Abstract
Heat shock protein 40s (Hsp40s) and heat shock protein 70s (Hsp70s) form chaperone partnerships that are key components of cellular chaperone networks involved in facilitating the correct folding of a broad range of client proteins. While the Hsp40 family of proteins is highly diverse with multiple forms occurring in any particular cell or compartment, all its members are characterized by a J domain that directs their interaction with a partner Hsp70. Specific Hsp40–Hsp70 chaperone partnerships have been identified that are dedicated to the correct folding of distinct subsets of client proteins. The elucidation of the mechanism by which these specific Hsp40–Hsp70 partnerships are formed will greatly enhance our understanding of the way in which chaperone pathways are integrated into finely regulated protein folding networks. From in silico analyses, domain swapping and rational protein engineering experiments, evidence has accumulated that indicates that J domains contain key specificity determinants. This review will critically discuss the current understanding of the structural features of J domains that determine the specificity of interaction between Hsp40 proteins and their partner Hsp70s. We also propose a model in which the J domain is able to integrate specificity and chaperone activity.
Keywords: J domain, DnaJ, Hsp70, specificity determinants
Hsp40 and Hsp40-like proteins
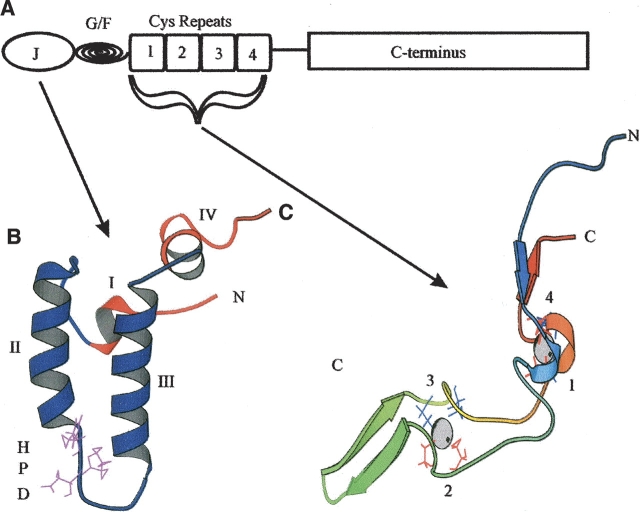
The heat shock protein 70 family (Hsp70) of molecular chaperones is a major component of the cellular chaperone network and the stress response. Hsp70 proteins are regulated by several co-chaperones, in particular the heat shock protein 40 (Hsp40) family, which stimulates Hsp70 ATP hydrolysis, thereby regulating Hsp70 client protein interactions. The Hsp40 family of proteins, including so-called Hsp40-like proteins, are defined by the presence of the J domain, a 70-amino-acid domain with similarity to the initial 73 amino acids of the Escherichia coli Hsp40 called DnaJ (Pellecchia et al. 1996). In addition to the J domain, Hsp40 and Hsp40- like proteins may have certain other structural features that are conserved from E. coli DnaJ (Ohki et al. 1986; Cheetham and Caplan 1998). E. coli DnaJ is comprised of four canonical domains, a J domain, a Gly/Phe-rich region, four cysteine repeats, and an uncharacterized C terminal region. A schematic of the domains present in E. coli DnaJ is given in Figure 1 ▶.
Figure 1.
Structural motifs found in E. coli DnaJ. (A) A diagrammatic representation of the domains present in E. coli DnaJ. J represents the J domain, G/F stands for the Gly/Phe-rich region, and cysteine repeats represents the four zinc-finger-like motifs. The first three domains correspond to approximately half DnaJ. (B) A ribbon representation of the J domain (1XBL) (Pellechia et al. 1996). The conserved HPD motif is depicted in purple. The four helices are labeled. (C) A ribbon representation of the cysteine repeats (1EXK) (Martinez-Yamout et al. 2000). Cysteine residues are depicted in red (repeats 1 and 2) and blue (repeats 3 and 4). Repeats 1 and 4 form zinc center 1 and repeats 2 and 3 form zinc center 2 (Linke et al. 2003). The coordinated zinc atoms are shown in CPK format. This diagram is not to scale. The figures were generated using Molscript (Kraulis 1991).
The J domain
The J domain is believed to be the major binding site and the minimal region required for interaction between Hsp40 and Hsp40-like proteins with their partner Hsp70s (Corsi and Schekman 1997; Landry 2003; Wittung-Stafshede et al. 2003). This domain is normally found at the N terminus of proteins, although this is not always the case (Cyr et al. 1994). The structures of the J domain from seven Hsp40 and Hsp40-like proteins have been determined using nuclear magnetic resonance (NMR) or X-ray crystallography. These are from E. coli DnaJ (Pellecchia et al. 1996; Huang et al. 1998), human HDJ1 (Qian et al. 1996), human Kiaa0730 (N. Kobayashi, S. Koshiba, T. Kigawa, and S. Yokoyama, unpubl.), E. coli Hsc20 (Cupp-Vickery and Vickery 1997, 2000), the large T antigen from murine polyomavirus (Berjanskii et al. 2000), the large T antigen from SV40 in conjunction with the retinoblastoma tumor suppressor (Kim et al. 2001), and bovine auxilin (Jiang et al. 2003; Gruschus et al. 2004b). J domain structures are depicted in Figure 2 ▶. The structures contain four α-helices (helices I–IV), with a loop region containing a highly conserved tripeptide of histidine, proline, and aspartic acid (HPD motif) located between helices II and III (Qian et al. 1996). The HPD motif is present in all known J domains, with the exception of the ring infected erythrocyte surface antigen (RESA) proteins of Plasmodium falciparum (Bork et al. 1992), the E. coli DjlB/DjlC family of proteins (Kluck et al. 2002), and the yeast protein Tim16/Pam16 (Walsh et al. 2004). Binding inhibition studies using peptides suggested that the minimal Hsp70-binding site in the J domain of the yeast Hsp40, Ydj1, was between amino acids 2 and 35, which included helices I and II, and the HPD motif (Tsai and Douglas 1996; Greene et al. 1998). The auxilin J domain, like that of Sec63 and Scj1, contains an extra loop region between helices I and II, which is proposed to act as an extended interface for interaction with Hsc70 during clathrin uncoating (Jiang et al. 2003). The J domain also appears to be flexible in structure, and an induced-fit mechanism has been proposed, with the HPD motif aiding in the alteration of the orientation of the charged residues in helix II, such that helix II can interact correctly with the ATPase domain of a partner Hsp70 (Huang et al. 1999; Berjanskii et al. 2002; Landry 2003).
Figure 2.
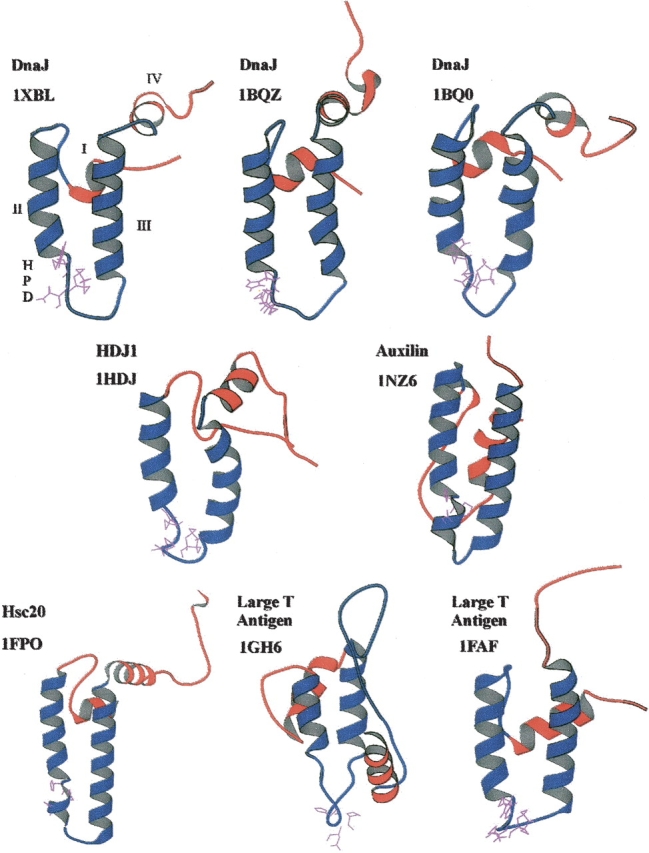
Ribbon representation of the structures of J domains from various Hsp40 and Hsp40-like proteins. The structures for E. coli DnaJ (1XBL, 1BQZ, 1BQ0) (Pellecchia et al. 1996; Huang et al. 1998), human HDJ1 (1HDJ) (Qian et al. 1996), and the murine polyomavirus T antigen (1FAF) (Berjanskii et al. 2000) J domains were determined using NMR. The structure of E. coli Hsc20 (1FPO) (Cupp-Vickery and Vickery 2000), SV40 T antigen (1GH6) (Kim et al. 2001), and bovine auxilin (1NZ6) (Jiang et al. 2003) J domains were determined using X-ray crystallography. The structures were visualized using Molscript (Kraulis 1991). The HPD motif is depicted in purple. Helices II and III are in blue, with the more mobile helices I and IV in red. The helices and HPD motif are labeled on the DnaJ 1XBL J domain structure. Only the J domain region is shown even if additional structural regions were determined.
While the J domain is the specific feature that defines a protein as an Hsp40 or Hsp40-like protein, the presence of a J domain does not imply that the protein is a full homolog of E. coli DnaJ. Many Hsp40-like proteins contain regions that are not found in E. coli DnaJ (Cheetham and Caplan 1998), and the presence of other domains may allow a particular Hsp40-like protein to fulfill a specific function. It appears that the J domain has been recruited by unrelated proteins over time to help fulfill a number of functions. An example of such an adaptation of function would be the presence of the J domain in the large and small T antigen proteins of viruses (Cheetham et al. 1992; Brodsky and Pipas 1998). This T antigen J domain can functionally replace the J domain of E. coli DnaJ in an in vivo complementation assay (Kelley and Georgopoulos 1997). Another example is the mammalian protein ERdj5/JPD1 (endoplasmic reticulum DnaJ protein 5/J protein domain 1), which contains a J domain, and a protein disulphide isomerase-like domain (or thioredoxin domain) (Cunnea et al. 2003; Hosoda et al. 2003). ERdj5 may use these domains to facilitate the formation of appropriate disulphide bonds during the folding of ER proteins, both via its ability to promote the formation of disulphide bonds and its ability to sequester the ER-based Hsp70, BiP.
Subdivision and nomenclature of Hsp40 and Hsp40-like proteins
The classification of the large and diverse groupings of Hsp40-like proteins is a challenge, and there have been several attempts to subdivide them to make analysis easier. One categorization proposed the division of the family into two groups, depending on the presence or absence of the Gly/Phe region (Kelley 1998). An alternative categorization involved the division of the family into three groups depending on the presence or absence of the Gly/Phe region and the cysteine repeats (Cheetham and Caplan 1998). In this system, Type I Hsp40-like proteins contain similarity to E. coli DnaJ over all the domains present, namely the J domain, the Gly/Phe region, and the cysteine repeats. Type II proteins have a J domain and the Gly/Phe-rich region. Type III proteins only have a J domain in common with E. coli DnaJ. Recently the term “J-protein family” was formally coined for the first time (Walsh et al. 2004). This paper proposed that the J domain should be strictly defined on the basis of its four helices and the HPD motif, so as to be able to distinguish true J proteins from J-like proteins. The letter J is used in the nomenclature of a number of other systems unrelated to Hsp40 (e.g., DJ-1 or ApoJ) and could lead to confusion. Therefore, for the purposes of this review we will utilize Hsp40 protein to refer to full homologs of E. coli DnaJ containing all four canonical domains (Type I proteins), and Hsp40-like to refer to proteins that do not have all four domains but contain at least the J domain or a J-like domain (Type II and III proteins).
Hsp40s integrate the ATPase and chaperone activities of Hsp70s
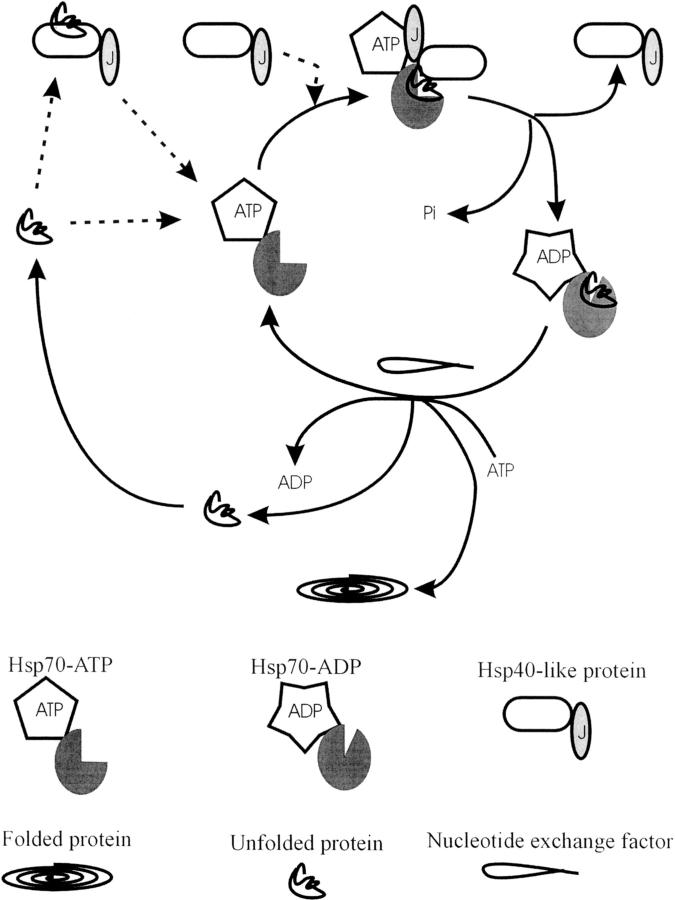
Hsp70s in general have low basal ATPase activity involving ATP binding and hydrolysis and nucleotide exchange. Hsp40 and Hsp40-like proteins stimulate the ATPase activity of Hsp70 by specifically enhancing the rate of ATP hydrolysis (Szabo et al. 1994; Russell et al. 1999). Nucleotide exchange factors also enhance the ATPase activity of Hsp70 by increasing the rate of nucleotide exchange (Kabani et al. 2003; Shomura et al. 2005). The so-called Hsp70 ATPase cycle is tightly coupled to its cycle of client protein association, and has been well characterized in prokaryotic systems such as E. coli, but also in eukaryotes such as yeast and mammalian systems. A schematic model of the interaction of Hsp40 and Hsp40-like proteins with Hsp70s is shown in Figure 3 ▶.
Figure 3.
Schematic showing the protein folding cycle involving the interaction of Hsp40 and Hsp40-like proteins with partner Hsp70s. Pi is inorganic phosphate. Dotted lines indicate the two different paths by which client proteins and Hsp40 or Hsp40-like proteins can enter the cycle. Client proteins are either recognized by Hsp70, following which an Hsp40 or Hsp40-like protein enters the cycle, or are presented to Hsp70 by an Hsp40 or Hsp40-like protein. ATP hydrolysis, stimulated by an Hsp40 or Hsp40-like protein, causes a conformational shift in the peptide binding domain, locking in the client protein. Nucleotide exchange then reverses the conformational shift, allowing for the release and subsequent folding of the client protein. Alternatively the client protein can re-enter the cycle.
The Hsp70 ATPase cycle and associated protein folding cycle in prokaryotes
E. coli DnaJ stimulates the in vitro ATP hydrolysis by the E. coli Hsp70 homolog DnaK, thereby stimulating the DnaK ATPase activity. The in vitro ATPase activity of E. coli DnaK is further enhanced in the presence of the nucleotide exchange factor GrpE (Liberek et al. 1991), and maximal enhancement occurs in the presence of a client protein (Laufen et al. 1999; Suh et al. 1999). However, while maximal enhancement of the in vitro ATPase activity of E. coli DnaK by DnaJ alone occurs at stoichiometric amounts of DnaJ, optimal in vitro refolding activity of E. coli DnaK occurs at substoichiometric levels of DnaJ (Liberek et al. 1991; Laufen et al. 1999). Therefore, it is important to note that the ATPase cycle of DnaK is stoichiometrically coupled to the cycle of client protein binding and release, and that DnaJ integrates ATP hydrolysis with the protein folding chaperone cycle (Pierpaoli et al. 1997; Davis et al. 1999). In addition, DnaJ has a relatively low-affinity association with ATP-bound DnaK, such that the association is transient and dynamic in nature (Russell et al. 1999). This dynamic low-affinity association makes sense in the cellular context, as it would prevent nonproductive hydrolysis of ATP by DnaK in the absence of client proteins.
In E. coli, the protein folding cycle has been proposed to start when DnaJ presents a client protein to DnaK (Szabo et al. 1994; McCarty et al. 1995; Russell et al. 1999) or binds to an already existing DnaK–client protein complex (Pierpaoli et al. 1997, 1998; Gisler et al. 1998; Nagata et al. 1998). It is likely that the nature of the amino acid sequence of the client protein dictates which mechanism is employed for entry of the client protein and DnaJ into the cycle (Pierpaoli et al. 1998), and that, in general, different partner Hsp40 or Hsp40- like proteins may deliver different client proteins to an Hsp70 (Misselwitz et al. 1998). The interaction of E. coli DnaJ with DnaK allows DnaK to attain a high client protein-affinity conformational state after the hydrolysis of ATP (Szabo et al. 1994), and in the process generates a DnaK–client–DnaJ ternary complex (Han and Christen 2003). Thus, upon hydrolysis of ATP a conformational shift occurs in DnaK, locking the client protein into its peptide binding cleft (Moro et al. 2003). DnaJ leaves the DnaK–client protein complex, and GrpE then stimulates the dissociation of ADP from the DnaK ATPase domain, allowing replacement with ATP. This triggers the release of the client protein, which may now fold correctly via a folding intermediate form, or rebind to DnaK and restart the cycle, or associate with other chaperone systems such as the chaperonin GroEL/ES system.
The Hsp70 ATPase and protein folding cycle in eukaryotes
A similar ATPase cycle occurs for eukaryotic Hsp70s, in the presence of Hsp40 or Hsp40-like proteins and nucleotide exchange factors. GrpE homologs do not appear to be present in the cytosol of eukaryotes (Schumacher et al. 1996), although they have been shown to be present in mitochondria (Bolliger et al. 1994). However, other nucleotide exchange factors (Bag1 and HspBP1) have been identified in the eukaryotic cytosol that appear to have different effects on the function of Hsp70s (Kabani et al. 2003; Shomura et al. 2005). In the eukaryotic cytosol, the Hsp70 ATPase and client protein folding cycles are embedded within a complex network of protein folding pathways regulated by specialized co-chaperones. An Hsp40 or Hsp40-like protein stimulates the ATPase activity of Hsp70, causing the formation of a client protein-bound Hsp70 as described for the E. coli system. The Hsp70–client protein conformation can then be further stabilized by the Hsc70-interacting protein (Hip). Hip is involved in the stabilization of the ADP-bound form of Hsp70 in eukaryotic systems (Höhfeld et al. 1995). This allows further time for the unfolded polypeptide to attain a native or native-like conformation. The client protein may be released from Hsp70 to fold correctly on its own or it may be passed onto other chaperone systems (e.g., the chaperonin system or the Hsp90 chaperone machinery) (Schumacher et al. 1996).
Binding sites and binding determinants within the Hsp70–J domain pairing
Binding sites and determinants on Hsp70s
There is considerable genetic, biochemical, and NMR structural data suggesting that DnaK has a binding surface for DnaJ on the underside cleft of its ATPase domain. Evidence has been derived from studies of the in vitro and in vivo interactions of full-length E. coli DnaK with full-length DnaJ (Gässler et al. 1998; Greene et al. 1998; Suh et al. 1998), and the in vitro interaction of the ATPase domain of DnaK with the J domain of DnaJ (Landry 2003; Wittung-Stafshede et al. 2003). In particular, genetic studies have led to the identification of several DnaK mutations in the underside cleft region that suppress the effects of mutations in the critical HPD motif of DnaJ. For example, an R167H substitution in DnaK fully suppressed the functional defects of DnaJ-D35N (Greene et al. 1998). Protein–protein binding studies using full-length proteins showed that while DnaK could no longer bind to DnaJ-D35N, DnaKR167H and DnaK-R167A bound DnaJ-D35N with affinities comparable to the affinity of DnaK for DnaJ (Suh et al. 1998). By contrast, DnaK-R167H and DnaKR167A had significantly reduced affinities for DnaJ compared to the affinity of DnaK for DnaJ. However, analysis of the interactions of the isolated domains gave surprisingly different results; the DnaK ATPase domain had a significantly higher affinity for J domain-D35N compared to its affinity for the unmodified J domain, while the R167A substitution in the DnaK ATPase domain did not significantly affect the domain’s affinities for J domain-D35N and the unmodified J domain (Landry 2003). The fact that (1) the R167A substitution had no significant effect on affinity when considering the interaction of the domains, and (2) that there were contrasting results for the full-length proteins versus the domains when considering the effects of substitutions at R167, suggests that R167 of DnaK does not interact directly with D35 of DnaJ, but that it may be directly or indirectly affecting a DnaK–DnaJ interaction involving the peptide binding domain. Landry (2003) suggests that the D35N mutation may overstabilize a subset of J domain conformations that approximate the induced fit conformation adopted by the J domain when binding to the ATPase domain in the presence of client protein. This would therefore prevent the conformational fluctuations necessary to allow bending of the J domain for the coupling of ATP hydrolysis with a conformational change in DnaK. Furthermore, D167A and D167H substitutions in DnaK may suppress the defect caused by the D35N substitution in DnaJ by destabilizing the induced fit conformational change. Alternatively, these results could be interpreted in the context of the proposal that DnaK may have two distinct binding sites for DnaJ (Karzai and McMacken 1996; Mayer et al. 1999; Suh et al. 1999); certain substitutions at D35 of the J domain may stabilize binding of DnaJ to the ATPase domain of DnaK (through a stabilized induced fit conformation), while certain substitutions at R167 may stabilize binding to the peptide binding domain of DnaK, with both situations resulting in nonfunctional interactions through uncoupling of the stimulation of ATP hydrolysis from the conformational changes needed to trap the client protein.
In vitro analysis of the C-terminal region of rat Hsc70, including the α-helical lid region, indicated that this region also possessed a binding site for Hsp40 (Freeman et al. 1995; Demand et al. 1998). However, for DnaK two observations suggest that the region N-terminal to this proposed C-terminal binding site, including but not limited to the ATPase domain, probably contains the major site of interaction with DnaJ. First, a truncated E. coli DnaK mutant lacking the last 100 residues at the C terminus was still capable of having its ATPase activity stimulated by E. coli DnaJ and of binding to E. coli DnaJ, albeit at a slightly reduced level compared to the binding of full-length DnaK (Gässler et al. 1998; Suh et al. 1999). Second, under conditions where full-length DnaK functionally interacted with DnaJ, the ATPase core of DnaK in isolation (residues 1–385) was not able to bind to DnaJ, nor have its ATPase activity stimulated by DnaJ (Gässler et al. 1998). Blocking of the peptide binding domain of DnaK also abrogated ATPase stimulation by DnaJ. Thus, when Val436 in the peptide binding region was mutated to phenylalanine, a mutation that inhibited the binding of client protein to the peptide binding domain, DnaJ was unable to stimulate the ATPase activity as efficiently (Laufen et al. 1999). These data imply that binding of a client protein and/or DnaJ to the peptide binding site of DnaK is important for ATPase stimulation.
Binding sites on Hsp40-like proteins
The amino acids on Hsp40 and Hsp40-like proteins that are involved in the binding to a partner Hsp70 are less precisely defined. While there is evidence to suggest that the Gly/Phe region may be important for substrate specificity and binding to Hsp70 (Yan and Craig 1999; Fan et al. 2004), it is now well established that Hsp40 and Hsp40-like proteins interact with Hsp70s primarily through the J domain and in particular the HPD motif. Substitutions of the HPD residues abolish the stimulation of the Hsp70 ATPase activity (Tsai and Douglas 1996). However, other residues and regions outside the HPD motif, especially helix II, are gradually being implicated in the interaction of Hsp40 and Hsp40- like proteins with Hsp70 (Lu and Cyr 1998; Genevaux et al. 2002, 2003; Hennessy et al. 2005). For example, the ability of peptides corresponding to various portions of the Ydj1 J domain to compete with full-length Ydj1 for interaction with Hsp70 (Lu and Cyr 1998) showed that helix II and the HPD motif were almost as effective as the full-length J domain in perturbing Hsp70–Ydj1 interactions. However, helix III and the HPD motif were also shown to be almost as capable of affecting the interaction, but a competing peptide of helix I was not (Lu and Cyr 1998). Neither helix II nor helix III was as effective as the full-length J domain. Therefore, while the minimal region for the interaction of Ydj1 with Hsp70 was found to be located in helix II and the loop region of the J domain, other parts of the J domain were also required for complete binding.
A QKRAA motif has been identified in helix IV of the J domain in E. coli DnaJ, and QKRAA-containing peptides have been shown to be recognized by DnaK and to prevent DnaJ binding to DnaK (Auger and Roudier 1997). The lysine and arginine residues present in that motif are conserved in most J domains (Hennessy et al. 2000), and mutations of these residues in the J domain of E. coli DnaJ caused partial abrogation of the interaction of the J domain with DnaK (Suh et al. 1999). Furthermore, Suh et al. (1999) proposed that the QKRAA motif might be a binding site for the interaction of DnaJ with DnaK by transiently interacting with the peptide binding domain.
There is strong evidence that the binding of E. coli DnaJ to DnaK is bipartite in nature, with two distinct DnaJ binding sites on DnaK with differing affinities (Karzai and McMacken 1996; Mayer et al. 1999; Suh et al. 1999). In addition, the nature of this bipartite interaction is central to the mechanism by which DnaJ targets client proteins to DnaK for effective assisted protein folding. While the presence of client protein and an Hsp40 or Hsp40-like protein are required for maximal stimulation of Hsp70 ATP hydrolysis activity, the mechanism of assisted protein folding relies on the J domain binding to Hsp70 in such a manner that ensures that ATP hydrolysis is tightly coupled to client protein binding (Jordan and McMacken 1995; Laufen et al. 1999; Wittung-Stafshede et al. 2003).
Specificity of Hsp40–Hsp70 interaction
It is known that different Hsp40 and Hsp40-like proteins are not completely interchangeable with respect to their interaction with distinct Hsp70s. For example in vitro assays investigating the stimulation of the ATPase activity of Hsp70s by Hsp40-like proteins have also revealed significant differences in the level of the stimulation attained. MmDjC7, a newly identified murine Hsp40-like protein, stimulates the ATPase activity of DnaK, human Hsc70, and murine BiP to different levels (Kroczynska and Blond 2001). As the J domain in MmDjC7 is approximately half of the entire protein, and there is limited similarity to other Hsp40-like proteins in the remainder of the protein, the regulation of stimulation very likely occurs at the level of the J domain. Equally, E. coli DnaJ is capable of stimulating theATPase activity of mammalian Hsc70, whereas mammalian Hdj1 is incapable of stimulating the ATPase activity of DnaK (Minami et al. 1996). Hence, there may be a device within the J domain that mediates the specificity of binding between Hsp70s and partner Hsp40 or Hsp40-like proteins, such that a productive interaction results.
J domain swapping experiments
Substitution of a J domain from one protein by a J domain from another has been a fruitful area in terms of establishing the elements of specificity of interaction. Results from various experiments are shown in Table 1. One of the first experiments conducted showed that the J domain from the yeast mitochondrial Hsp40 protein Mdj1 (Type I) could effectively substitute for the J domain of E. coli DnaJ (Deloche et al. 1997). Equally, the J domain from the Type III E. coli Hsp40-like protein DjlA could effectively substitute for E. coli DnaJ’s J domain (Genevaux et al. 2001). However, both DnaJ and DjlA interact with the same Hsp70, namely DnaK. The J domain from another Type III E. coli Hsp40-like protein, DjlC (which interacts with heat shock cognate protein C/62 kDa [HscC/Hsc62]) was not able to replace the J domain of E. coli DnaJ in in vivo complementation assays, implying that it was unable to interact with DnaK (Kluck et al. 2002).
Table 1.
Summary of J domain swapping experiments
| J domain source | C terminus source | |||||
| Proteina | Type | Protein | Type | Function | Method | Reference |
| Sis1 | II | Sec63 | III | No | Complementation assays in yeast cells not producing Sec63 | Schlenstedt et al. 1995 |
| Scj1 | I | Yes | ||||
| Mdj1 | I | No | ||||
| Mdj1 | I | DnaJ | I | Yes | Complementation for lack of E. coli DnaJ and CbpA | Deloche et al. 1997 |
| DjlA | III | DnaJ | I | Yes | Complementation for lack of E. coli DnaJ and CbpA | Genevaux et al. 2001 |
| DjlC | III | DnaJ | I | No | Inability to replace E. coli DnaJ in vivo | Kluck et al. 2002 |
| Large T Agb | III | DnaJ | I | Yes | Complementation for lack of E. coli DnaJ and CbpA | Kelley and Georgopoulos 1997 |
| Ydj1 | I | Yes | ||||
| DnaJ | I | Large T Ag | III | No | Inability to form infectious virions; ineffective DNA replication; inability to stimulate ATPase activity | Sullivan et al. 2000b |
| Ydj1 | I | No | ||||
| Hdj1 | I | Large T Ag | III | Yes | Mediating reduction in levels of phosphorylated p130 protein | Stubdal et al. 1997 |
| Hsj1 | II | Yes | ||||
| P58IPK | III | DnaJ | I | Yes | Complementation for lack of E. coli DnaJ and CbpA; substitute for lack of Ydj1 in yeast | Yan et al. 2002 |
| Ydj1 | I | Yes | ||||
| Hsj1 | II | Large T Ag | III | Yes | Allows efficient viral DNA replication | Campbell et al. 1997 |
| DnaJA2 | I | Yes | ||||
| Hdj1 | I | DnaJ | I | Yes | Complementation for lack of E.coli DnaJ and CbpA | Genevaux et al. 2002 |
| Hsj1 | II | T antigen | III | Yes | Functional in T Ag mediated cellular transformation | Zalvide et al. 1998 |
| T Ag | III | Ydj1 | I | Yes | Substitutes for lack of Ydj1 in yeast | Fewell et al. 2002 |
a Sis1, Scj1, Mdj1, and Ydj1 are from Saccharomyces cerevisiae; DnaJ, DjlA, and DjlC are from E. coli; Hdj1, Hsj1, DnaJA2, and P58IPK are from Homo sapiens.
bThe large T antigen J domains from SV40, JC virus, and BK virus were used in this experiment.
The situation becomes more complicated when looking at eukaryotic proteins. An analysis of yeast Hsp40 and Hsp40-like proteins showed that neither the Sis1 J domain nor the Mdj1 J domain could functionally substitute for the J domain from Sec63, whereas the J domain from Scj1 could (Schlenstedt et al. 1995). Sis1 is a cytosolic Type II Hsp40-like protein and Mdj1 is a mitochondrial Type I Hsp40 protein, whereas Sec63 and Scj1 are both located in the endoplasmic reticulum (ER) with their J domains facing the lumen of the ER. Additional substitutions (Q13R, K17S, K42V) in the J domain of the Sis1J–Sec63 chimera resulted in a functional chimeric protein (Schlenstedt et al. 1995), implying that these amino acids are important determinants that would affect the interaction of these proteins with their partner Hsp70s. Scj1 and Sec63 both interact with the ER Hsp70, Kar2p, whereas Sis1 interacts with a cytosolic Hsp70, and Mdj1 with a mitochondrial Hsp70.
Domain swapping experiments have also been performed using the J domains from the viral large T antigen proteins. J domains from these proteins used to substitute for the J domains from E. coli DnaJ and yeast Ydj1 gave fully functional chimeric DnaJ and Ydj1 proteins (Kelley and Georgopoulos 1997). The converse, however, did not apply. Substitutions of either the DnaJ or the Ydj1 J domains in the large T antigen only gave partially functional chimeras, with the inability to perform some of their cellular roles (Sullivan et al. 2000b). SV40 T antigen chimeras containing the J domains from Hdj1 or Hsj1 were able to function in the reduction of p130 and phosphorylation of p130 (Stubdal et al. 1997) which was important for T antigen mediated cellular transformation. Large T antigen proteins containing the J domains from Hsj1 or DnaJ2 (also known as Hsj2/Hdj2) could promote viral replication (Campbell et al. 1997). Recently a mammalian J domain was shown to be able to substitute for the J domains from E. coli DnaJ and yeast Ydj1 (Yan et al. 2002). The J domain from P58IPK, a mammalian protein that also contains tetratricopeptide repeat motifs, could functionally replace the DnaJ and Ydj1 J domains in vivo using complementation assays. Mutations in the HPD motif of the chimeric proteins prevented successful complementation for the lack of DnaJ in E. coli or Ydj1 in yeast knockout strains (Yan et al. 2002). This is the only published example of a J domain from a mammalian Type III Hsp40-like protein that is able to functionally replace the J domain in a yeast and a prokaryotic Type I Hsp40 protein. However, the meaning of these findings for J domain specificity is debatable, since the J domain of P58IPK does not appear to be critical for any of the functions of P58IPK. To date no systematic analysis has been conducted on the interchangeability of J domains between all the Type I, II, and III Hsp40 and Hsp40-like proteins from any one cell type, compartment, or system.
Specificity of the Hsp40–Hsp70 Interaction in E. coli
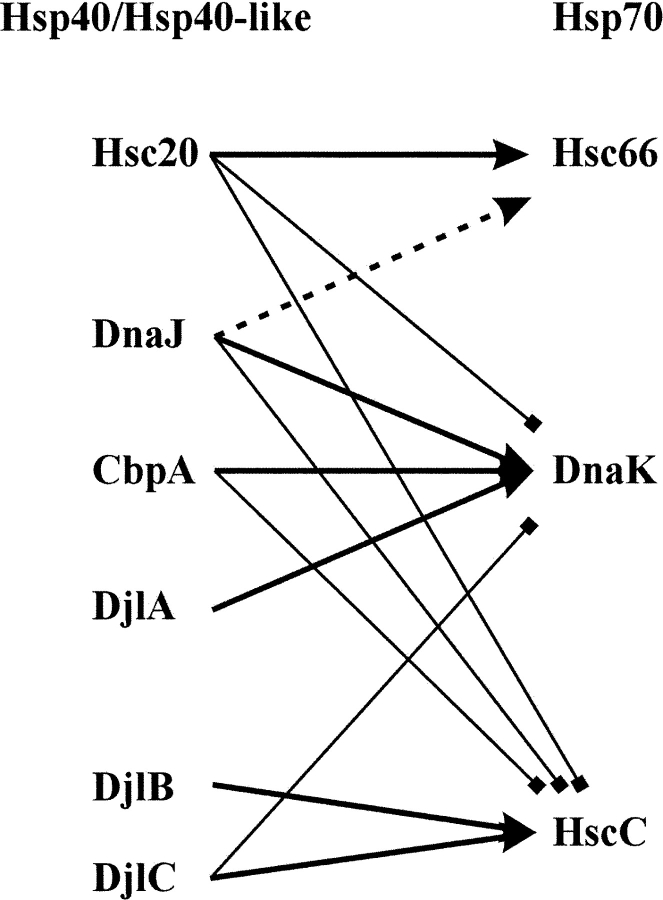
As E. coli has no compartments within the cytoplasm, proteins containing J domains can potentially interact with all Hsp70s in the cell. This could allow for a situation whereby the levels of nonspecific Hsp40–Hsp70 interactions could interfere with productive interactions. E. coli has three identified Hsp70s, DnaK, HscA/Hsc66 (heat shock cognate protein A/66 kDa) (Lelivelt and Kawula 1995; Vickery et al. 1997; Silberg and Vickery 2000), and HscC/Hsc62 (Kluck et al. 2002; Yoshimune et al. 2002). It also has several identified Hsp40 and Hsp40-like proteins, DnaJ, CbpA (Ueguchi et al. 1994), DjlA (Clarke et al. 1996), Hsc20 (Silberg et al. 1998), DjlB, and DjlC/Hsc56 (Kluck et al. 2002; Yoshimune et al. 2002). All these Hsp40 and Hsp40-like and Hsp70 proteins have defined partnerships (Fig. 4 ▶).
Figure 4.
Specificity of interaction between Hsp70 and Hsp40 or Hsp40-like proteins in E. coli. Thick arrows indicate known partnerships. Dotted lines and arrows indicate lower levels of ATP hydrolysis stimulation in vitro. Thin lines with squares replacing arrowheads indicate no detectable levels of ATPase stimulation and no known partnership. “Hsp40/Hsp40-like” indicates the list of different E. coli Hsp40 or Hsp40-like proteins; “Hsp70” indicates the list of different E. coli Hsp70 proteins.
E. coli DnaJ, CbpA (Ueguchi et al. 1995; Wegrzyn et al. 1996), and DjlA (Genevaux et al. 2001) interact with DnaK, Hsc20 (HscB) interacts withHsc66 (HscA) (Vickery et al. 1997; Silberg et al. 1998), and DjlB and DjlC interact with HscC (Kluck et al. 2002; Yoshimune et al. 2002). DnaJ can stimulate the ATPase activity of Hsc66 to a certain extent, whereas Hsc20 does not stimulate the ATPase activity of DnaK (Silberg et al. 1998). DnaJ, CbpA, and Hsc20 do not stimulate the ATPase activity of HscC (Kluck et al. 2002), and DjlC does not stimulate the ATPase activity of DnaK. This apparent specificity is most likely due to the glutamic acid substitution in the HPD motifs in DjlB and DjlC, both of which contain an HPE motif. DnaJ cannot stimulate the ATPase activity of the mammalian ER Hsp70, BiP (Chevalier et al. 2000). Conversely, as discussed at the start of this section, DnaJ can stimulate the ATPase activity of the mammalian Hsc70, whereas Hdj1 cannot stimulate the activity of DnaK (Minami et al. 1996).
General binding and specificity determinants
Several lines of evidence suggest that the major site of interaction of an Hsp40 or Hsp40-like protein with its partner Hsp70 resides in the J domain. First, the J domain alone has been shown to be sufficient to bind and stimulate a partner Hsp70 (Wittung-Stafshede et al. 2003), and substitutions in the HPD motif of the J domain abolish functional interactions with partner Hsp70s (Mayer et al. 1999; Kluck et al. 2002). Second, there are Hsp40-like proteins that consist almost solely of a J domain (e.g., the tiny T antigen; Riley et al. 1997) or have a J domain region isolated within a cellular compartment (e.g., the Sec63 J domain, which protrudes into the ER lumen; Schlenstedt et al. 1995), implying that any ability of these Hsp40-like proteins to interact with a partner Hsp70 must reside primarily in the J domain. Third, the fact that only the J domain residues show conservation across all Hsp40 and Hsp40-like proteins suggests that the J domain is the key component in a conserved mechanism of interaction between these proteins and partner Hsp70s. However, these arguments only apply to a general binding mechanism applicable to most Hsp40–Hsp70 interactions, as opposed to a mechanism for specificity determination.
It is important to distinguish between general binding determinants that are important in the majority of Hsp40–Hsp70 partnerships, and specificity determinants, which are important in specific Hsp40–Hsp70 partnerships. The inability of all Hsp40 and Hsp40-like proteins to interact with all Hsp70s implies a level of mechanistic binding discrimination. These discriminatory binding determinants could reside in the poorly conserved regions of Hsp40 and Hsp40-like proteins that occur beyond the J domain. However, the same lines of evidence that suggest that general binding determinants reside in the J domain can also be used to argue that specificity determinants reside in the J domain. In addition, if one considers that affinity plays a role in specificity, certain of the general binding determinants could also be involved in specificity determination. J domain-based specificity determination is not exclusive, and mechanisms for cellular regulation of specific interactions could occur at the level of colocalization of partnerships to specific organelles or tissues (in eukaryotes) or at the level of coexpression under certain special conditions. In addition, one cannot exclude the possibility that there exist proteins whose functions are to keep Hsp70 and an Hsp40 or Hsp40-like partner protein in a complex, and that this clamp provides specificity (e.g., in Thermus thermophilus; Motohashi et al. 1994, 1996).
Substitutions in the J domain and a model for specificity
The majority of substitutions performed on the J domain have involved mutations in the HPD motif (Cheetham and Caplan 1998). However, recent work has investigated substitutions in other sections of the J domain (Fewell et al. 2002; Genevaux et al. 2002; Hennessy et al. 2005). A list of published amino acid substitutions in the J domain and the functional consequences of the changes are summarized in Table 2. From an analysis of conserved J domain residues (Hennessy et al. 2005), several residues can be defined as being important for J domain function and can be divided into two main categories, namely, charged residues/ motifs and hydrophobic residues (Fig. 5 ▶). Highly conserved charged residues and motifs include the positively charged residues located on helix II, negatively charged residues on helix IV, the KFK motif on helix III (Hennessy et al. 2000), and the QKRAA motif on helix IV (Auger and Roudier 1997). Highly conserved hydrophobic residues include conserved leucines on helices I and III, aromatic residues on helix I, and an alanine residue on helix III (Hennessy et al. 2000). The highly conserved charged residues/motifs are likely to be important in J domain structure and/or function, whereas the conserved hydrophobic residues are likely to be mainly critical for maintaining the structural integrity of the J domain. Some of these residues are discussed in more detail below.
Table 2.
Effect of substitutions performed in the J domain
| Mutation | Helix | Protein | Effect | Reference |
| L13M; L13I; L14V | I | T Ag | Functional | Li et al. 2001 |
| L13V | I | T Ag | Decrease in viral DNA synthesis | Li et al. 2001 |
| L17K | I | Large T antigen | Nonfunctional—reduction of levels of p130 | Stubdal et al. 1997 |
| Y7A; L10A | I | A. tumefaciens DnaJ | Lack of complementation | Hennessy et al. 2005 |
| A33R; Y34K | II | Large T antigen | Functional—reduction of levels of p130 | Stubdal et al. 1997 |
| RE(19,20)AA; RK(22,23)AA; R27A; L28A; A29G; A29G; MK(30,31)AA; Y32A | II | E. coli DnaJ | No effect | Genevaux et al. 2002 |
| E20A; K22A; K27A; Y32A | II | A. tumefaciens DnaJ | No effect | Hennessy et al. 2005 |
| RK(22,23)AA/KR(26,27)AA; Y25A, K26A; KR(26,27)AA; K26E | II | E. coli DnaJ | Lack of complementation | Genevaux et al. 2002 |
| R26A; RK(26,27)AA | II | A. tumefaciens DnaJ | Lack of complementation | Hennessy et al. 2005 |
| W24R; L29P; M30T; Y34N; A37V; C38R | II | T Ag-Ydj1 chimera | Lack of complementation | Fewell et al. 2002 |
| Y24A; RR(25,26)AA | II | Hdj1-DnaJ chimera | Lack of complementation | Genevaux et al. 2002 |
| YKR(25-27)AAA | II | E. coli DnaJ | Toxic | Genevaux et al. 2002 |
| A181T | III | Sec63 | Translocation defects | Lyman and Schekman 1995 |
| F45A | III | Hdj1-DnaJ chimera | Lack of complementation | Genevaux et al. 2002 |
| F46L | III | Ydj1 | Lack of complementation | Johnson and Craig 2000 |
| F47A | III | E. coli DnaJ | Lack of complementation | Genevaux et al. 2002 |
| K51E; K53R; M55T; N56D; Y59N | III | T Ag-Ydj1 chimera | Lack of complementation | Fewell et al. 2002 |
| KE(41,42)AA; E44A; K46A; KE(48,49)AA; KE(51,52)AA; Y54A; E55A; T58A; TD(58-59)AA | III | E. coli DnaJ | No effect | Genevaux et al. 2002 |
| K46T; F47L; K48T; A53S | III | A. tumefaciens DnaJ | No effect | Hennessy et al. 2005 |
| L57S; D59A | IV | A. tumefaciens DnaJ | Lack of complementation | Hennessy et al. 2005 |
| K62A; R63A; K62A,R63A | IV | E. coli DnaJ | Diminished interaction with DnaK by SPR | Laufen et al. 1999 |
| R63A | IV | A. tumefaciens DnaJ | Variable ability to complement | Hennessy et al. 2005 |
| SQ(60,61)AA; KR(62,63)AA; DQ(66,67)AA | IV | E. coli DnaJ | No effect | Suh et al. 1999 |
| D59N; K62A; Y66A; D67A; D67N | IV | A. tumefaciens DnaJ | No effect | Hennessy et al. 2005 |
| D35N | Loop | E. coli DnaJ J domain | Diminished interaction with DnaK | Wittung-Stafshede et al. 2003 |
| D44N | Loop | T Ag | Nonfunctional | Sullivan et al. 2000a |
| H233Q; D235N | Loop | DjlA-DnaJ chimera | Lack of complementation | Genevaux et al. 2001 |
| H32Q | Loop | Hsp40 | Inhibition of Hsp70 luciferase refolding | Michels et al. 1999 |
| H33Q | Loop | Ydj1 | No stimulation of ATPase | Tsai and Douglas 1996 |
| H33Q | Loop | E. coli DnaJ | Lack of complementation | Kelley and Georgopoulos 1997 |
| H33Q | Loop | E. coli DnaJ | Lack of interaction with DnaK by SPR | Mayer et al. 1999 |
| H33Q | Loop | E. coli DnaJ | Stimulation of DnaK lost | Laufen et al. 1999 |
| H33Q; ΔH33; ΔP34; P34F; ΔD35 | Loop | E. coli DnaJ | Lack of complementation | Genevaux et al. 2002 |
| H42Q; D44N | Loop | T Ag | Nonfunctional | Zalvide et al. 1998 |
| H42Q; D44N | Loop | Large T antigen | Nonfunctional—reduction of levels of p130 | Stubdal et al. 1997 |
| H42R; D48G | Loop | T Ag-Ydj1 chimera | Lack of complementation | Fewell et al. 2002 |
| H43Q; D45A | Loop | Csp | No stimulation of Hsc70 ATPase | Chamberlain and Burgoyne 1997 |
| H89Q | Loop | Mtj1 J domain | No stimulation of BiP ATPase: Poor interaction | Chevalier et al. 2000 |
| HPD-AAA | Loop | P58IPK | No stimulation of Hsc70 ATPase | Yan et al. 2002 |
| K35G | Loop | Hdj1-DnaJ chimera | Marked effects on function | Genevaux et al. 2002 |
| N36G | Loop | Hdj1-DnaJ chimera | No effect | Genevaux et al. 2002 |
| R36G; N37G; Q38G | Loop | E. coli DnaJ | Partial complementation | Genevaux et al. 2002 |
| RNQ (36-38) GGG | Loop | E. coli DnaJ | Lack of complementation | Genevaux et al. 2002 |
| H33Q; HD(33,35)YY; D35E | Loop | A. tumefaciens DnaJ | Lack of complementation | Hennessy et al. 2005 |
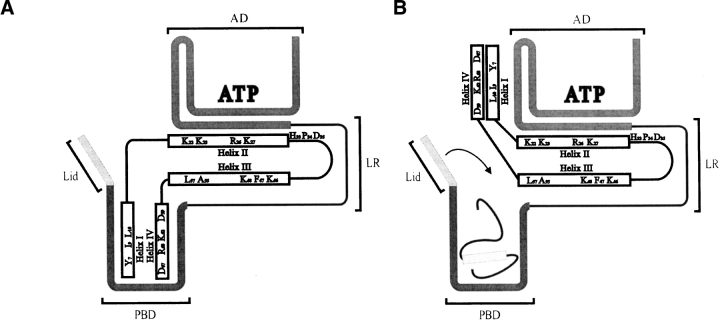
Figure 5.
Model for the interaction of the J domain with Hsp70. Residues important in J domain–based interactions with Hsp70 are shown using the single letter code. The J domain has four helices labeled I–IV. Charged residues in helices II and III interact with the underside of the Hsp70 ATPase domain (AD). (A) helices I and IV possibly act as a client protein mimic and interact transiently with the Hsp70 peptide binding domain (PBD). (B) The correct client protein at the correct concentration is able to displace helices I and IV and bind in the PBD; this interaction is stabilized by subsequent ATP hydrolysis and PBD lid closure (indicated by the downward arrow). The Hsp70 linker region is designated LR.
Recently it has been shown that substitution of certain key residues in helix II, such as K26 in E. coli DnaJ (Genevaux et al. 2002), R26 in Agrobacterium tumefaciens DnaJ (Hennessy et al. 2005), and helix IV (D59 and R63 of the QKRAA motif of A. tumefaciens DnaJ; Hennessy et al. 2005), disrupt J domain-based interactions with Hsp70. Interestingly, in a modeled complex between auxilin and Hsc70, the auxilin residue (R867) equivalent to E. coli DnaJ K26 has been predicted to make contacts with D206 of the Hsc70 ATPase domain (Gruschus et al. 2004a). The D206 residue is highly conserved in the Hsp70 family, forming part of the ATP binding site, and Gruschus et al. (2004a) propose that the positively charged J domain could pull the negatively charged phosporyl group of ATP toward it, thereby enhancing ATP hydrolysis. Based on our recent studies (Hennessy et al. 2005) and the extensive findings of others (Szyperski et al. 1994; Greene et al. 1998; Suh et al. 1998), we propose that helix II (and possibly helix III) with its strong positively charged character is the primary site on the J domain that interacts with the negatively charged undercleft of the ATPase domain of Hsp70. Helix IV has been suggested for certain Hsp40s to form a secondary site of contact with Hsp70 (Suh et al. 1999). We propose that not only helix IV, but also the hydrophobic helix I, may together bind to the peptide binding domain of Hsp70 as a weakly binding “client protein mimic”, thereby changing the affinity of binding and acting as a specificity determinant (Fig. 5 ▶). This is similar to the “client protein” function initially envisaged for the Gly/Phe-rich region (Karzai and McMacken 1996). This proposal is partly consistent with the modeled auxilin–Hsc70 complex (Gruschus et al. 2004a), which predicted that sequences near helix I of the J domain (helix 2 of auxilin) interacted with the peptide binding domain of Hsc70.
We envisage that these J domain contacts with Hsp70 are highly dynamic interactions and that the nature of binding changes in the presence of client protein. An appropriate client protein at the right concentration may enter the system by direct interaction with Hsp70 or through presentation by an Hsp40 or Hsp40-like partner protein. In either scenario, we propose that the client protein would have to have a high enough affinity for Hsp70, and be at the right concentration, before it can compete effectively with the J domain of an Hsp40 or Hsp40-like partner protein, so as to result in a protein triad that fully and appropriately couples the stimulation of ATP hydrolysis and the conformational changes necessary for client protein capture by Hsp70. Therefore the specificity of the system resides at the level of both the client protein and the J domain. We propose that an inappropriate client protein with a low capacity to be chaperoned (e.g., peptides from protein degradation) may not be able to compete with the J domain and will be excluded from the Hsp40–Hsp70 system, and only once an appropriate client protein and an Hsp40 or Hsp40-like partner protein are present simultaneously will a productive interaction occur with a particular Hsp70. This proposed model for the specificity of J domain function has implications for the mechanism of J domain action, especially when one considers the direct dynamic binding of the J domain to two different sites on an Hsp70 (ATPase and peptide binding domains). In the absence of substrate there are potentially two independent binding interactions occurring simultaneously between two different surfaces of the J domain with two different surfaces of an Hsp70, thereby creating dynamic strain/tension in the system that is delicately poised to facilitate the coupling of ATP hydrolysis with conformational change, should a suitable client protein enter the system. Once a suitable client protein enters the system, binding of the J domain to the ATPase domain could be favored, perhaps through conformational bias of an induced fit conformation, and in the process ATP hydrolysis would be coupled to the conformational changes necessary for the Hsp70 to capture the client protein in its peptide binding domain. Consequently, amino acid substitutions (e.g., D35N on the J domain or changes to the ATPase binding cleft residues of an Hsp70) that stabilize a J domain conformation or set of conformations that favor binding to one of the binding sites on an Hsp70, will disrupt the train/tension in the Hsp40–Hsp70 system required for a fully functional Hsp40–Hsp70–client protein interaction.
To fully define the specificity features of the J domain a systematic approach of rational mutagenesis and domain swapping experiments is required. For example, a comparative analysis of the ER-based Hsp40 and Hsp40-like proteins (ERj1–5) with the cytosolic Hsp40 and Hsp40-like proteins of mammalian or yeast systems would be a worthwhile and informative study. Ultimately the acquisition of the three-dimensional structures of Hsp40–Hsp70 complexes, perhaps using modified versions of Hsp40 or Hsp40-like proteins that stably associate with a partner Hsp70 (e.g., a J–D35N– DnaK ATPase domain complex), will help to resolve the molecular basis of the Hsp40–Hsp70 interaction.
Acknowledgments
This research was funded in part by a Volkswagen Programme of Partnerships grant (Germany, No. 1/77 191) awarded to G.L.B. and R.Z., a Wellcome Trust grant (UK, No. 066705) awarded to G.L.B., and a National Research Foundation grant (NRF, South Africa, GUN No. 2053542) awarded to G.L.B. F.H. was awarded a NRF grant-holder doctoral bursary, and W.S.N. a NRF postdoctoral fellowship. We are very grateful for the constructive feedback received from the anonymous referees during the review of our manuscript.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051406805.
References
- Auger, I. and Roudier, J. 1997. A function for the QKRAA amino acid motif: Mediating binding of DnaJ to DnaK. Implications for the association of rheumatoid arthritis with HLA-DR4. J. Clin. Invest. 99 1818–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjanskii, M.V., Riley, M.I., Xie, A., Semenchenko, V., Folk, W.R., and Van Doren, S.R. 2000. NMR structure of the N-terminal J domain of murine polyomavirus T antigens. J. Biol. Chem. 275 36094–36103. [DOI] [PubMed] [Google Scholar]
- Berjanskii, M.V., Riley, M.I., and Van Doren, S.R. 2002. Hsc70-interacting HPD loop of the J domain of polyomavirus T antigens fluctuates in ps to ns and μs to ms. J. Mol. Biol. 321 503–516. [DOI] [PubMed] [Google Scholar]
- Bolliger, L., Deloche, O., Glick, B.S., Georgopoulos, C., Jenö, P., Kronidou, N., Horst, M., Morishima, N., and Schatz, G. 1994. A mitochondrial homologue of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. EMBO J. 13 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P., Sander, C., Valencia, A., and Bukau, B. 1992. A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem. Sci. 17 129. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L. and Pipas, J.M. 1998. Polyomavirus T antigens: Molecular chaperones for multiprotein complexes. J. Virol. 72 5329–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, K.S., Mullane, K.P., Aksoy, I.A., Stubdal, H., Zalvide, J., Pipas, J.M., Silver, P.A., Roberts, T.M., Schaffhausen, B.S., and DeCaprio, J.A. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes & Dev. 11 1098–1110. [DOI] [PubMed] [Google Scholar]
- Chamberlain, L.H. and Burgoyne, R.D. 1997. The molecular chaperone function of the secretory vesicle cysteine string protein. J. Biol. Chem. 272 31420–31426. [DOI] [PubMed] [Google Scholar]
- Cheetham, M.E. and Caplan, A.J. 1998. Structure, function and evolution of DnaJ: Conservation and adaption of chaperone function. Cell Stress Chap. 3 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham, M.E., Brion, J-P., and Anderton, B.H. 1992. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochemistry 284 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, M., Rhee, H., Elguindi, E.C., and Blond, S.Y. 2000. Interaction of murine BiP/Grp78 with the DnaJ homologue MTJ1. J. Biol. Chem. 275 19620–19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, D.J., Holland, I.B., and Jacq, A. 1996. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol. Microbiol. 20 1273–1286. [DOI] [PubMed] [Google Scholar]
- Corsi, A.K. and Schekman, R. 1997. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnea, P.M., Miranda-Vizuete, A., Bertoli, G., Simmen, T., Damdimopoulos, A.E., Hermann, S., Leinonen, S., Huikko, M.P., Gustafsson, J.-Å., Sitia, R., et al. 2003. ERdj5, an endoplasmic reticulum ER-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 278 1059–1066. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery, J.R. and Vickery, L.E. 1997. Crystallization and preliminary X-ray crystallographic properties of Hsc20: A J-motif co-chaperone protein from Escherichia coli. Protein Sci. 6 2028–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Crystal structure of Hsc20: A J-type co-chaperone from Escherichia coli. J. Mol. Biol. 304 835–845. [DOI] [PubMed] [Google Scholar]
- Cyr, D.M., Langer, T., and Douglas, M.G. 1994. DnaJ-like proteins: Molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19 176–181. [DOI] [PubMed] [Google Scholar]
- Davis, J.E., Voisine, C., and Craig, E.A. 1999. Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase-and peptide-binding domains. Proc. Natl. Acad. Sci. 96 9269–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche, O., Kelley, W.L., and Georgopoulos, C. 1997. Structure-function analyses of Ssc1p, Mdj1p and Mge1p Saccharomyces cerevisiae mitochondrial proteins in Escherichia coli. J. Bacteriol. 179 6066–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand, J., Lüders, J., and Höhfeld, J. 1998. The carboxy-terminal domain of Hsc70 provides binding sited for a distinct set of chaperone co-factors. Mol. Cell. Biol. 18 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C.Y., Lee, S., Ren, H.Y., and Cyr, D.M. 2004. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell 15 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell, S.W., Pipas, J.M., and Brodsky, J.L. 2002. Mutagenesis of a functional chimeric gene in yeast identifies mutations in the simian virus 40 large T antigen J domain. Proc. Natl. Acad. Sci. 99 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, B.C., Myers, M.P., Schumacher, R., and Morimoto, R.I. 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gässler, C.S., Buchberger, A., Laufer, T., Mayer, M.P., Schröder, H., Valencia, A., and Bukau, B. 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. 95 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, P., Wawrzynáw, A., Zylicz, M., Georgopoulos, C., and Kelley, W.L. 2001. DjlA is a third DnaK co-chaperone of Escherichia coli, and DjlA-mediated induction of colanic acid capsule requires DjlA–DnaK interaction. J. Biol. Chem. 276 7906–7912. [DOI] [PubMed] [Google Scholar]
- Genevaux, P., Schwager, F., Georgopoulos, C., and Kelley, W.L. 2002. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ Hsp40 J domain. Genetics 162 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, P., Lang, F., Schwager, F., Vartikar, J.V., Rundell, K., Pipas, J.M., Georgopoulos, C., and Kelley, W.L. 2003. Simian virus 40 T antigens and J domains: Analysis of Hsp40 cochaperone functions in Escherichia coli. J. Virol. 77 10706–10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler, S.M., Pierpaoli, E.V., and Christen, P. 1998. Catapult mechanism renders the chaperone action of Hsp70 unidirectional. J. Mol. Biol. 279 833–840. [DOI] [PubMed] [Google Scholar]
- Greene, M., Makos, K., and Landry, S.J. 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. 95 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschus, J.M., Greene, L.E., Eisenberg E., and Ferretti J.A. 2004a. Experimentally biased model structure of the hsc70/auxilin complex: Substrate transfer and interdomain structural change. Protein Sci. 13 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschus, J.M., Han, C.J., Greener, T., Ferretti, J.A., Greene, L.E., and Eisenberg, E. 2004b. Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles. Biochemistry 43 3111–3119. [DOI] [PubMed] [Google Scholar]
- Han, W. and Christen, P. 2003. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J. Biol. Chem. 278 19038–19043. [DOI] [PubMed] [Google Scholar]
- Hennessy, F., Cheetham, M.E., Dirr, H.W., and Blatch, G.L. 2000. Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chap. 5 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy, F., Boshoff, A., and Blatch, G.L. 2005. Rational mutagenesis of the J domain identifies residues critical to the in vivo function of the Agrobacterium tumefaciens DnaJ. Int. J. Biochem. Cell Biol. 37 177–191. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J., Minami, Y., and Hartl, F.-U. 1995. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 83 589–598. [DOI] [PubMed] [Google Scholar]
- Hosoda, A., Kimata, Y., and Kohno, K. 2003. JPD1: A novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 278 2669–2676. [DOI] [PubMed] [Google Scholar]
- Huang, K., Flanagan, J.M., and Prestegard, J.H. 1998. The influence of C-terminal extension on the structure of the J domain in E. coli DnaJ. Protein Sci. 8 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., Ghose, R., Flanagan, J.M. and Prestegard, J.H. 1999. Backbone dynamics of the N-terminal domain in E. coli DnaJ determined by 15N-and 13CO-relaxation measurements. Biochemistry 38 10567–10577. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Taylor, A.B., Prasad, K., Ishikawa-Brush, Y., Hart, P.J., Lafer, E.M., and Sousa, R. 2003. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry 42 5748–5753. [DOI] [PubMed] [Google Scholar]
- Johnson, J.L. and Craig, E.A. 2000. A role for the Hsp40 YDJ1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 20 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, R. and McMacken, R. 1995. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J. Biol. Chem. 270 4563–4569. [DOI] [PubMed] [Google Scholar]
- Kabani, M., Beckerich, J.-M., and Brodsky, J.L. 2003. The yeast Sls1p and Fes1p proteins define a new family of Hsp70 nucleotide exchange factors. Curr. Genom. 4 263–273. [Google Scholar]
- Karzai, A.W. and McMacken, R. 1996. A bipartite signalling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271 11236–11246. [DOI] [PubMed] [Google Scholar]
- Kelley, W.L. 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23 222–227. [DOI] [PubMed] [Google Scholar]
- Kelley, W.L. and Georgopoulos, C. 1997. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl. Acad. Sci. 94 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.-Y., Ahn, B.-Y., and Cho, Y. 2001. Structural basis for the inactivation of retinoblastoma tumor supressor by SV40 large T antigen. EMBO J. 20 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluck, C.J., Patzelt, H., Genevaux, P., Brehmer, D., Rist, W., Schneider- Mergener, J., Bukau, B., and Mayer, M.P. 2002. Structure-function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J. Biol. Chem. 277 41060–41069. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. 1991. Molscript: A programme to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Kroczynska, B. and Blond, S.Y. 2001. Cloning and characterization of a new soluble murine J-domain protein that stimulates BiP, Hsc70 and DnaK ATPase activity with different efficiencies. Gene 273 267–274. [DOI] [PubMed] [Google Scholar]
- Landry, S.J. 2003. Structure and energetics of an allele-specific interaction between dnaJ and dnaK: Correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry 42 4926–4936. [DOI] [PubMed] [Google Scholar]
- Laufen, T., Mayer,M.P., Beisle, C., Klostermeier, D.,Mogk, A., Reinstein, J., and Bukau, B. 1999. Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. 96 5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt, M.J. and Kawula, T.H. 1995. Hsc66, an Hsp homologue in Escherichia coli, is induced by cold shock and not by heat shock. J. Bacteriol. 177 4900–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Söderbärg, K., Houshmand, H., You, Z.-Y., and Magnusson, G. 2001. Effect on polyomavirus T-antigen function of mutations in a conserved leucine-rich segment of the DnaJ domain. J. Virol. 75 2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek, K., Marszalek, J., Ang, D., Georgopoulos, C., and Zylicz, M. 1991. Escherichia coli DnaJ and GrpE heat proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. 88 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke, K., Wolfram, T., Bussemer, J. and Jakob, U. 2003. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 278 44457–44466. [DOI] [PubMed] [Google Scholar]
- Lu, Z. and Cyr, D.M. 1998. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone YDJ1 assist Hsp70 in protein folding. J. Biol. Chem. 273 5970–5978. [DOI] [PubMed] [Google Scholar]
- Lyman, S.K. and Schekman, R. 1995. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J. Cell Biol. 131 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Yamout, M., Legge, G.B., Zhang, O., Wright, P.E. and Dyson, H.J. 2000. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol. 300 805–818. [DOI] [PubMed] [Google Scholar]
- Mayer, M.P., Laufen, T., Paal, K., McCarty, J.S., and Bukau, B. 1999. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289 1131–1144. [DOI] [PubMed] [Google Scholar]
- McCarty, J.S., Buchberger, A., Reinstein, J., and Bukau, B. 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249 126–137. [DOI] [PubMed] [Google Scholar]
- Michels, A.M., Kanon, B., Bensaude, O., and Kampinga, H.H. 1999. Heat shock proteins Hsp40 mutants inhibit Hsc70 in mammalian cells. J. Biol. Chem. 274 36757–36763. [DOI] [PubMed] [Google Scholar]
- Minami, Y., Höhfeld, J., Ohtsuka, K., and Hartl, F.-U. 1996. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homologue Hsp40. J. Biol. Chem. 271 19617–19624. [DOI] [PubMed] [Google Scholar]
- Misselwitz, B., Staeck, O., and Rapoport, T.A. 1998. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2 593–603. [DOI] [PubMed] [Google Scholar]
- Moro, F., Fernández, V., and Muga, A. 2003. Interdomain interaction through helices A and B of DnaK peptide binding helices. FEBS Lett. 533 119–123. [DOI] [PubMed] [Google Scholar]
- Motohashi, K., Taguchi, H., Ishii, N., and Yoshida, M. 1994. Isolation of the stable hexameric DnaK–DnaJ complex from Thermus thermophilus. J. Biol. Chem. 269 27074–27079. [PubMed] [Google Scholar]
- Motohashi, K., Yohda, M., Endo, I., and Yoshida, M. 1996. A novel factor required for the assembly of the DnaK and DnaJ chaperones of Thermus thermophilus. J. Biol. Chem. 271 17343–17348. [DOI] [PubMed] [Google Scholar]
- Nagata, H., Hansen, W.J., Freeman, B., and Welch, W.J. 1998. Mammalian cytosolic DnaJ homologues affect the hsp70 chaperone-substrate reaction cycle, but do not interact directly with nascent or newly synthesized proteins. Biochemistry 37 6924–6938. [DOI] [PubMed] [Google Scholar]
- Ohki, M., Tamura, F., Nishimura, S., and Uchida, H. 1986. Nucleotide sequence of the Escherichia coli dnaJ gene and purification of the gene product. J. Biol. Chem. 261 1778–1781. [PubMed] [Google Scholar]
- Pellecchia, M., Szyperski, T., Wall, D., Georgopoulos, C., and Wüthrich, K 1996. NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol. 260 236–250. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, E.V., Sandmeier, E., Baici, A., Schönfeld, H.-J., Gisler, S., and Christen, P. 1997. The power stroke of the DnaK/DnaJ/GrpE molecular chaperone system. J. Mol. Biol. 269 757–768. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, E.V., Gisler, S., and Christen, P. 1998. Sequence-specific rates of interaction of target peptides with the molecular chaperones DnaK and DnaJ. Biochemistry 37 16741–16748. [DOI] [PubMed] [Google Scholar]
- Qian, Y.Q., Patel, D., Hartl, F.-U., and McColl, D.J. 1996. Nucleic Magnetic Resonance solution structure of the human Hsp40 HDJ-1 J domain. J. Mol. Biol. 260 224–235. [DOI] [PubMed] [Google Scholar]
- Riley, M.I., Yoo, W., Mda, N.Y., and Folk, W.R. 1997. Tiny T antigen: An autonomous polyomavirus T antigen amino-terminal domain. J. Virol. 71 6068–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R., Karzai, A.W., Mehl, A.F., and McMacken, R. 1999. DnaJ dramatically stimulates ATP hydrolysis by DnaK: Insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry 38 4165–4176. [DOI] [PubMed] [Google Scholar]
- Schlenstedt, G., Harris, S., Risse, B., Lill, R., and Silver, P.A. 1995. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 129 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, R.J., Hansen, W.J., Freeman, B.C., Alnemri, E., Litwack, G., and Toft, D.O. 1996. Cooperative action of Hsp70, Hsp90 and DnaJ proteins in protein renaturation. Biochemistry 35 14889–14898. [DOI] [PubMed] [Google Scholar]
- Shomura, Y., Dragovic, Z., Chang, H.-C., Tzvetkov, N., Young, J.C., Brodsky, J.L., Guerriero, V., Hartl, F.-U., and Bracher, A. 2005. Regulation of Hsp70 by HspBP1. Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17 367–379. [DOI] [PubMed] [Google Scholar]
- Silberg, J.J. and Vickery, L.E. 2000. Kinetic characterization of the ATPase cycle of the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem. 275 7779–7786. [DOI] [PubMed] [Google Scholar]
- Silberg, J.J., Hoff, K.G., and Vickery, L.E. 1998. The Hsc66–Hsc20 chaperone system in Escherichia coli: Chaperone activity and interactions with the DnaK–DnaJ–GrpE system. J. Bacteriol. 180 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubdal, H., Zalvide, J., Campbell, K.S., Schweitzer, C., Roberts, T.M., and DeCaprio, J.A. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of Simian Virus 40 Large T antigen. Mol. Cell. Biol. 17 4979–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, W.-C., Burkholder, W.F., Lu, C.Z., Zhao, X., Gottesman, M.E., and Gross, C.A. 1998. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. 95 15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, W.-C., Lu, C.Z., and Gross, C.A. 1999. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 274 30534–30539. [DOI] [PubMed] [Google Scholar]
- Sullivan, C.S., Cantalupo, P., and Pipas, J.M. 2000a. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20 6233–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, C.S., Tremblay, J.D., Fewell, S.W., Lewis, J.A., Brodsky, J.L., and Pipas, J. 2000b. Species-specific elements in the large T antigen J domain are required for cellular transformation and DNA replication by Simian virus 40. Mol. Cell. Biol. 20 5749–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., Langer, T., Schröder, H., Flanagan, J., Flanagan, J., Bukau, B., and Hartl, F.-U. 1994. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system—DnaK, DnaJ and GrpE. Proc. Natl. Acad. Sci. 91 10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski, T., Pellecchia, M., Wall, D., Georgopoulos, C., and Wuthrich, K. 1994. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: Secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc. Natl. Acad. Sci. 91 11343–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J. and Douglas, M.G. 1996. A conserved HPDsequence of the J-domain is neccessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271 9347–9354. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Kakeda, M., Yamada, H., and Mizuno, T. 1994. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. 91 1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., Shiozawa, T., Kakeda, M., Yamada, H., and Mizuno, T. 1995. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J. Bacteriol. 177 3894–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery, L.E., Silberg, J.J., and Ta, D.T. 1997. Hsc66 and Hsc20: A new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 6 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, P., Bursac, D., Law, Y.C., Cyr, D., and Lithgow, T. 2004. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 5 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn, A., Taylor, K., and Wegrzyn, G. 1996. The cbpA chaperone gene function compensates for dnaJ in plasmid replication during amino acid starvation of Esherichia coli. J. Bacteriol. 178 5847–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittung-Stafshede, P., Guidry, J., Horne, B.E., and Landry, S.J. 2003. The J domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry 42 4937–4944. [DOI] [PubMed] [Google Scholar]
- Yan, W. and Craig, E.A. 1999. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 19 7751–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W., Gale, M.J.J., Tan, S.-L., and Katze, M.G. 2002. Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone P58 IPK does not require J domain function. Biochemistry 41 4938–4945. [DOI] [PubMed] [Google Scholar]
- Yoshimune, K., Yoshimura, T., Nakayama, T., Nishino, T., and Esaki, N. 2002. Hsc62, Hsc56 and GrpE, the third Hsp70 chaperone system of Escherichia coli. Biochem. Biophys. Res. Commun. 293 1389–1395. [DOI] [PubMed] [Google Scholar]
- Zalvide, J., Stubdal, H., and DeCaprio, J.A. 1998. The J domain of Simian virus 40 Large T Antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18 1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]