Abstract
Topology predictions for integral membrane proteins can be substantially improved if parts of the protein can be constrained to a given in/out location relative to the membrane using experimental data or other information. Here, we have identified a set of 367 domains in the SMART database that, when found in soluble proteins, have compartment-specific localization of a kind relevant for membrane protein topology prediction. Using these domains as prediction constraints, we are able to provide high-quality topology models for 11% of the membrane proteins extracted from 38 eukaryotic genomes. Two-thirds of these proteins are single spanning, a group of proteins for which current topology prediction methods perform particularly poorly.
Keywords: topology prediction, transmembrane protein, domain assignment, prediction constraints
α-Helical transmembrane proteins constitute about 20% of all proteins encoded by most genomes (Krogh et al. 2001), and are responsible for several vital processes in the cell. In addition, the medical importance of membrane bound receptors, channels, and pumps as targets for drugs is well established. Still, for the large majority of membrane proteins, the structure or even the topology, i.e., the positions and in/out orientations of all transmembrane helices, is not known experimentally. The continuously growing amount of sequence data, in combination with the limited amount of structural data available, highlight the need for better and more accurate theoretical structure prediction methods, particularly for the annotation of membrane proteins.
Protein domains are modular, independently evolving, and structurally similar amino acid segments, which may exist alone in single-domain proteins, or may combine to form multidomain proteins. Although covalent combinations between transmembrane domains, (i.e., domains with one or more membrane spanning regions) rarely occur, covalent combinations between soluble domains and transmembrane domains are observed frequently (Liu et al. 2004). Moreover, domains are often compartment-specific, and information about domain occurrence can be used to predict the subcellular localization of soluble proteins (Mott et al. 2002).
Here, we explore the possibility that the presence of compartment-specific extra-membranous protein domains in transmembrane protein sequences might be used as a constraint in a subsequent topology prediction step, in much the same way that experimentally determined “anchor points” have been used to constrain topology predictions (Kim et al. 2003; Rapp et al. 2004; Daley et al. 2005). Unconstrained topology predictions are correct for only ~55%–60% of all membrane proteins (Melén et al. 2003), while, as shown below, compartment-specific domains that are always located on just one side of a membrane (facing, e.g., the extracellular space or the cytosol) can be identified with high reliability. If such a domain is found in a membrane protein, that particular segment in the protein sequence can be fixed to the corresponding side of the membrane before applying a sequence-based topology prediction algorithm on the rest of the sequence. Here, we show that domains of this kind are found in at least 11% of many eukaryotic proteomes, and that a significant improvement in topology prediction can be achieved by using these domains as prediction constraints.
Results
Our basic approach consists of three steps:
Domain selection. Identify compartment-specific domains that always reside on either the inside or outside of the membrane. Each domain is represented by a profile Hidden Markov Model (HMM).
Domain assignment. For each query sequence, try to find one or more of the domains identified in the first step and fix those residues to the corresponding side of the membrane.
Topology prediction. Use a sequence-based method to predict the topology of the remaining part of the protein sequence, with the domain(s) found in the previous step constrained to either the inside or out-side of the membrane.
Domain selection
SMART (Letunic et al. 2004) is a database of well-annotated protein domains, represented as profile- HMMs, and is divided into four main categories: extracellular, nuclear, signaling, and others. In general, we considered domains annotated in SMART 4.0 as “extracellular” to reside outside of the membrane (i.e., on the noncytoplasmic side), and domains annotated as “signaling” to reside on the inside of the membrane (i.e., on the cytoplasmic side). This assumption is, for the most part, correct, and in agreement with, e.g., Mott et al. (2002).
However, we made one general exception to this rule. All domains were assigned to the 78,371 putative membrane protein sequences (see below), and the domain hits were compared to the topologies predicted by PRO-TMHMM (Viklund and Elofsson 2004), which uses the TMHMM 2.0 architecture (Krogh et al. 2001). If a domain was found to contain one or more predicted transmembrane helices, it was removed from the domain collection. Only four out of 372 domains were discarded this way.
Estimation of error frequency of domain assignments
In order to assess the validity of our domain selection method, the domains were assigned to 297 homology reduced sequences of membrane proteins with experimentally known topologies. This resulted in 48 domain hits, contained in 29 (10%) of the sequences. Out of all domain hits, 47 (98%) were in agreement with the topology. One domain (TarH) was in conflict with a known topology, and was thus removed from the domain collection. Although the test set is small, we consider our domain collection as highly reliable.
The final domain list used for placing constraints on the topology predictions consisted of 367 domains, of which 146 were “IN-domains” (i.e., appear only on the cytoplasmic side of the membrane), and 221 were “OUT-domains” (i.e., appear only on the non-cytoplasmic side of the membrane) (see Supplemental Material S1).
Unconstrained topology predictions
A total of 553,974 protein sequences from 38 eukaryotic genomes (Supplemental Material S2) was downloaded from the SUPERFAMILY Web site (Gough et al. 2001). In an initial topology prediction step, 24% of the sequences were predicted by TMHMM to be membrane proteins, which is in agreement with earlier estimates (Krogh et al. 2001). After a second topology prediction step using PRO-TMHMM (Viklund and Elofsson 2004) and homology reduction (see Materials and Methods), 78,371 putative membrane protein sequences remained for further analysis. These sequences, together with their predicted topologies, are available as Supplemental Material S3 both for the full and homology-reduced data sets.
Constrained topology predictions
The IN/OUT location for the final list of 367 domains was used as constraint for the topology prediction; in other words, we considered the domain assignments to be entirely correct. Of all 78,371 predicted membrane proteins, 8703 (11%) contained one or more of the 367 domains, which is consistent with the fraction of membrane proteins with known topology that contain at least one of the domains (10%; see above). Of these domain hits, 4126 (34%) were in conflict with the unconstrained topology predictions, which is much higher than the same figure for proteins with known topology (Table 1). This discrepancy is not surprising, since we are now dealing with topology predictions as opposed to known topologies, but rather suggests that in those cases where the domain assignments and topology prediction are in conflict, the latter is most likely incorrect. In fact, the fraction of conflicting domain hits is consistent with earlier reported error frequencies of TMHMM topology predictions (Krogh et al. 2001), further supporting this idea.
Table 1.
Fraction of sequences with at least one domain hit in membrane proteins with known topology and those with predicted topology
| Fraction of sequences with at least one domain hit | Fraction of domain hits in conflict with topology | |
| MPs with known top | 10% | 2% |
| MPs with predicted top | 11% | 34% |
For membrane proteins with predicted topology, the fraction of topology-conflicting domain hits is consistent with earlier reported error frequencies of TMHMM (Krogh et al. 2001).
All proteins with at least one domain hit then had their topologies repredicted, but now with the assigned part(s) of the sequence constrained to the corresponding side of the membrane (see Supplemental Material S3).
Domains are more frequent in single-spanning membrane proteins
Based on the constrained predictions, the topologies of the 8703 proteins containing at least one domain were analyzed. Sixty-six percent were single-spanning proteins (Fig. 1 ▶), compared to just 37% in the complete set of predicted membrane proteins, suggesting that our method will have particular impact on single-spanning proteins. Single-spanning proteins are often mispredicted by the current topology prediction methods, mostly due to an inversion of the predicted topology such that the TM-segment is correctly located but the overall orientation is wrong. Large extra-membranous domains carry little or no orientational information in the current predictors, and our domain-based method thus solves a major weakness in these methods.
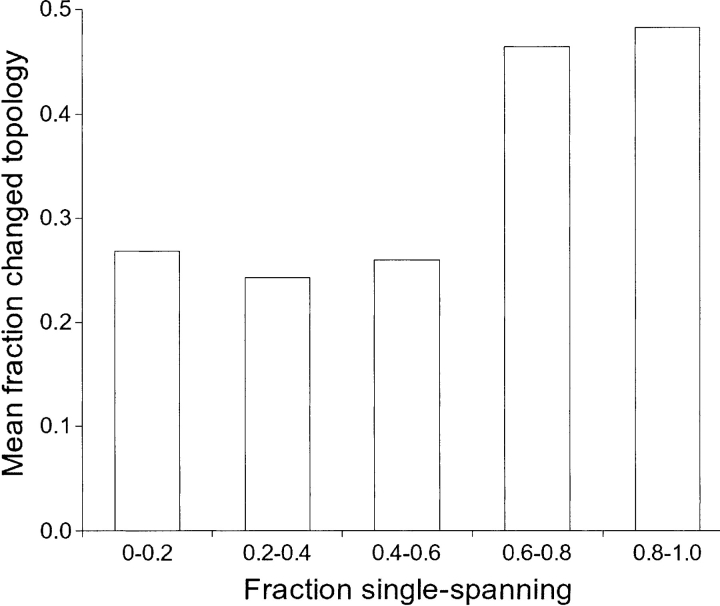
Figure 1.
Mean value of fraction of hits that are in conflict with the unconstrained topology prediction plotted against fraction of hits in single spanning proteins, divided into intervals. Domains with at least 60% single spanning hits are more often in conflict with the unconstrained prediction. Statistics are based on domains with at least 10 different hits. Intervals are exclusive for lower limits and inclusive for upper limits.
Frequency of single domains and domain pairs
For each of the domains, the total number of hits in the 8703 predicted membrane protein sequences was recorded. The large majority of the domains were only found a few times, whereas a few domains were much more prevalent; for instance, the top 15 domains in terms of number of hits represent 44% of the total number of domain hits (Table 2).
Table 2.
The most common IN/OUT-domains found in the predicted membrane protein sequences
| SMART ID | Description | IN/OUT | No. of hits | % Single span |
| S_TKc | Serine/Threonine protein kinases, catalytic domain | IN | 691 | 50 |
| IG | Immunoglobulin | OUT | 522 | 66 |
| TyrKc | Tyrosine kinase, catalytic domain | IN | 487 | 54 |
| RING | Ring finger | IN | 410 | 45 |
| IGc2 | Immunoglobulin C-2 type | OUT | 301 | 67 |
| CA | Cadherin repeats | OUT | 271 | 53 |
| FN3 | Fibronectin type 3 domain | OUT | 246 | 66 |
| CLECT | C-type lectin (CTL) or carbohydrate-recognition domain (CRD) | OUT | 235 | 85 |
| LRRCT | Leucine rich repeat C-terminal domain | OUT | 213 | 74 |
| t_SNARE | Helical region found in SNAREs | IN | 210 | 99 |
| C2 | Protein kinase C conserved region 2 (CalB) | IN | 179 | 68 |
| cNMP | Cyclic nucleotide- monophosphate binding domain | IN | 178 | 2 |
| EGF_CA | Calcium-binding EGF-like domain | OUT | 175 | 67 |
| IGc1 | Immunoglobulin C-Type | OUT | 171 | 55 |
| GPS | G-protein–coupled receptor proteolytic site domain | OUT | 169 | 1 |
The percentage of domain hits in single-spanning proteins, as determined by the constrained predictions, is also indicated.
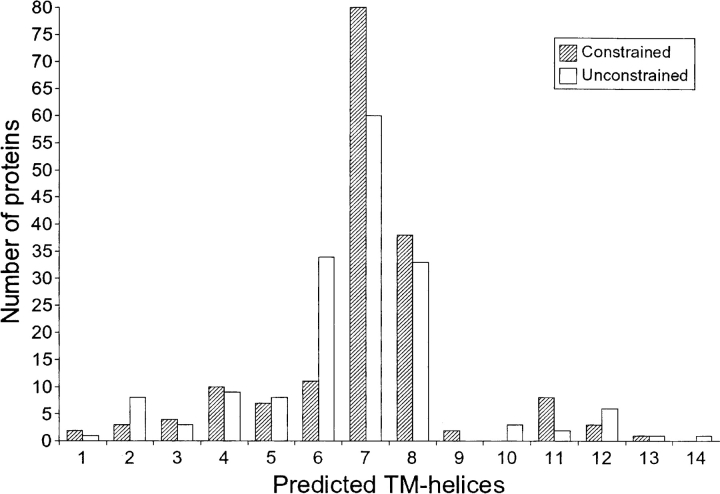
Kinase domains, which are common in various types of membrane bound receptors, are the most prevalent in our data set. This is reflected in their relative ubiquity in single spanning proteins, a property that is shared by most of the domains in Table 2. As an example, the t_SNARE domain is almost exclusively found in single-spanning proteins, which is consistent with experimental data suggesting that most SNAREs have a single TM-helix at their C-terminal end (Ungar and Hughson 2003). In contrast, the number of TM-helices in proteins containing the GPS-domain found in certain G-protein– coupled receptors (GPCRs) peaks at seven (Fig. 2 ▶), which conforms with the 7TM-helix topology characteristic of GPCRs. In this case, the main difference between the unconstrained and constrained predictions is that, for a number of proteins, the topology prediction changes from six TM-helices to seven. It is notable that the SignalP program (Dyrløv-Bendtsen et al. 2004) predicts the presence of a cleavable, N-terminal signal peptide overlapping the most N-terminal predicted TM-helix in 47% of the GPCRs with eight predicted TM-helices but only in 1% of those with seven predicted TM-helices. Cleavable signal peptides are often mistakenly predicted as TM-helices by TMHMM (Krogh et al. 2001; Käll et al. 2004), and are frequently found in GPCRs with large N-terminal domains, but not in those with shorter N-terminal tails (Wallin and von Heijne 1995). Although the GPS domain occurs mainly in the 7TM latrophilin family, it is also found in certain other cell surface receptors such as polycystin-1 (Ponting et al. 1999) that do not share the common 7TM topology of most GPCRs, explaining why a few proteins in Figure 2 ▶ do not have a 7TM or 8TM topology.
Figure 2.
Distribution of the number of predicted TM-helices for proteins containing the GPS-domain, which is found in GPCRs. Fixation of the GPS-domain to the outside of the membrane mainly resulted in a change in topology prediction for a number of proteins from a 6TM-topology to the 7TM-topology characteristic of GPCRs.
Multidomain proteins
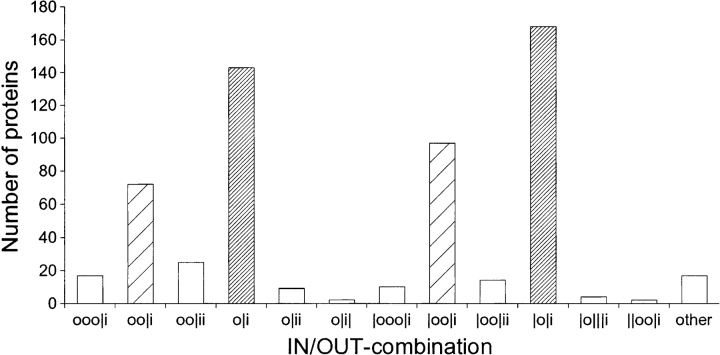
The majority of the 8703 proteins had only one domain hit, but in 2013 (23%) of the cases, more than one domain was found. The 15 most common pair combinations of domains are listed in Table 3. Immunoglobulin domains, which are found in, e.g., antibodies, often appeared together in our data set. The FN3/TyrKc, IG/TyrKc, and Igc2/TyrKc domain pairs mainly represent receptor tyrosine kinases, which constitute a major class of cell surface receptors. In 580 cases, domains were present on both sides of the membrane, i.e., at least one IN-domain and at least one OUT-domain were found in the same protein sequence. Interestingly, these proteins are similar in their IN/OUT combination of domains (Fig. 3 ▶). Denoting an OUT-domain by “o,” an IN-domain by “i,” and a TM-helix by “|,” the two most prevalent IN/OUT combinations are |o|i and o|i (counting from N-to-C terminus), followed by |oo|i and oo|i. In 99% of the cases, the domain closest to the N terminus is an OUT-domain, and the one closest to the C terminus is an IN-domain.
Table 3.
The most common domain pairs and their IN/OUT-position relative to the membrane
| SMART ID | Description | IN/OUT | No. of hits |
| IG | Immunoglobulin | OUT | 156 |
| IGc2 | Immunoglobulin C-2 type | OUT | |
| FN3 | Fibronectin type 3 domain | OUT | 123 |
| IGc2 | Immunoglobulin C-2 type | OUT | |
| EGF | Epidermal growth factor-like domain | OUT | 106 |
| EGF_CA | Calcium-binding EGF-like domain | OUT | |
| LRRCT | Leucine rich repeat C-terminal domain | OUT | 104 |
| LRR_TYP | Leucine-rich repeats, typical (most populated) subfamily | OUT | |
| FN3 | Fibronectin type 3 domain | OUT | 84 |
| IG | Immunoglobulin | OUT | |
| FN3 | Fibronectin type 3 domain | OUT | 75 |
| TyrKc | Tyrosine kinase, catalytic domain | IN | |
| B_lectin | Bulb-type mannose-specific lectin | OUT | 74 |
| S_TKc | Serine/Threonine protein kinases, catalytic domain | IN | |
| B_lectin | Bulb-type mannose-specific lectin | OUT | 64 |
| PAN_AP | Divergent subfamily of APPLE domains | OUT | |
| IG | Immunoglobulin | OUT | 62 |
| TyrKc | Tyrosine kinase, catalytic domain | IN | |
| PSI | Domain found in Plexins, Semaphorins, and Integrins | OUT | 62 |
| Sema | Semaphorin domain | OUT | |
| ACR | ADAM cysteine-rich domain | OUT | 60 |
| DISIN | Homologs of snake disintegrins | OUT | |
| IGc2 | Immunoglobulin C-2 type | OUT | 56 |
| TyrKc | Tyrosine kinase, catalytic domain | IN | |
| LRRNT | Leucine rich repeat N-terminal domain | OUT | 51 |
| LRR_TYP | Leucine-rich repeats, typical (most populated) subfamily | OUT | |
| FN3 | Fibronectin type 3 domain | OUT | 46 |
| PTPc | Protein tyrosine phosphatase, catalytic domain | IN | |
| LRRCT | Leucine rich repeat C-terminal domain | OUT | 43 |
| LRRNT | Leucine rich repeat N-terminal domain | OUT |
Only combinations of different domain types were considered.
Figure 3.
IN/OUT-combinations for proteins with domains on both sides of the membrane. Part of the |o|i-proteins may, in fact, be of the o|i type (narrowly striped bars), with a signal peptide erroneously predicted as a TM-helix. Analogously, |oo|i-proteins may be of the oo|i type (widely striped bars). o=OUT-domain; i=IN-domain; |=TM-helix.
Many of the proteins with |o|i and |oo|i IN/OUT combinations might, in fact, be type Ia single-spanning proteins with an N-terminal signal peptide (see above). If that is the case here, the majority of proteins with domains on both sides of the membrane in reality belong to the o|i and oo|i IN/OUT combinations, i.e., they are single-spanning membrane proteins of type Ia. Since type II proteins, i.e., single-spanning with a cytoplasmic N terminus, often have the TM stretch close to the N terminus, it is not surprising that we find very few i|o proteins. Nevertheless, the bias in favor of type Ia proteins provides further evidence that an IN/OUT assignment of certain domains is indeed valid.
To be certain that the trend observed was not just an artifact of the domain composition, such that the proteins with domains on both sides of the membrane were, e.g., closely related, we looked further into which domains were present in those proteins. No such artifacts were found; for instance, 58 different domain types are represented in the IN/OUT combinations in Figure 3 ▶, and no domain represents> 17% of the total number of domain hits.
Discussion
It has been shown previously that membrane protein topology predictions can be considerably improved if one or the more residues or segments in a protein can be constrained to lie on one or the other side of the membrane prior to running the predictor (Melén et al. 2003). Such information can be obtained experimentally on a proteomewide scale (Daley et al. 2005); here, we show that certain extramembranous protein domains from the SMART database (Letunic et al. 2004) can also be used as prediction constraints.
In a large collection of 78,371 redundancy-reduced proteins from fully sequenced eukaryotic genomes, 11% contain domains that, when found in soluble proteins, have compartment-specific localization. At least two-thirds of these 8703 proteins are single-spanning, and overall, we can correct the unconstrained topology prediction for 34% of the 8703 domain-containing proteins.
Although the coverage of compartment-specific domain hits is limited, this figure will increase as more domains are characterized and included in the SMART database. In fact, domains from the Pfam database (Bateman et al. 2004) were found in >90% of the 297 known membrane proteins analyzed here (data not shown), although the predictive value of those domains remains to be investigated. Although in this paper we have focused only on soluble domains that are devoid of TM-helices, a possible further use of domain information in topology prediction is to attempt to define conserved partial topologies (Nilsson et al. 2002) for protein domains that contain one or more TM-helix and use these as constraints in a subsequent topology prediction step.
In conclusion, domain-based topology constraints provides a solution to a major weakness in current topology prediction schemes, which in general, gain little information from large extramembranous domains.
Materials and methods
Unconstrained topology predictions
In order to extract integral membrane protein (IMP) sequences from the complete set of 553,974 eukaryotic protein sequences in our initial collection, the TMHMM predictor (Krogh et al. 2001) was used and yielded 132,631 sequences with at least one predicted TM-region. As a refinement step, a more computationally demanding topology prediction algorithm employing sequence profiles, PRO-TMHMM (Viklund and Elofsson 2004), was applied to the TMHMM set, generating 100,603 sequences which could more certainly be classified as membrane proteins, i.e., as having at least one TM-region. Finally, to filter out duplicates and close homologs, the sequences were homology-reduced at 90% threshold using the CD-HIT algorithm (Li et al. 2002) (word-size 5), which left us with 78,371 putative IMP sequences lacking any close internal homology, together with their predicted topologies. All topology predictions were performed using the modhmm topology prediction package (Viklund and Elofsson 2004).
Membrane proteins with experimentally known topology were used to test the accuracy of the domain assignment method. Sequences and topologies from three different sources, Mptopo (Jayasinghe et al. 2001), TMpdb (Ikeda et al. 2003), and the Möller database (Möller et al. 2000), were combined, and homology reduced at 40% threshold using the CD-HIT algorithm (Li et al. 2002) (word-size 2). This produced 297 nonredundant membrane protein sequences with experimentally known topologies.
Domain selection
All predicted membrane protein sequences were searched for SMART 4.0 domains (Letunic et al. 2004) annotated as “extracellular” or “signaling,” using an E-value cutoff of 10−6. In order to avoid artifacts arising from domain repeats, only the most significant domain hit for each sequence was retained. Conflicting domain assignments were resolved so that the assignment with the lowest E-value was regarded first, and then any nonconflicting assignments were added in order of increasing E-values. For each domain, the predicted partial topologies (i.e., the topology within the region of the domain hit) of all proteins assigned with this domain were examined, and the total fraction of residues predicted as containing a predicted TM-region was calculated. If this fraction was above 10%, the domain was considered to actually contain TM-regions, and was removed from the domain collection.
As a test of the accuracy of our method, the remaining domains were searched for in the 297 membrane proteins with known topologies. Out of 48 domain hits, one hit was in conflict with the experimentally known topology, and this domain (TarH) was removed from the domain collection.
Constrained topology predictions
All proteins with at least one domain hit had their topologies repredicted using the PRO-TMHMM prediction algorithm (Viklund and Elofsson 2004), with the domain region fixed to the corresponding side of the membrane. The IN/OUT-fixation of a certain residue is achieved by setting the corresponding state probability in the HMM equal to 1.0, and is straightforward using the modhmm package (Viklund and Elofsson 2004). As a precaution not to interfere with any TM-regions, since the positions of both the predicted domain and any predicted TM-helices might be somewhat imprecise, only the core part (i.e., the middle 50%) of the domain assignment was fixed. Conflicting domain assignments were resolved as described above.
SignalP predictions
Predictions were performed using SignalP-HMM for the 70 most N-terminal residues of the sequence. If the probability for a signal peptide exceeded 0.5, and if there was an overlap of at least 10 residues between the signal peptide and the most N-terminal predicted TM-helix, this was taken as an indication that an actual signal peptide might have been mistaken for a TM-helix by TMHMM.
Electronic supplemental material
S1 is a list of SMART domains fixed to an IN/OUT position relative to the membrane. S2 is a list of the 38 eukaryotic species analyzed. S3 contains sequences and predicted topologies for the 78,371 putative eukaryotic membrane proteins analyzed. A redundant version, including all 100,603 non-homology reduced sequences and predicted topologies, is also included.
Acknowledgments
This work was supported by grants from the European Commission (BioSapiens), the Swedish Foundation for Strategic Research, and the Swedish Research Council to GvH.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051395305.
Supplemental material: see www.proteinscience.org
References
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32 D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, D.O., Rapp, M., Granseth, E., Melén, K., Drew, D., and von Heijne, G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science, in press. [DOI] [PubMed]
- Dyrløv-Bendtsen, J., Nielsen, H., von Heijne, G., and Brunak, S. 2004. Improved prediction of signal peptides—SignalP3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R., and Chothia, C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313 903–919. [DOI] [PubMed] [Google Scholar]
- Ikeda, M., Arai, M., Okuno, T., and Shimizu, T. 2003. TMPDB: A database of experimentally-characterized transmembrane topologies. Nucleic Acids Res. 31 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe, S., Hristova, K., and White, S.H. 2001. MPtopo: A database of membrane protein topology. Protein Sci. 10 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll, L., Krogh, A., and Sonnhammer, E.L. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kim, H., Melén, K., and von Heijne, G. 2003. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and prediction. J. Biol. Chem. 278 10208–10213. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. 2001. Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32 D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Jaroszewski, L., and Godzik, A. 2002. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18 77–82. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Gerstein, M., and Engelman, D.M. 2004. Transmembrane protein domains rarely use covalent domain recombination as an evolutionary mechanism. Proc. Natl. Acad. Sci. 101 3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melén, K., Krogh, A., and von Heijne, G. 2003. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327 735–744. [DOI] [PubMed] [Google Scholar]
- Möller, S., Kriventseva, E., and Apweiler, R. 2000. A collection of well-characterised integral membrane proteins. Bioinformatics 16 1159–1160. [DOI] [PubMed] [Google Scholar]
- Mott, R., Schultz, J., Bork, P., and Ponting, C.P. 2002. Predicting protein cellular localization using a domain projection method. Genome Res. 12 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, J., Persson, B., and von Heijne, G. 2002. Prediction of partial membrane protein topologies using a consensus approach. Protein Sci. 11 2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P., Hofmann, K., and Bork, P. 1999. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr. Biol. 9 R585–R588. [DOI] [PubMed] [Google Scholar]
- Rapp, M., Drew, D.E., Daley, D.O., Nilsson, J., Carvalho, T., Melén, K., de Gier, J.W., and von Heijne, G. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13 937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, D. and Hughson, F.M. 2003. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19 493–517. [DOI] [PubMed] [Google Scholar]
- Viklund, H. and Elofsson, A. 2004. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 13 1908–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin, E. and von Heijne, G. 1995. Properties of N-terminal tails in G-protein coupled receptors—A statistical study. Protein Eng. 8 693–698. [DOI] [PubMed] [Google Scholar]