Biol. Chem. 247 5312–5321. [PubMed] [Google Scholar]
- Thoden, J.B., Holden, H.M., Wesenberg, G., Raushel, F.M., and Rayment, I. 1997. Structure of carbamoyl phosphate synthetase: A journey of 96 A from substrate to product. Biochemistry 36 6305–6316. [DOI] [PubMed] [Google Scholar]
- Thoden, J.B., Huang, X., Raushel, F.M., and Holden, H.M. 1999a. The small subunit of carbamoyl phosphate synthetase: Snapshots along the reaction pathway. Biochemistry 38 16158–16166. [DOI] [PubMed] [Google Scholar]
- Thoden, J.B., Raushel, F.M., Benning, M.M., Rayment, I., and Holden, H.M. 1999b. The structure of carbamoyl phosphate synthetase determined to 2.1 Å resolution. Acta. Crystallogr. D. Biol. Crystallogr. 55 8–24. [DOI] [PubMed] [Google Scholar]
- Wimmer, M.J., Rose, I.A., Powers, S.G., and Meister, A. 1979. Evidence that carboxyphosphate is a kinetically competent intermediate in the carbamyl phosphate synthetase reaction. J. Biol. Chem. 254 1854–1859. [PubMed] [Google Scholar]
- Wood, T. 1961. A procedure for the analysis of acid-soluble phosphorus compounds amd related substances in muscle and other tissues. J. Chromatogr. 6 142–154. [Google Scholar]