Abstract
Escherichia coli thioredoxin is a small monomeric protein that reduces disulfide bonds in cytoplasmic proteins. Two cysteine residues present in a conserved CGPC motif are essential for this activity. Recently, we identified mutations of this motif that changed thioredoxin into a homodimer bridged by a [2Fe-2S] iron–sulfur cluster. When exported to the periplasm, these thioredoxin mutants could restore disulfide bond formation in strains lacking the entire periplasmic oxidative pathway. Essential for the assembly of the iron–sulfur was an additional cysteine that replaced the proline at position three of the CGPC motif. We solved the crystalline structure at 2.3 Å for one of these variants, TrxA(CACA). The mutant protein crystallized as a dimer in which the iron–sulfur cluster is replaced by two intermolecular disulfide bonds. The catalytic site, which forms the dimer interface, crystallized in two different conformations. In one of them, the replacement of the CGPC motif by CACA has a dramatic effect on the structure and causes the unraveling of an extended α-helix. In both conformations, the second cysteine residue of the CACA motif is surface-exposed, which contrasts with wildtype thioredoxin where the second cysteine of the CXXC motif is buried. This exposure of a pair of vicinal cysteine residues apparently allows thioredoxin to acquire an iron–sulfur cofactor at its active site, and thus a new activity and mechanism of action.
Keywords: thioredoxin, iron–sulfur cluster, crystal structure, disulfide bond, periplasm
An important step in the folding of newly synthesized proteins is the formation of native disulfide bonds between the thiol groups of two cysteine residues (Collet and Bardwell 2002). In prokaryotes, disulfide bond formation occurs in the periplasm and is catalyzed by a protein called DsbA (Bardwell et al. 1991). DsbA is a small soluble protein, which has a catalytic CXXC motif present in a thioredoxin-like fold. The two cysteines of DsbA are found to be predominantly oxidized in vivo (Kishigami et al. 1995). The disulfide bond of DsbA is very unstable (Zapun et al. 1993) and can be rapidly transferred to newly translocated proteins. After the transfer of its disulfide bond to target proteins, DsbA is reduced and is then reoxidized by the membrane protein DsbB. DsbB itself is kept oxidized by the electron transport chain (Bader et al. 1999). Both DsbA and DsbB are required to form disulfide bonds in vivo. Inactivation of either the dsbA or dsbB gene abolishes the oxidation of secreted proteins. One striking example of the importance of disulfide bonds for the correct folding of secreted proteins is the Escherichia coli flagellar protein FlgI, a component of the bacterial flagellum. The folding of FlgI requires a disulfide bond. When this disulfide is not introduced in FlgI, FlgI cannot be properly folded and a functional flagellum cannot be assembled. This results in the complete loss of bacterial motility (Dailey and Berg 1993).
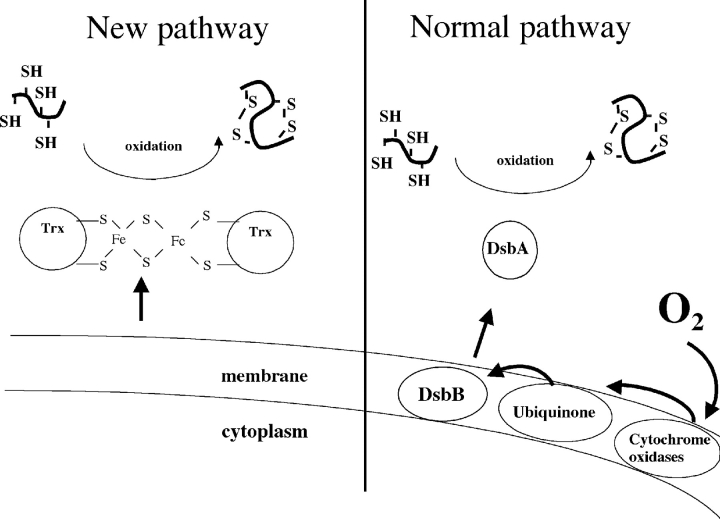
Recently, we designed a new pathway for the formation of disulfide bonds in the periplasm that completely bypasses the need for both DsbA and DsbB (Fig. 1 ▶) (Masip et al. 2004). By imposing evolutionary pressure, we isolated mutants of E. coli thioredoxin (TrxA) that can catalyze oxidative protein folding in the bacterial periplasm independently of the action of DsbA and DsbB (Masip et al. 2004). TrxA is a small monomeric protein that has a CXXC catalytic motif (CGPC) and a thioredoxin fold, like DsbA. In contrast to DsbA, however, TrxA catalyzes disulfide bond reduction and it does so in the cytoplasm, not the periplasm. Formation of disulfide bonds by the TrxA mutants represents a reversal of the physiological action of this protein. The hallmark of the isolated mutants is the presence of a third cysteine in the catalytic site of the protein, replacing the CXXC motif with CXCC. We characterized the TrxA mutants that could rescue disulfide bond formation and showed that they are dramatically different from the wild-type thioredoxin: They are dimeric proteins bridged by a [2Fe-2S] iron–sulfur cluster. We also showed that the first and the second cysteine residues of the CXCC motif are involved in cluster coordination: A TrxA mutant with a CACA active site sequence [TrxA(CACA)] is also a dimeric protein bridged by an iron–sulfur cluster.
Figure 1.
A new pathway for the formation of disulfide bonds in the periplasm. In the normal pathway, disulfide bonds are introduced in reduced secreted proteins by DsbA. DsbA is then recycled by the inner-membrane protein DsbB, which is reoxidized by molecular oxygen in a ubiquinone- and cytochrome oxidase-dependent reaction. We recently developed a new pathway for disulfide bond formation: A thioredoxin mutant with a [2Fe-2S] cluster is exported from the cytoplasm to the periplasm where it can catalyze disulfide bond formation independently of the action of DsbA and DsbB.
Why did the simple addition of a cysteine into the catalytic site of thioredoxin cause such a dramatic modification in the function of this enzyme? To address this question, we decided to examine the structure of the mutant protein using X-ray crystallography. Because we observed that the iron–sulfur cluster in TrxA(CACA) is more stable than the one in TrxA mutants with a CXCC motif, TrxA(CACA) was used in this study.
The structure of the TrxA(CACA) has been solved at 2.3 Å resolution. The mutant protein crystallized as a disulfide-linked dimer, each monomer had an active site that was present in a different conformation. In one of them, the replacement of the CGPC motif by CACA has a dramatic effect on the structure and causes the unraveling of an extended α-helix. In both conformations, the second cysteine residue of the CACA motif is surface- exposed. Based on this structure, we propose a model for the formation of an iron–sulfur cluster in this novel thioredoxin.
Results
Description of the structure
We solved the structure of a mutant of thioredoxin that had been selected by its ability to catalyze disulfide bond formation and had been subsequently shown to have acquired a [2Fe-2S] iron–sulfur cluster. The TrxA(CACA) mutant protein crystallized in the monoclinic P21 space group (Table 1). This differs from wildtype TrxA, which crystallizes in the C2 space group (Holmgren et al. 1975; Katti et al. 1990). Though unusual, other thioredoxin mutants are known to crystallize in a space group different from that of the wild-type protein. For instance, TrxA(CVWC) crystallized in a tetragonal P43212 space group (Schultz et al. 1999) and TrxA(CGSC) in the triclinic P1 space group (Rudresh et al. 2002).
Table 1.
Crystallographic data statistics
| Data collection statistics | |
| Space group | P21 |
| Unit cell | a = 34.7 Å, b = 46.3 Å, c = 126.0 Å, β = 93.1° |
| Dmin (Å) | 50–2.3 |
| No. of measurements | 60,618 |
| No. of unique reflections | 16,159 |
| Completeness (%)a | 87.8 (89.8) |
| I/σa | 13 (3.5) |
| Rsym (%)a,b | 7.3 (27.7) |
| Refinement statistics (native dataset) | |
| No. of reflections (working/test) | 13,702/1,509 |
| No. of nonhydrogen atoms | 3,164 |
| No. of water molecules | 65 |
| Resolution (Å) | 43.4–2.3 |
| Rcryst/Rfree (%)C | 22.9/28.1 |
| Bond length deviation (Å) | 0.006 |
| Bond angle deviation (°) | 1.3 |
| Average B-factor of model (Å2) | 25.5 |
| Estimated minimal error (Å) | 0.039 |
| Estimated maximal error (Å) | 0.277 |
a Values in parentheses are for the highest resolution bin.
bRsym= ∑h∑i|Ii(h) − 〈I(h)〉|/ ∑h〈I(h)〉, where Ii(h) is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
cR= ∑ (|Fobs| − K|Fcalc|)/∑|Fobs|. Rfree is the R-value obtained for a test set of reflections that consisted of a randomly selected 5% subset of the diffraction data used during refinement of σA value calculations.
TrxA(CACA) has an overall structure very similar to that of wild-type TrxA: a central pleated β-sheet with five parallel and anti-parallel β-strands, surrounded by four α-helices. There are four molecules (A, B, C, D) of TrxA(CACA) per asymmetric unit. These four TrxA (CACA) molecules are arranged as two dimers. One dimer is made up of chains A and B and the other of chains C and D. In each dimer, both chains have nearly identical structures except in the active site (see below). The first two residues of all four molecules are missing and assumed to be disordered.
The second α-helix crystallized in two different conformations
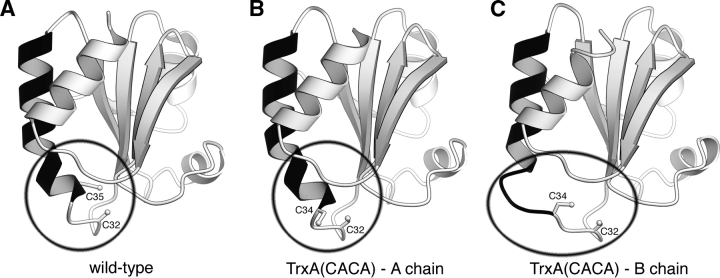
The active site of TrxA is located at the beginning of a long α-helix (helix B), which extends from residue 32 to residue 49 (Fig. 2 ▶). This α-helix is somewhat kinked due to the presence of a proline at residue 40. In molecules A and D, the structure of this region resembles that of the catalytic site of wild-type TrxA (conformation 1). In molecules B and C, however, we observe a dramatic modification of the structure: The first helical turn (32–35) of the helix B completely unravels to adopt an extended conformation while the ensuing helical turn (35–40) unbends itself at the level of Pro40 (conformation 2). As a result, helix B is no longer kinked. This conformational change causes Cys34 to swing out in the catalytic site.
Figure 2.
The unraveling active site. Three ribbon renderings of TrxA monomers. An extended helix (residues 34–50) is colored black. (A) The structure of the wild-type TrxA (PDB accession code 1XOB). (B) The A chain of TrxA(CACA). (C) The B chain of TrxA(CACA). The A chain of the TrxA(CACA) dimer has nearly identical structure to the wild-type form. In the B chain, the beginning of the black extended helix has unraveled to allow the disulfide bond formation. Note that in the wild-type enzyme, only one cysteine (Cys32) is accessible to the solvent, whereas in Trx(CACA) two cysteines (Cys32 and Cys34) are accessible.
Both cysteine residues of the CXCX motif are surface-exposed
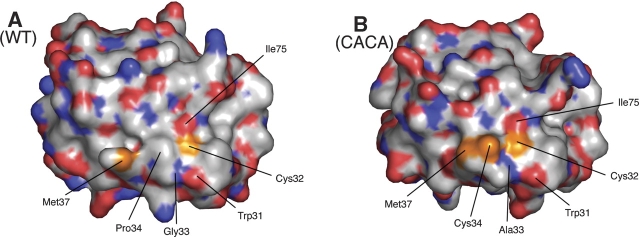
In wild-type TrxA, only the first cysteine of the CXXC motif (Cys32) is surface-exposed (Fig. 3 ▶). Cys32 has two microscopic pKa values (≈7.5 and 9.2) depending on the protonated state of another residue, Asp26 (Chivers et al. 1997). When Asp26 is protonated, Cys32 has a relatively low pKa of 7.5 and is essentially present as a thiolate at physiological conditions. The deprotonation of Cys32 and its location on the surface of the protein allows this residue to perform a nucleophilic attack on a wide variety of substrates, which results in the formation of a mixed-disulfide. The mixed-disulfide is resolved by attack of the second cysteine residue of the CXXC motif (Cys35), which leads to the formation of a disulfide bond between Cys32 and Cys35. In the wildtype protein, Cys35 is buried in the protein structure and kept protonated. It is activated by general base catalysis by Asp26. To determine the location of the two cysteines (Cys32 and Cys34) in TrxA (CACA), we computed the surface of the mutant protein using Pymol (Delano Scientific). In contrast to the situation in the wild-type protein, both active site cysteines are surfaceexposed in the mutant (Fig. 3 ▶).
Figure 3.
Surface accessibility of the catalytic site cysteines. A solvent-accessible surface representation of the wild-type (left) and CACA mutant (right) forms of TrxA (chain A). The surface is colored red for oxygen, blue for nitrogen, white for carbon, and orange for sulfur (cysteines and methionines). In the wild-type enzyme, only Cys32 can be seen on the surface while Cys35 is completely buried. In the CACA mutant, both Cys32 and Cys34 are surface-exposed.
The iron–sulfur cluster is replaced by two disulfide bonds
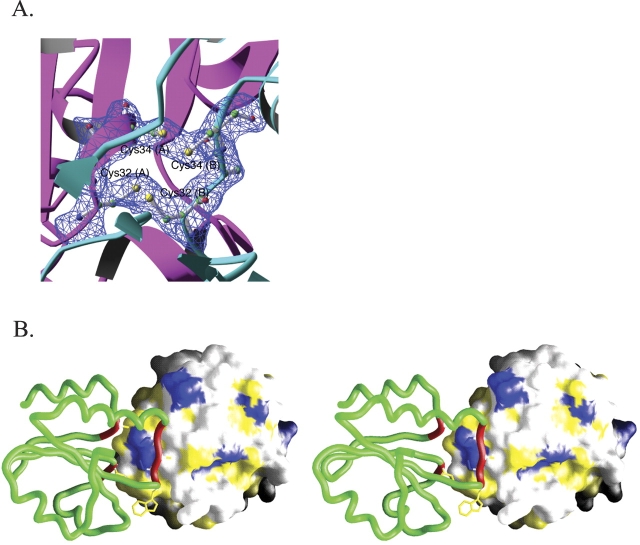
The protein used for the crystallization contained iron and sulfur in a 1:1 ratio. The ratio of iron to thioredoxin monomer was also close to 1:1. We crystallized TrxA (CACA) at 4°C to increase the stability of the cluster. Although the crystals we obtained were brown, which is expected for an iron–sulfur cluster containing crystals, no electron density corresponding to an iron–sulfur cluster could be observed in the structure. We instead found two intermolecular disulfide bonds bridging the catalytic site cysteines (Fig. 4A ▶). Cysteines 32 and 34 of molecule A or D are connected to the corresponding residues of molecule B or C. We suspect that the iron– sulfur cluster was likely degraded over time and free irons were deposited in the solvent channel of the crystal reflecting the brown color of the crystal. Because these free irons do not occupy specific locations in the crystals, they do not contribute to X-ray diffraction. The formation of the disulfide bonds occurs upon break down of the iron–sulfur cluster and had previously been observed in this mutant (Masip et al. 2004).
Figure 4.
The dimer interface. (A) Electron density showing the presence of a pair of disulfide bonds at the TrxA(CACA) dimer interface. A σA-weighted 2|Fo|− |Fc|electron density map (contoured at 1.2 σ) are superimposed on the refined model of TrxA(CACA). For clarity, only residues 32–34 from chain A (magenta) and B (cyan) are shown in atomic details with associated electron density. (B) A stereo view of the TrxA(CACA) dimer is shown. One monomer is represented as a green tube colored red for the residues involved in the dimer interface. The other is represented as a solvent–accessible surface colored blue for charged residues and yellow for hydrophobic residues.
Dimer interface
TrxA(CACA) crystallized as a biological dimer. The region around the catalytic site constitutes the dimer interface, the catalytic site of chain A and C facing the catalytic site of chain B and D, respectively. Using the program CNS (Brunger et al. 1998), we calculated that the surface involved in intermolecular contacts at the level of the dimer interface represents 14% (774 Å2 on 5528 Å2) of the total solvent-accessible surface of the TrxA(CACA) protein, a significant proportion of the total surface area. This value is in the range expected for intersubunit contacts in oligomeric proteins (as little as 5% for larger proteins and up to 20% for smaller ones) (Janin and Chothia 1990).
Many interactions are present at the dimer interface. As shown in Figure 4B ▶, most of them involve hydrophobic residues. Several backbone atom hydrogen bonds also contribute to the interface. For the dimer constituted of chains A and B, this includes interactions between the amide of IleA75 to the carbonyl of TrpB31, the amide of IleB75 to the carbonyl of TrpA31, the amide of AlaA33 to the carbonyl of IleB75, the amide of AlaB33 to the carbonyl of IleA75, and finally the amide of AlaA93 to the carbonyl of AlaB33 (the symmetry related hydrogen bond between AlaB93 and AlaB33 is lost due to the disulfide bond formation breaking the symmetry).
Discussion
Comparison with other TrxA mutants
Several TrxA mutants have been previously crystallized (Nikkola et al. 1993; Schultz et al. 1999; Rudresh et al. 2002). Although the mutations affected residues in close proximity of the catalytic CXXC motif, the structures of these mutants were very similar to that of the wild-type thioredoxin, including the catalytic site. A striking example of this is found in the structure of the P40S TrxA mutant in which Pro40, located in the interior of the α-helix that contains the catalytic site (helix B) and is responsible for introducing a kink in this helix, is replaced by a serine (Nikkola et al. 1993; Rudresh et al. 2002). Interestingly, the elimination of this proline did not affect the overall structure of the helix, which remained kinked. In contrast, the catalytic site of TrxA (CACA) presents some striking differences with the wildtype TrxA. First, it is the first time that TrxA crystallized as a dimer with the catalytic site being the dimer interface. Second, the two cysteine residues at the catalytic site are surface-exposed, which allows them to participate in either iron–sulfur cluster or disulfide bond formation. Finally, in chains B and C, helix B has unraveled, resulting in the alteration of the catalytic site structure. This change in the structure is likely to be a consequence of the mutations introduced at the catalytic site. However, we cannot rule out the possibility that the formation of an intermolecular disulfide bond strained the structure and caused the unraveling of the helix.
Why is an iron–sulfur cluster introduced into TrxA(CACA)?
The replacement of two amino acids in the catalytic site of TrxA changed thioredoxin into a dimeric protein bridged by a [2Fe-2S] iron–sulfur cluster. Iron–sulfur clusters are usually assembled on selected proteins by complex machinery, but they can also form spontaneously. In both cases, four accessible cysteine residues are required for coordination of the cluster. We propose that the presence of two cysteine residues (Cys32 and Cys34) on the surface of the mutant protein is key to the assembly of an iron–sulfur cluster in thioredoxin. However, it is not enough to have solvent exposed cysteines. These cysteines must also be spatially proximate to allow cluster coordination. Therefore, one has to assume that TrxA can form weak dimer in vivo. Formation of a dimer has been reported for one TrxA mutant (Schultz et al. 1999) but not to our knowledge for the wild-type protein.
Both the extent and the nature of the contacts taking place at the dimer interface reinforce the hypothesis of dimer formation by TrxA. As explained above, the size of the contact area and the percentage of hydrophobic residues present at the interface are within the range of what is observed for biological dimer. In fact, most of the residues involved in these contacts have already been proposed to be part of a substrate-binding area by Eklund and coworkers (Eklund et al. 1984). We propose therefore that the residues that are involved in substrate binding by TrxA might also serve to dimerize TrxA in vivo. We cannot exclude the possibility that the formation of some of the interactions present in our structure might be the result of the conformational change observed in chains B and C. For this reason, the dimers formed by TrxA could differ from the dimers formed by TrxA(CACA). However, the requirement to bring the four cysteine residues together to coordinate the iron–sulfur cluster strongly suggests that, in both cases, the catalytic site serves as the dimer interface.
A model for the formation of an iron–sulfur cluster in TrxA(CACA)
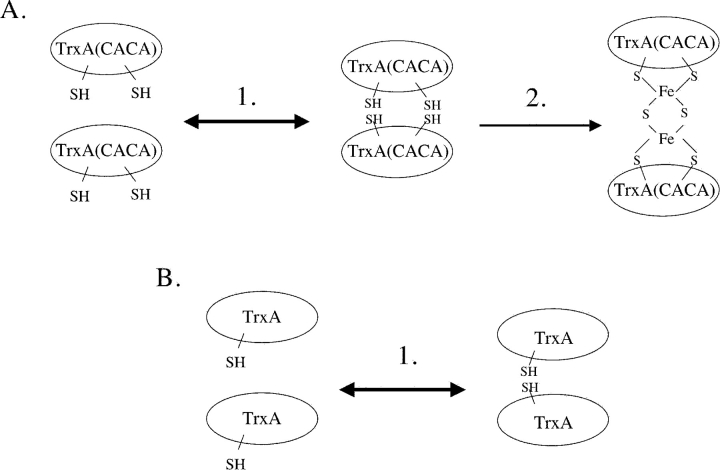
We propose that monomeric TrxA is in equilibrium with dimeric TrxA in vivo (Fig. 5 ▶). In the case of TrxA (CACA), dimer formation brings Cys32 and Cys34 of one subunit in close proximity to Cys32 and Cys34 of a second one. The proximity of these four cysteine residues allows the coordination of an iron–sulfur cluster. This cluster either assembles spontaneously or is introduced by the iron–sulfur cluster machinery. Our structure suggests that the [2Fe-2S] cluster can be fit into the four-cysteine dimer interface with only minor conformational adjustment.
Figure 5.
Model for the formation of a thioredoxin dimer bridged by a [2Fe-2S] iron–sulfur cluster. (A) Monomeric TrxA(CACA) is in equilibrium with a dimeric form of the protein (step 1). The catalytic sites constitute the dimer interface. The presence of four surface-exposed cysteine residues in the dimer interface allows the synthesis of an iron– sulfur cluster by the iron–sulfur cluster machinery (step 2). (B) Monomeric wild-type TrxA also forms weak dimers in vivo (step 1). However, only one cysteine residue is exposed in the catalytic site, which prevents the formation of an iron–sulfur cluster.
Materials and methods
Purification of TrxA(CACA)
BL21(DE3) pLysS bacteria harboring the pET15b plasmids containing the coding DNA sequence of the TrxA(CACA) thioredoxin mutant fused to a N-terminal His-tag were grown aerobically in 1 L of LB medium at 37°C until an A600 of 0.8–1.0 was reached. Isopropylthiogalactoside (IPTG) was added to a final concentration of 0.5 mM and shaking was continued for 5 h to allow protein expression. Cultures were then centrifuged at 5000 rpm and 4°C for 15 min. From this point on all the steps were performed at 4°C. Bacteria were resuspended in 25 mL of buffer A (50 mM sodium phosphate [pH 8.0], 300mMNaCl) containing 1 mMphenylmethylsulfonyl fluoride, and a tablet of Complete protease inhibitor cocktail (Roche) was also added. Bacterial cells were disrupted by French Press and lysates were centrifuged at 14,000 rpm for 30 min. The resulting supernatant (~20mL) was diluted threefold with buffer A and applied onto a 5-mL Hi-Trap Ni++-affinity column (Amersham). The column was washed with 15 mL of buffer A and 15 mL of bufferA containing 20mMimidazole successively. The His-tagged thioredoxin mutant was then eluted with buffer A containing 150 mM imidazole. The purified protein was then concentrated by ultrafiltration and desalted by gel filtration on PD-10 columns in 20 mM Hepes (pH 7.5).
After gel filtration, TrxA(CACA) was incubated with thrombin overnight at 4°C to cleave off the His-tag sequence. The digestion mixture was then loaded onto a second Hi-Trap Ni++-affinity column to purify away the cleaved His-tag and uncleaved protein. The flow-through from this column was then concentrated, desalted as described above, and applied onto a Q-Sepharose Hi-trap column. The column was washed with 25 mL of buffer B (sodium phosphate 25 mM [pH 8]) and protein was eluted with a NaCl gradient (0–400 mM in 100 mL of buffer B).
Protein crystallization and data collection
Diffraction quality crystals of TrxA(CACA) were grown by the sitting drop method. The protein was crystallized at 25 mg/mL in 30%PEG 4000, 0.1MTris (pH 8.5), 0.2MMgCl2, 4% acetonitrile at 4°C. Long, plate-like crystals grew in 1–2 wk. Crystals were cryoprotected by immersion for 1 min in 35% PEG 4000, 0.1MMgCl2, 0.1MTris (pH 8.5), and 4% acetonitrile, followed by flash cooling in liquid nitrogen. The crystals belonged to the monoclinic space group P21 and had unit cell dimensions a=34.7 Å, b=46.3 Å, c=126.0 Å, β =93.1 Å, and contained four molecules of TrxA per asymmetric unit. A 2.3 Å data set was collected at the DND-CAT beamline 5-ID at the Advanced Photon Source. Data were processed with DENZO and SCALEPACK (Otwinowski and Minor 1997).
Structure determination
Initial phases were obtained through the molecular replacement method using the wild-type thioredoxin as a starting model. Four monomers were placed using the program MOLREP from the CCP4 package (Vagin and Teplyakov 2000). All subsequent refinement was performed in CNS with all of the data from 45–2.3 Å, with bulk solvent correction, and with progressmeasured using cross-validation (free R-factor calculation) with a 5% randomly selected test set (Brunger et al. 1998). Initial refinement consisted of rigid body refinement to better place the four monomers, which were then used to generate the non-crystallographic symmetry (NCS) relationships between the monomers. Several cycles of refinement then proceeded with strict NCS imposed on all four molecules. One cycle of refinement consisted of conjugated gradient minimization, grouped B-factor refinement, torsion angle dynamics simulated annealing using the maximum likelihood target function (MLF), followed by model rebuilding in O (Jones et al. 1991). Over the course of refinement, it was realized that the four molecules in the asymmetric unit adopt two substantially different conformations at the catalytic sites. Therefore, the NCS constraints were reduced to restraints between conformationally distinct molecules in the later rounds of refinement. In the final rounds, individual atomic B-factors for the model were refined with no NCS restraints imposed between conformationally distinct molecules and weak restraints between conformationally-identical molecules. The coordinates and structure factors have been deposited into the Protein Data Bank (PDB ID code 1ZCP).
Acknowledgments
We thank J. Stuckey for maintaining the X-ray facility at the University of Michigan Medical School and Life Sciences Institute; K. Yoshino and J.L. Pan for assistance at the early stage of the project; and Z. Wawrzak for access and help at APS DND-CAT beamline 5-ID. DND-CAT is supported by the E.I. DuPont de Nemours & Co., the Dow Chemical Company, the U.S. National Science Foundation through Grant DMR-9304725, and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. This work was supported by an NIH grant to Z.X. (R01-GM60997) and J.C.B. and the University of Michigan Biological Scholar Program. Z.X. is a PEW Scholar in Biomedical Sciences. J.F.C. is supported by a grant from the Belgian Interuniversity Attraction Poles Program.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051464705.
References
- Bader, M., Muse, W., Ballou, D.P., Gassner, C., and Bardwell, J.C. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98 217–227. [DOI] [PubMed] [Google Scholar]
- Bardwell, J.C., McGovern, K., and Beckwith, J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67 581–589. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse- Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Chivers, P.T., Prehoda, K.E., Volkman, B.F., Kim, B.M., Markley, J.L., and Raines, R.T. 1997. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry 36 14985–14991. [DOI] [PubMed] [Google Scholar]
- Collet, J.F. and Bardwell, J.C. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44 1–8. [DOI] [PubMed] [Google Scholar]
- Dailey, F.E. and Berg, H.C. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. 90 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, H., Cambillau, C., Sjoberg, B.M., Holmgren, A., Jornvall, H., Hoog, J.O., and Branden, C.I. 1984. Conformational and functional similarities between glutaredoxin and thioredoxins. EMBO J. 3 1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, A., Soderberg, B.O., Eklund, H., and Branden, C.I. 1975. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 Å resolution. Proc. Natl. Acad. Sci. 72 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin, J. and Chothia, C. 1990. The structure of protein–protein recognition sites. J. Biol. Chem. 265 16027–16030. [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Katti, S.K., LeMaster, D.M., and Eklund, H. 1990. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J. Mol. Biol. 212 167–184. [DOI] [PubMed] [Google Scholar]
- Kishigami, S., Akiyama, Y., and Ito, K. 1995. Redox states of DsbA in the periplasm of Escherichia coli. FEBS Lett. 364 55–58. [DOI] [PubMed] [Google Scholar]
- Masip, L., Pan, J.L., Haldar, S., Penner-Hahn, J.E., DeLisa, M.P., Georgiou, G., Bardwell, J.C., and Collet, J.F. 2004. An engineered pathway for the formation of protein disulfide bonds. Science 303 1185–1189. [DOI] [PubMed] [Google Scholar]
- Nikkola, M., Gleason, F.K., Fuchs, J.A., and Eklund, H. 1993. Crystal structure analysis of a mutant Escherichia coli thioredoxin in which lysine 36 is replaced by glutamic acid. Biochemistry 32 5093–5098. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Rudresh, Jain, R., Dani, V., Mitra, A., Srivastava, S., Sarma, S.P., Varadarajan, R., and Ramakumar, S. 2002. Structural consequences of replacement of an α-helical Pro residue in Escherichia coli thioredoxin. Protein Eng. 15 627–633. [DOI] [PubMed] [Google Scholar]
- Schultz, L.W., Chivers, P.T., and Raines, R.T. 1999. The CXXC motif: Crystal structure of an active-site variant of Escherichia coli thioredoxin. Acta Crystallogr. D Biol. Crystallogr. 55 1533–1538. [DOI] [PubMed] [Google Scholar]
- Vagin, A. and Teplyakov, A. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56 1622–1624. [DOI] [PubMed] [Google Scholar]
- Zapun, A., Bardwell, J.C., and Creighton, T.E. 1993. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32 5083–5092. [DOI] [PubMed] [Google Scholar]