Abstract
Eukaryotic membrane proteins cannot be produced in a reliable manner for structural analysis. Consequently, researchers still rely on trial-and-error approaches, which most often yield insufficient amounts. This means that membrane protein production is recognized by biologists as the primary bottleneck in contemporary structural genomics programs. Here, we describe a study to examine the reasons for successes and failures in recombinant membrane protein production in yeast, at the level of the host cell, by systematically quantifying cultures in high-performance bioreactors under tightly-defined growth regimes. Our data show that the most rapid growth conditions of those chosen are not the optimal production conditions. Furthermore, the growth phase at which the cells are harvested is critical: We show that it is crucial to grow cells under tightly-controlled conditions and to harvest them prior to glucose exhaustion, just before the diauxic shift. The differences in membrane protein yields that we observe under different culture conditions are not reflected in corresponding changes in mRNA levels of FPS1, but rather can be related to the differential expression of genes involved in membrane protein secretion and yeast cellular physiology.
Keywords: membrane proteins, expression systems, miniarray, protein production, yeast
Membrane proteins in general, and those from eukaryotes in particular, have always been the “high-hanging fruit” of structural biology, with most groups traditionally working on a single protein or protein family that they have found to be highly abundant in native cellular membranes and amenable to structural, and more usually, functional analysis in their hands. With the advent of structural genomics, however, the need to develop tools to study the structural biology of membrane proteins as a class has come into sharp focus. Since it is clear that the vast majority of membrane proteins are not available in sufficient quantities from their natural sources, the lack of suitable systems to overproduce recombinant membrane proteins is a substantial bottleneck in the pipeline from gene to structure that must be relieved. The development of generic production systems that can be applied to a wide range of membrane proteins, rather than those that have previously been found to be effective on a case-by-case basis, is thus an area of research of the utmost importance.
Since Grisshammer and Tate’s seminal 1995 review (Grisshammer and Tate 1995) is still regarded as the primary reference for membrane protein production, it is clear that the field as a whole has not advanced as rapidly as might have been hoped. Although more recent articles have covered the use of bacteria (Miroux and Walker 1996; Ward et al. 2000; Kunji et al. 2003), yeasts (Bill 2001), insect cells (Bosman et al. 2003), mammalian cells (Lundstrom 2003), and cell-free systems (De Keyzer et al. 2002) as production hosts for membrane proteins (Sarramegna et al. 2003), no generic strategies for milligram-scale membrane protein production have been published. The widely applicable solution for soluble protein—the pET-based system of vectors from Novagen—has not been applicable to membrane proteins (Wang et al. 2003), and no alternative membrane protein–specific panacea has been discovered.
In an attempt to develop membrane protein–specific strategies, several groups have focused on yeast species, especially Pichia pastoris and Saccharomyces cerevisiae (Reilander and Weiss 1998; Bill 2001; Sarramegna et al. 2003). For S. cerevisiae in particular, a sequenced genome, the value of well-understood protein quality-control mechanisms in the endoplasmic reticulum, the ability to increase the secretory capacity, and the effective coexpression of chaperones all make this yeast species an attractive host for recombinant membrane protein production (see http://www.esf.org/eurocores). Despite the fact that focusing on promoters and expression tags is not sufficient to overcome the complex problem of inserting a recombinant membrane protein into the membrane of a heterologous host, this is still the most widely used strategy to improve protein production. Only recently have researchers exploited specific pathways to tailor the host cell itself for membrane protein production. Oesterhelt’s group (Griffith et al. 2003), for example, has exploited the unfolded protein response (UPR) pathway of S. cerevisiae by tuning the production level of the protein to avoid or minimize UPR induction, resulting in an increased functional quorum. This system is illustrative of the effectiveness of taking a more systematic approach to optimizing host response, but unfortunately, is not generally applicable, since not all proteins induce the UPR pathway (Griffith et al. 2003), and high yields are not explicit.
In this study, we have taken an even broader view to try to quantify how the host cell responds to high-yielding and low-yielding production experiments. We have therefore analyzed production yield as a function of culture conditions for a eukaryotic membrane protein with a fixed promoter and tag combination (constitutive TPI promoter, (HA)3 tag) in high-performance bioreactors under tightly-defined growth regimes. We have chosen to study the well-characterized (Bill et al. 2001; Tamas et al. 2003; Hedfalk et al. 2004), eukaryotic glycerol facilitator, Fps1p, a unique member of the six-transmembrane helix aquaporin family of water and polyol transporters, which based on previous experience, is a nontrivial production target for further structural study (Hedfalk et al. 2000). Endogenous Fps1p is estimated to be present at 900 copies/cell, and attempts to produce the heterologous protein typically give low yields in the tens-of-microgram range. In a new approach, we quantify the relationship between protein yield, RNA levels, and culture conditions, and relate this to the genomic profile of the cultures. We show that optimal production conditions do not support the most rapid growth of the culture, the phase of harvest is critical, and that low-membrane protein yields can be related to defects in yeast’s secretory pathway and its cellular physiology.
Results
The growth characteristics of yeast cultures producing recombinant Fps1p can be described quantitatively
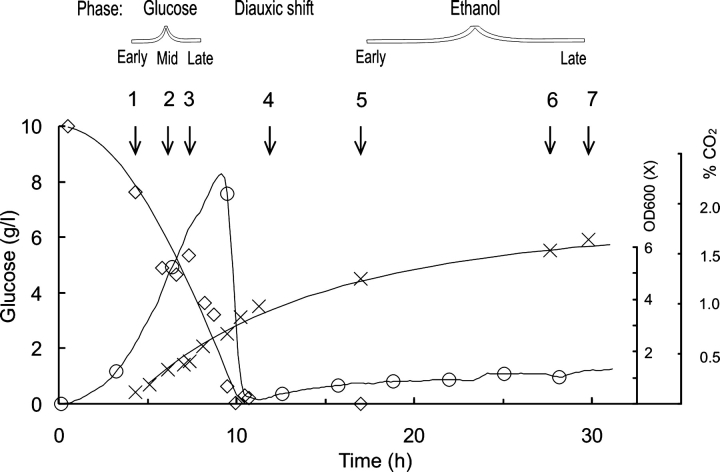
We began our study by analyzing the profile of our cultures (Fig. 1 ▶), thereby defining the points at which we would harvest cells for further analysis (Fig. 1 ▶, points 1–7). The retention of plasmid in the cells was validated by colony counting, the number of colonies on yeast nitrogen base (YNB) plates compared with yeast extract/peptone/dextrose (YPD) consistently being >70%.
Figure 1.
Culture profile for a transformant grown aerobically on glucose at 30°C (pH 5). Glucose concentration (⋄), OD600 (×) and CO2 level (○) vs. time is shown for growth under these “normal” conditions. The phase of sampling is indicated with an arrow, and each sample is also given a number for ease of identification.
Table 1 summarizes the duration of the glucose phase for the five fermentation conditions studied. All glucose was consumed within 13 h for cells cultured at 30°C (pH 5), 35°C (pH 5), and 35°C (pH 7), while at 30°C (pH 7) or 20°C (pH 5), the glucose was still not consumed after 40 h. The final dry weights for all experiments (point 7) ranged from 1 g/L at 35°C (pH 7) to 2 g/L at 20–35°C (pH 5), corresponding to ~2.5–5 g wet cells per liter of medium. The ethanol concentration in the cultures reached a maximum level of 2.5% at the end of the glucose growth phase, while the glycerol concentration reached a plateau value in the middle of the ethanol growth phase of 0.2–0.3 g/L (pH 5 and pH 3.5, for which data are not shown here), whereas a higher concentration of 0.5–0.6 g/L of glycerol was seen at pH 7.
Table 1.
Duration of glucose phase and glucose consumption rate for the five fermentation conditions studied
| Temperature (°C) | pH | Duration of glucose phase (h) | Glucose consumption rate mmolGlc L−1 h−1 |
| 30 | 5 | 13.1 (0.02) | 6.8 (0.07) |
| 35 | 5 | 10.7 (0.21) | 8.9 (0.08) |
| 20 | 5 | 42.5 (0.11) | 2.7 (0.08) |
| 30 | 7 | 43.9 (0.11) | 2.8 (0.08) |
| 35 | 7 | 13.1 (0.11) | 6.1 (0.08) |
Growth was monitored using manual OD measurements as well as on-line monitoring of the addition of 1 M NaOH, which can be used as an on-line parameter for the estimation of biomass formation. The duration of the glucose phase and the glucose consumption rate were derived from the growth curve. Duplicate fermentations were used to calculate the standard error of the mean, in parentheses, for the cultures at 30°C (pH 5) and 35°C (pH 5); values in italic font are the standard error of the mean for the other growth conditions, estimated from the calculated error for 30°C (pH 5) and 35°C (pH 5).
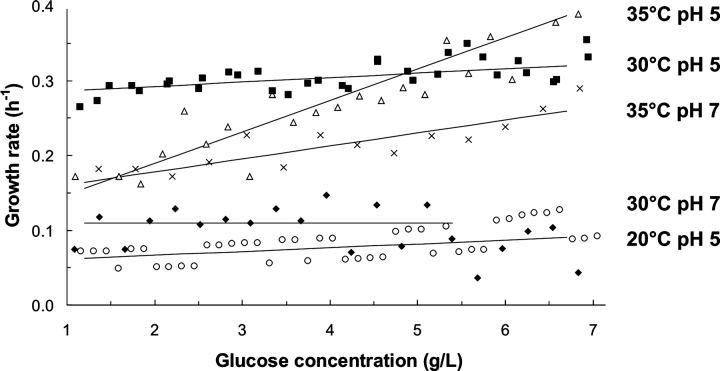
Growth characteristics were calculated for the different fermentations from both optical density (OD600) measurements and base addition. These methods yielded the same specific growth rate (μ), but the manual OD600 method was much less reproducible and labor intensive compared with the on-line base addition method. A plot of μ at different pHs and temperatures was prepared as a function of residual glucose concentration, with a least squares linear fit of μ between 1.5 and 7 g/L glucose (Fig. 2 ▶). Figure 2 ▶ shows clearly that the best growth conditions of those tested were 30°C (pH 5) and 35°C (pH 5) with μ=0.3 h−1 at 5 g/L glucose in both cases. Although μ at 35°C (pH 5) was 20% higher at 7 g/L glucose, this declined substantially at lower glucose concentrations and at 2 g/L, μ was only 0.18 h−1. The growth rate was lowest for 20°C (pH 5) (Fig. 2 ▶).
Figure 2.
Growth rates for cultures defined by base addition. Specific growth rate, μ (h−1) was computed using the amount of base added in two adjacent 20-min segments in the 1.5–7 g/L glucose range. Conditions are as follows: 30°C (pH 5) (squares); 35°C (pH 5) (triangles); 35°C (pH 7) (crosses); 30°C (pH 7) (diamonds); 20°C (pH 5) (circles).
Culture conditions affect the yield of recombinant Fps1p
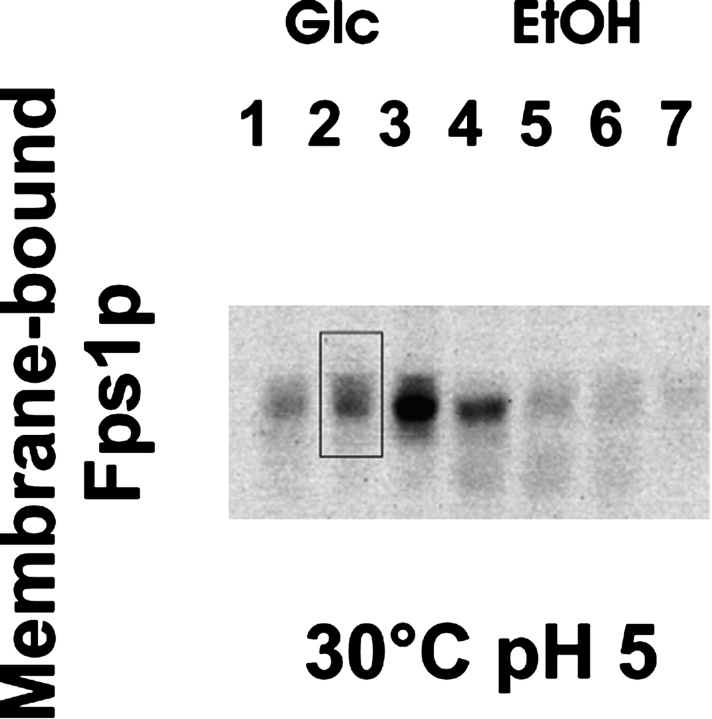
After harvesting samples as shown in Figure 1 ▶, protein yields were quantified by immunoblot. Both the total yield of Fps1p and the membrane-bound fraction were quantified by densitometry. In order to compare the samples, each blot was scaled to a standard, namely sample 2, from the growth at 30°C (pH 5) (Fig. 3 ▶).
Figure 3.
Yield of membrane-bound Fps1p as a function of growth phase. In subsequent calculations, the amount of membrane-bound Fps1p was quantified from immunoblot using the integration window shown. “-Glc” and “-EtOH” refer to samples extracted during the glucose and ethanol growth phases, respectively, as illustrated in Figure 1 ▶.
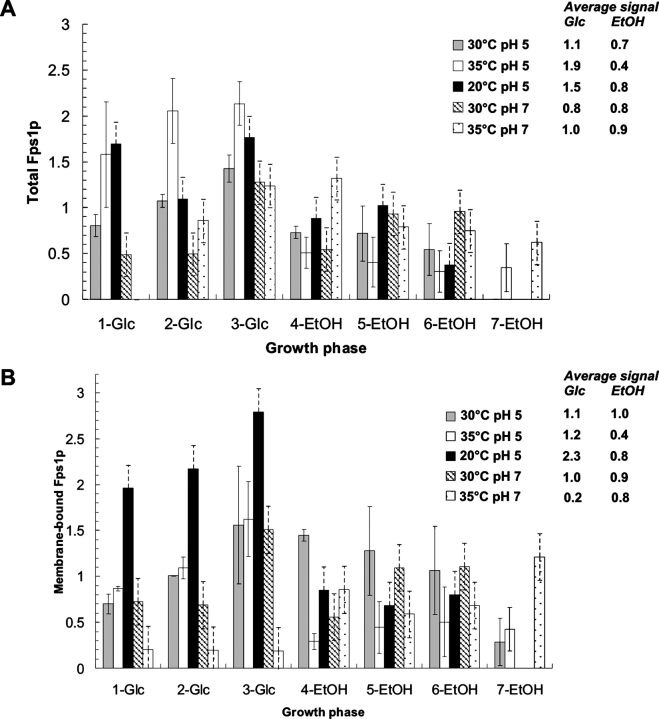
The data for the total Fps1 protein are shown in Figure 4A ▶. When the yields were compared under different growth conditions, it was clear that both the amount of protein and the profile of production were different. The highest yield was not obtained from cells cultured under the best growth conditions of 30°C (pH 5) (see above). For example, the yield of total Fps1p could be increased both by lowering and increasing the temperature to 20°C and 35°C, respectively. This change in temperature also changed the overall production profile, high yields being obtained even in the early log phase, with average values being 1.5 and 1.9 compared with 1.1 for the best growth conditions (Fig. 4A ▶). However, only at the lower temperature was the yield of membrane-bound Fps1 protein increased; the highest yield of membrane-bound Fps1p occurred when the temperature was lowered to 20°C, conditions under which good yields of protein were uniquely obtained throughout the glucose phase (Fig. 4B ▶). Consequently, the amount of protein obtained in the total extract was not a good indicator of the yield of functional membrane-bound Fps1 protein: Cells grown at 35°C (pH 5) gave the best total yield (Fig. 4A ▶), which was not reflected in the membrane-bound yield (Fig. 4B ▶).
Figure 4.
Quantitation of relative Fps1 protein yields. Immunoblots were performed and quantified on both the total (A) and membrane-bound (B) fractions for samples cultured under the five conditions shown, and harvested as indicated in Figure 1 ▶. “-Glc” and “-EtOH” refer to samples extracted during the glucose and ethanol growth phases, respectively. The immunoblot signals were quantified in the ImageGauge program. Either total extract or the membrane-bound fraction isolated from Sample 2 of the 30°C (pH 5) culture was used as an internal standard. All signals were below saturation and related to the signal of the internal standard. Duplicate fermentations (solid bars) were used to calculate the standard error of the mean at 30°C (pH 5) and 35°C (pH 5). For all other conditions, the error (hatched bars) was estimated based on the values for 30°C (pH 5) and 35°C (pH 5). Average values were calculated for total Fps1 protein in both the glucose and ethanol phases to give an indication of yield variation within and between different growth conditions.
Across a given growth curve, protein yields in the membrane fraction varied by a factor of two- to sixfold. For example, at 20°C (pH 5), there was a fourfold difference between the highest and the lowest yield. Under all conditions, regardless of the variation, the maximum yield was obtained prior to glucose exhaustion, before the diauxic shift (Fig. 1 ▶, sample 3 for four conditions), with yield typically dramatically falling off after that maximal point. The exception to this general trend was for cells grown at 35°C (pH 7). Here, the production profile was altered so that although reasonable levels of Fps1p were produced in the total protein fraction, the proportion of membrane-bound Fps1 protein was substantially reduced. Under these conditions, the yield of membrane-bound Fps1 protein was also greater later in the growth curve, in contrast to the other conditions tested.
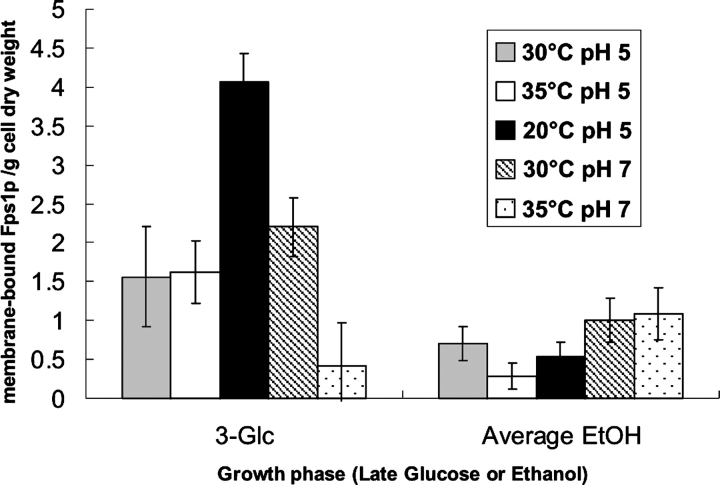
In order to extract the effect of culture conditions on the yield of protein per se, we calculated membrane-bound Fps1 protein yield (Fig. 4B ▶) as a function of dry weight (Fig. 5 ▶). It was clear that in the glucose phase, the yield of protein per dry weight is severalfold higher than in the ethanol phase, and in the case of the best production conditions of 20°C (pH 5), there is a sevenfold improvement in yield. Under these conditions, the improvement in yield over the standard conditions of 30°C (pH 5) was threefold.
Figure 5.
Quantitation of the yield of membrane-bound Fps1p as a function of dry weight and growth phase. The amount of membrane-bound Fps1p was quantified from immunoblot (Fig. 4 ▶) and related to the appropriate dry weight for the time point (point 3 for “3-Glc” and the average of points 5–7 for “Average EtOH”). In all cases, except at 35°C (pH 7), the maximum yield was obtained just prior to the diauxic shift (3-Glc). In the ethanol phase, yields were fairly constant throughout that part of the growth curve, as indicated by the averages given.
Transcript levels do not correlate with protein yield under different growth conditions
Real time quantitative PCR (Q-PCR) was used to quantify the levels of plasmid-borne FPS1 to determine whether this was correlated with the protein yields observed in Figure 4 ▶. Levels of TPI1, endogenous FPS1, and three standards—IPP1, ACT1, and PDA—were also measured (Table 2). The absolute values obtained could be directly compared between experiments.
Table 2.
Analysis of RNA
| Culture conditions | |||||
| RNA analyzed | 30°C pH 5 | 30°C pH 7 | 20°C pH 5 | 35°C pH 5 | 35°C pH 7 |
| FPS1-ectopic | |||||
| Glucose phase | 47 (16) | 941 (208) | 136 (40) | 103 (21) | 435 (64) |
| Ethanol phase | 55 (15) | 535 (69) | 115 (30) | 140 (35) | 174 (73) |
| FPS1-endogenous | |||||
| Glucose phase | 11 (1) | 81 (5) | N.D. | 10 (1) | 19 (3) |
| Ethanol phase | 13 (1) | 45 (6) | N.D. | 12 (1) | 11 (3) |
| TPI1 | |||||
| Glucose phase | 41 (12) | 76 (23) | 45 (3) | 23 (6) | 68 (6) |
| Ethanol phase | 25 (1) | 35 (9) | 21 (3) | 21 (4) | 18 (5) |
| IPP1 | |||||
| Glucose phase | 7 (1) | 4 (1) | 7 (1) | 6 (1) | 6 (1) |
| Ethanol phase | 4 (1) | 3 (1) | 4 (1) | 6 (1) | 5 (1) |
| ACT1 | |||||
| Glucose phase | 78 (11) | 83 (16) | 85 (9) | 68 (5) | 94 (4) |
| Ethanol phase | 71 (5) | 57 (10) | 91 (9) | 76 (8) | 68 (12) |
| PDA1 | |||||
| Glucose phase | 5 (1) | 6 (1) | 4 (0) | 5 (0) | 3 (0) |
| Ethanol phase | 5 (0) | 6 (1) | 4 (0) | 5 (1) | 3 (0) |
Analysis of mRNA was performed using real time Q-PCR, which yielded copies of mRNA/cell. The data were normalized using the reference genes IPP1, PDA1, ACT1, and the signal was scaled to mRNA copies/cell according to a SAGE study (Velculescu et al. 1997), in which copies of mRNA/cell of all the reference genes had previously been determined. Standard error of the mean, in parentheses, was calculated for the samples taken in the glucose (n=3; points 1, 2, 3) and ethanol (n=3 or 4; points 5, 6, 7, [8]) growth phases.
Within error, RNA levels for each of TPI1, endogenous FPS1, and three standards—IPP1, ACT1, and PDA—were comparable. The only exception was the level for endogenous FPS1 at 30°C (pH 7), where the level was more than four times higher than for the other conditions (Table 2).
Within a growth curve, the ectopic FPS1 RNA level (Table 2) did not correspond to the Fps1p yield (Fig. 4 ▶). In fact, the RNA level was unchanged within error between the logarithmic and respiratory phases, with the exception of 35°C (pH 7), where the level for the logarithmic phase was significantly higher than that in the respiratory phase (Table 2). Even in this case, the range in Fps1p yield (Fig. 4 ▶) could not be correlated with the RNA level. When comparing conditions, the best Fps1p yield (Fig. 4 ▶) did not correspond to the highest FPS1 RNA level (Table 2). Indeed, when cells were cultured at 35°C (pH 7), among the highest ectopic FPS1 RNA levels were obtained in the glucose phase, but this also corresponded to the lowest membrane protein yields. The highest pH levels were also associated with high FPS1 RNA levels, and for Fps1p, all nonstandard growth conditions had higher mean FPS1 RNA levels than the best growth conditions of 30°C (pH 5). In essence, then, there was no clear correlation between yield of Fps1p and the level of FPS1 RNA.
Genes encoding proteins involved in membrane protein secretion and yeast cellular physiology are implicated in the production of membrane-inserted Fps1p
We sought to understand the molecular reasons for the observations that in logarithmically growing cells (1) raising the temperature and pH to 35°C (pH 7) from 30°C (pH 5) gave reasonable yields of total Fps1 protein, but dramatically reduced membrane-bound Fps1 protein yields, and (2) raising the temperature to 35°C (pH 5) from 30°C (pH 5) gave the very best total Fps1 protein yields, but no improvement in membrane-bound Fps1 protein yields. We therefore performed an analysis using yeast miniarrays to deduce what was failing in the conversion of improved total Fps1 protein yields to membrane-inserted protein. Samples 2, 3, and 4 (Fig. 1 ▶) from the three conditions 30°C (pH 5), 35°C (pH 7), and 35°C (pH 5) were used, giving a total of nine miniarrays. A linear model was fitted to the logged expression values (Smyth 2004), with one effect measuring the difference between the 30°C (pH 5) and 35°C (pH 7) samples; another, the difference between the 30°C (pH 5) and 35°C (pH 5) samples; and a final, additive effect, the difference between the samples 4 and samples 2 or 3. At a 5% false discovery rate, 111 genes were found to be significantly changed when moving from 30°C (pH 5) to 35°C (pH7). A total of 84 genes were significantly changed when moving from 30°C (pH 5) to 35°C (pH 5), with 39 genes involved in membrane protein secretion and yeast cellular physiology common to both sets (Table 3).
Table 3.
Analysis of miniarray data
| Factor change | |||
| Gene | Function | 30°C pH 5 to 35°C pH 7 | 30°C pH 5 to 35°C pH 5 |
| Membrane Protein Secretory Pathway | |||
| Ribosome Biogenesis | |||
| RPP1A | Component of the ribosomal stalk, involved in the interaction between translational elongation factors and the ribosome | 0.30 | 0.17 |
| CGR1 | Pre-rRNA processing in ribosome biogenesis | 0.24 | 0.32 |
| BMS1 | GTP-binding protein required for ribosome assembly | 0.21 | 0.19 |
| Protein Synthesis | |||
| SEC62 | Membrane component of ER protein translocation apparatus | 0.26 | 0.24 |
| APM3 | Golgi to vacuole transport in plasma membrane protein endocytosis | 0.17 | 0.32 |
| VTC3 | Vacuole fusion and phosphate metabolism | 0.25 | 0.19 |
| SRP102 | Signal recognition particle receptor β subunit | 3.3 | 3.6 |
| ERG9 | Squalene synthase involved in ergosterol biosynthesis | 3.30 | 2.96 |
| HSP82 | Chaperone (Hsp90 family) required for nascent membrane protein integrity and in high concentrations for growth at high temperatures | ||
| Yeast Cellular Physiology | |||
| MFα1 | Mating factor α | 3.87 | 3.71 |
| HOR7 | Unknown; down-regulated by a factor | 0.12 | 0.21 |
| BIO2 | Biotin synthase | 5.29 | 5.49 |
| HFI1 | Global regulation of gene expression | 0.19 | 0.13 |
| SRB6 | RNA polymerase II transcription mediator of global gene expression | 0.25 | 0.27 |
| CDC54 | Initiation of DNA replication | 0.20 | 0.21 |
| RPO41 | Mitochondrial genome maintenance | 0.29 | 0.26 |
| RTS3 | Putative protein phosphatase | 0.31 | 0.39 |
| CPA1 | Carbamyl phosphate synthetase; generates an essential precursor in arginine biosynthesis | 0.30 | 0.28 |
| PCL8 | Control of glycogen biosynthesis | 0.17 | 0.18 |
| PHO89 | Na+ /Pi cotransporter, active in early growth phase | 0.10 | 0.04 |
| COX15 | Involved in cytochrome c oxidase biogenesis | 3.89 | 3.91 |
| MPS2 | Structural constituent of cytoskeleton | 9.08 | 10.24 |
| MDJ2 | A DnaJ homolog in the mitochondrial inner membrane | 0.26 | 0.29 |
| MSD1 | Mitochondrial aspartyl-tRNA ligase involved in protein biosynthesis | 0.27 | 0.31 |
Genes were tabulated if their expression was changed on going from both 30°C (pH 5) to 35°C (pH 7) and 30°C (pH 5) to 35°C (pH 5), as described in Materials and Methods. The change is expressed as a factor, where that factor is x when a gene expressed with intensity 1 at 30°C pH 5 is expressed with intensity x at the new condition. All t-tests were jointly adjusted for multiple testing using the false discovery rate method (disregarding correlations between genes): p-values were adjusted so that when selecting all genes with p-values less than a threshold q, a proportion of q false positives would be expected among these genes. The genes shown have p-values <0.05, and thus the expected false discovery rate is 5%. Only genes with known functions have been tabulated. In addition, 15 genes of unknown function with factor-changes between 0.09 and 0.37 were identified: SRF6, YPL206C, YKR040C, YOR333C, YCR018C-A, YPL216W, YLR162W, YLR202C, YOR389W, YFL066C, YLR149C, YMR290W-A, YDR444W, YOL098C, and YBL112C.
We also compared the 702 genes that were significantly changed during the glucose phase (samples 2 and 3) compared with the ethanol phase (sample 4) with the average of data from the Stanford Microarray Database 15.5, 18.5, and 20.5 h after the diauxic shift (http://genome-www5.stanford.edu/). A total of 79% of the 702 genes changed in the same direction (with values within a factor of 3) as those in the Stanford Microarray Database, while 20 genes changed in the opposite direction. All 20 genes were induced by a factor of <7.2 in the present study and repressed >0.4 in the Stanford Microarray Database. Of the annotated genes, six encoded regulatory proteins (CHS2, DCS2, GIC1, GIP2, HRR25, and UBA1), and the rest of the genes were either involved in zinc transport ZRT1 and ZRT2, or enzymatic activity (PUS1, FIS1, and EST2). Eleven genes had unknown function:, ERR2, ERR3, YKL061W, YMR323W, YKL202W, YML087C, YGR242W, YPL281C, YDL110C, YNL013C, and YMR130W.
Discussion
In a first step to taking a systematic, quantitative approach to membrane protein production in yeast, we went back to first principles to collect a data set for Fps1p in high-performance bioreactors under tightly-defined growth regimes. Our data show that the best condition of those chosen, which is also that most commonly used (i.e., 30°C at pH 5), is not the optimal production condition. Furthermore, the growth phase at which the cells are harvested is critical: For a successful membrane protein production experiment, we show that it is crucial to grow cells under tightly-controlled conditions and to harvest them prior to glucose exhaustion, i.e., just before the diauxic shift. Finally, we show that the differences in membrane protein yields that we observe under different culture conditions are not reflected in changed cellular mRNA levels of FPS1, but rather can be related to the differential expression of genes involved in membrane protein secretion and yeast cellular physiology.
Figure 4 ▶ shows clearly that protein in both the total extract and the membrane-bound fraction was predominantly produced in the glucose phase. The data highlight major differences in production throughout the growth curve under a single condition, mostly pronounced at 35°C (pH 5) (sevenfold in the total extract and fourfold in the membrane fraction). Comparing all tested growth conditions, the overall difference in production was 10-fold in the membrane fraction (35°C at pH 7 2-Glc and 3-Glc, [0.19] vs. 20°C at pH 5 3-Glc [2.8]). Importantly, it was clear that there is no correlation between the total production yield and the yield of membrane-localized protein, most pronounced at 35°C (pH 7). This result should be of particular interest to those setting up high-throughput platforms, since it is clear that a “quick and dirty” analysis of total extracts can be very misleading, since it is not representative of membrane-inserted protein yields.
Changes in temperature clearly affected the production time course, which had a different profile under different conditions, although it was very clear that the optimal time for harvest of active membrane-bound Fps1 protein was just before the diauxic shift in all cases except 35°C (pH 7) (Fig. 4B ▶). Our data, for the first time, provide quantitative evidence for the importance of controlling growth conditions and optimizing the time of harvest: The highest yields of Fps1p in S. cerevisiae grown in YNB medium and 1% glucose are obtained when the cells are cultured at 20°C (pH 5) and harvested when the glucose concentration is 1–2 g/L. Interestingly, the highest yield of total Fps1 protein was obtained under conditions at which the growth rate was the fastest, whereas the highest yield of membrane-bound Fps1p was obtained under the condition at which the growth rate was the slowest. This latter observation correlates with qualitative reports of cell growth slowing down or even ceasing during high levels of protein production. It remains to be seen how general this observation is, and indeed, if one can correlate increasing membrane protein enrichment in the plasma membrane with decreasing growth rate. In fact, the result is even clearer from Figure 5 ▶. Since protein production is dependent on both the total amount of cells and the yield of protein per cell, the yield of membrane-bound Fps1 protein per dry weight is a reliable indicator for comparing yields from different cultures.
In order to explain our membrane protein yield data, we quantified the corresponding transcript levels. When we performed real time quantitative PCR (Q-PCR) on TPI1 and the plasmid-borne FPS1 genes, it was clear that the observations seen in Figure 4 ▶ could not be explained by variations in the TPI1 or FPS1 transcripts. Even in cases where there is virtually no production or where stronger signals were observed on an immunoblot, there was no correlation with the mRNA levels for these conditions (Table 2). In fact, from our array analysis, it was clear that FPS1 mRNA gave one of the top 20 signals of all of the yeast genes, and hence, mRNA was not limiting. Consequently, high yields must be determined by co- or post-translational events (Joseph-Liauzun et al. 1995), as hypothesized by several groups working on membrane protein production (Arechaga et al. 2003; Butz et al. 2003; Niebauer et al. 2004).
Identification of genes (and their corresponding gene products) that are expressed or repressed under specific culture conditions leading to a given yield of functional membrane protein should allow for a better understanding of the critical parameters involved. To our knowledge, this approach has not been used to analyze any membrane protein production experiments: Only a microarray analysis of soluble LuxA production in Escherichia coli has been reported (Oh and Liao 2000). We therefore performed an analysis using yeast miniarrays to rationalize the observations that changing the culture conditions from 30°C (pH 5) to either 35°C (pH 7) or 35°C (pH 5) gave good yields of total Fps1 protein, but dramatically reduced membrane-bound Fps1 protein yields. In essence, we sought to understand what was failing in the conversion of improved total Fps1 protein yields to membrane-inserted protein under these conditions. In order to ensure consistency between our miniarray data, data we had generated using Q-PCR, and previously published SAGE data (Velculescu et al. 1997), we calculated signal intensities for the reference gene PDA1 normalized to an average of the intensities for IPP1 and ACT1. This was done for the three culture conditions, 30°C (pH 5), 35°C (pH 5) and 35°C (pH 7). From our miniarray analysis, the normalized PDA1 intensities were 0.2, 0.4, and 0.1, respectively; and from our real time Q-PCR they were 0.1, 0.1, and 0.3, respectively, both in good agreement with the ratio of 0.3 derived from the SAGE results (Velculescu et al. 1997).
We compared our nine miniarrays (samples 2, 3, and 4 at 30°C [pH 5], 35°C [pH 5], and 35°C [pH7]): 84 genes changed their expression level, going from 30°C (pH 5) to 35°C (pH 5), and 111 going from 30°C (pH 5) to 35°C (pH 7). Table 3 shows the 39 genes that varied on going from 30°C (pH 5) to 35°C (pH 5) and 3°C (pH 5) to 35°C (pH 7) with a confidence factor p<0.05. Results for the genes, MFα1 and HOR7 provide a useful validation of our data set, as it is has already been reported that up-regulation of MFα1 results in down-regulation of HOR7, a gene of unknown function (Seidel and Tanner 1997), as mirrored in Table 3.
Both the increase in total Fps1 protein production at 35°C (pH 5) compared with 30°C (pH 5), and the similar total Fps1 protein yield at 30°C (pH 5) compared with 35°C (pH 7) are not reflected in membrane-bound yields (Fig. 4 ▶). This is accompanied by down-regulation of three genes involved in ribosome biogenesis (RPP1A, CGR1, and BMS1) (Table 3); a membrane component of the ER protein translocation apparatus, SEC62; and two genes involved in vacuolar trafficking, APM3 and VTC3. Warner and coworkers (Mizuta and Warner 1994) have found that transcription of genes encoding both ribosomal proteins and rRNAs is repressed when the secretory pathway is defective, providing the cells with a mechanism for cellular stress adaptation (Schmidt et al. 1985). This hypothesis is also consistent with the observed down-regulation of SEC62 (Table 3), since yeast sec62 mutants are defective in the translocation of several secretory precursor proteins into the lumen of the endoplasmic reticulum, including α-factor precursors (Rothblatt et al. 1989) and certain membrane proteins (Stirling et al. 1992). Indeed, it has been known for a long time that the down-regulation of SEC62 triggers the accumulation of proteins in internal secretory vesicles (Deshaies and Schekman 1989). Interestingly, up-regulation of MFα1 also indicates that the protein secretory pathway is compromised (Caplan et al. 1991; Avaro et al. 2002). Indeed, the S. cerevisiae α-factor prepro-peptide leader sequence has been used to confer secretory competence to proteins such as insulin (Kjeldsen 2000), and constructed leaders have been developed for efficient secretory expression in P. pastoris.
Other genes related to defects in the secretory pathway can also be correlated to poor membrane-bound yields of Fps1p (Fig. 4B ▶). For example, SRP102 encodes the β-subunit of the signal recognition particle (SRP) receptor. Srp102p has been suggested to coordinate the release of the signal sequence from the SRP with the presence of the translocon (Pool 2003), and appears to regulate this process through a “switch cycle” of GTP/GDP binding (Schwartz and Blobel 2003). This and other evidence strongly support a role for yeast SRP in the translocation of molecules that are capable of post-translational membrane insertion (Stirling et al. 1992). The up-regulation of SRP102 during overproduction of a heterologous membrane protein might suggest an overloading of the translocation machinery on top of the normal 10- to 100-fold excess of ribosomes over SRP (Ogg and Walter 1995), consistent with a failure of correct localization of Fps1p in the membrane. Squalene synthase, encoded by ERG9, is also up-regulated in these cultures. ERG9 is an essential part of the complex regulatory circuit of sterol biosynthesis in S. cerevisiae; when up-regulated, the total sterol content of cells is increased (Kennedy and Bard 2001), a consistent response to heterologous membrane protein synthesis.
Two chaperones are also found to have altered expression patterns under the conditions tested: HSP82 encodes a member of the Hsp90 family required in high concentrations for growth at high temperatures (Borkovich et al. 1989) and membrane protein integrity (Youker et al. 2004); MDJ2 is expressed under normal conditions in the mitochondrial inner membrane (Westermann and Neupert 1997). As expected, HSP82 is up-regulated in cells cultured at 35°C (pH 5) and 35°C (pH 7) (Table 3). MDJ2, which has specialized functions in mitochondrial biogenesis, and is therefore unlikely to be involved in Fps1p production, is down-regulated.
The remaining genes in Table 3 have roles in key metabolic events, but not in protein quality control (Krebs et al. 2004); specifically, not in ubiquitination. Five are involved in the regulation of global gene expression (BIO2, HFI1, SRB6, CDC54, and RPO41), and the remainder in biosynthetic pathways. Interestingly, however, the reduced Fps1p yield in the ethanol growth phase compared with the glucose growth phase in the majority of the conditions investigated was found to correspond to an induction of UBA1 by a factor of 3.
We compared our data set of 39 genes to those published on the response of yeast cells to environmental changes (Gasch et al. 2000; Serrano et al. 2002), and specifically on moving cells from 27°C to 37°C (Gasch et al. 2000). Apart from the well-documented up-regulation of HSP82 on raising culture temperature, we found no overall correlation between the data sets, lending further support to the fact that the genomic profile we observe is linked to failure in membrane protein production, rather than merely resulting from an increase in temperature. We are in the process of gathering miniarray data for the cells cultured at 20°C (pH 5), and it will be instructive to see whether there are any markers of improved expression levels when comparing this condition with the data set for 30°C (pH 5).
In the search for generic membrane protein production systems, several solutions have been proposed. Highthroughput approaches—which involve trying many conditions chosen essentially at random—do not succeed in generating generic hosts, since proteins that are not produced using this format are discarded without understanding why. We suggest that a “smart-throughput” approach should enable a more focused, strategic method of recombinant eukaryotic membrane protein production through the identification and quantitation of the parameters critical for success, thereby allowing production on a milligram scale. In this study, we have specifically identified the importance of a functional secretion pathway in host cells grown under tightly-controlled conditions. This should ultimately contribute to understanding the critical parameters that define a successful membrane protein production experiment.
Materials and methods
Plasmids
FPS1, tagged at the 3′ end with a tag encoding the HA3 epitope for immunodetection, was expressed from the TPI promoter in the 2μpYX222 vector (Novagen; now discontinued), which contains the HIS3 selection marker. FPS1 was cloned into the BamHI and HindIII sites, such that the carboxy-terminal Thr-residue in Fps1p was replaced by SGRIFYPYDVPDYAGYPYDVPDYAGSYPYDVPDYA AQCGR; the 3×HA-tag sequence is underlined.
Yeast strains and growth conditions
The plasmid was transformed into S. cerevisiae strain 954 VW K70; MATα his3 SUC2 GAL MAL28c, which contains an endogenous copy of FPS1 (Dijken et al. 2000). Yeast cells were grown using YNB medium [1.7 g/L YNB (Q-Biogene), 5 g/L (NH4)2SO4] with 1% glucose; a concentration where cells could be grown without oxygen limitation. In this study, a selective marker for histidine was used on the expression vector, and hence, yeast transformants were grown in a minimal medium without histidine. Plasmid retention was verified by streaking cells on YNB and YPD plates. The YNB-agar plate, from which a shake flask culture was started, was incubated for 4 d at 30°C. Batch fermentation was performed in a 2.5-L volume and was inoculated 1/20 from an overnight shake culture (OD600 1.5). pH was kept constant using 1 M NaOH. The fermentors used in this study had on-line control of stirring, aeration, temperature, and pH, as well as monitoring of oxygen pressure in the medium and analysis of carbon dioxide and oxygen in the outlet gas. The stirring and aeration were set to 600 rpm and 0.3 vol/vol/min, respectively, which ensured a dissolved oxygen concentration above 65% air saturation. In a preliminary screen of growth conditions, all combinations of temperatures (20°, 30°, or 35°C) and pH (3.5, 5, or 7) were used before selecting the conditions presented in this study (data not shown). We have previously developed a series of simple functional in vivo assays for aquaporins in yeast (Bill 2001), and hence, were able to confirm that Fps1p was active under these conditions (data not shown).
Growth rate (μ) calculations using NaOH addition
Growth was monitored using manual OD measurements as well as on-line monitoring of the addition of 1 M NaOH. The growth rate (μ) was calculated from the amount of base added over a series of 20-min segments. The μ was then extracted from two adjacent time segments. When using NH4 as sole nitrogen source, there is a 1:1 relation between proton extrusion and nitrogen accumulation (Huth et al. 1990; Larsson and Gustafsson 1993). Hence, base addition can be used as an on-line parameter for estimation of biomass formation. A general plot at different pH and temperature, not dependent on duration of the glucose phase, was therefore prepared by converting the X-axis from time to glucose concentration. The lines in Figure 2 ▶ correspond to the region around which data points are a least squares fit for each different growth condition. Duplicate fermentations at 30°C (pH 5) and 35°C (pH 5) were used to estimate the standard error of the mean.
Protein, RNA, dry weight, and extracellular substrate determination
Samples were withdrawn at comparable OD600 values during the glucose and the ethanol phases (Fig. 1 ▶). In a preliminary screen, all culture conditions were repeated at least twice and the reproducibility of the production profile in the whole-cell extract was verified qualitatively on an immunoblot. The culture conditions of 30°C (pH 5) and 35°C (pH 5) were then repeated twice, and their quantitative reproducibility was verified by analysis of the total extract and the membrane fraction on an immunoblot, as described below. The error from these experiments was used to estimate the error for the other culture conditions. Samples were withdrawn regularly for measuring OD600, dry-weight, and the extracellular substrates glucose, glycerol, and ethanol. The sampling for protein and mRNA (see below) analysis was set up so that three samples were taken during the glucose growth phase and three or four samples during the ethanol growth phase (Fig. 1 ▶, samples 1–7). In YNB plus 1% glucose, OD was a reliable indicator of when samples should be harvested. Samples (200 mL) from the glucose growth phase were harvested at OD600 0.5 and 1.0, and (100 mL) at OD600 1.5. The cells were collected by centrifugation for immunoblot analysis. In the ethanol growth phase, up to four samples of 50-mL volume were harvested at 7–12-h intervals. Samples (2×1 mL) for extra cellular substrate analysis were harvested and collected by centrifugation for 2 min at 15,000g. The supernatants were stored at −20°C prior to analysis using kits from Boehringer Mannheim GmbH. Dry-weight determinations were performed by collecting two samples of 5 mL by centrifugation for 5 min at 5000g. The cells were washed once in 5 mL water, dried for 24 h at 105°C, and stored in a desiccator for 24 h before being weighed.
Membrane preparation and immunoblots
Analyses of the protein production levels were performed on both crude cell extracts and total membrane fractions. We assumed that membrane-inserted Fps1p was correctly folded and that the remaining protein was aggregated along the membrane protein secretory pathway. For protein fractionation, cells were suspended in Cell Resuspension Buffer (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 0.5 mM EDTA, 5% glycerol) and mixed with glass beads at a 1:1:1 ratio. Cells were shaken in a Fast Prep (speed 6.5, 3×20 sec, with incubation on ice between pulses). Unbroken cells were collected at 500g for 10 min at 4°C. A sample of the 500g supernatant was reserved for analysis of the crude extract. The remaining 500g supernatant was then clarified by centrifugation at 10,000g for 30 min at 4°C. The total membrane fraction was collected from this clarified supernatant at 100,000g for 90 min at 4°C. The protein concentration was measured using a Bio-Rad DC Protein Assay with bovine serum albumin as standard. For immunoblotting, 35 μg crude extract or 75 μg total membrane fraction were loaded per lane of a 7.5% acrylamide gel and separated by SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane (Hybond-ECL, Amersham Pharmacia Biotech). Even loading and transfer of the proteins were confirmed by Ponceau S staining. The membrane was blocked with PBS/5% milk before incubation with the mouse monoclonal antibody, anti-HA (clone 12CA5; Roche), at a 1:1000 dilution in PBS/5%milk for 90 min. After a wash in blocking solution, the membrane was incubated with the secondary antibody, goat anti-mouse IgG HRP (Promega), at a 1:2500 dilution in PBS/5% milk for 1 h. The membrane was washed in blocking solution, PBS/0.1% Tween, and finally PBS. Western blot membranes were developed using ECL Plus Western Blotting Detection Reagent (Amersham Biosciences) at 0.12 mL/cm2 membrane, and visualized using the Image Reader LAS-100 (Fujifilm). The signal from each lane was quantified using the ImageGauge 3.46 program (Fujifilm) and the integration window shown in Figure 3 ▶. Each signal was related, as appropriate, to either the total cell extract standard or the membrane-localized standard, both of which were isolated from sample 2 of the culture grown at 30°C (pH5). The appropriate standard was loaded on each gel. Duplicate fermentations were used to estimate the standard error of the mean.
RNA preparation and real time quantitative PCR
Yeast cells (2 mL) were harvested and frozen in liquid nitrogen. Total RNA was then prepared using the RNeasy kit from Qiagen, following the manufacturer’s instructions. Analysis of mRNA was performed using real time quantitative PCR (Q-PCR). A total of 0.5 μg RNA was used in the cDNA reaction using the Superscript II kit (Invitrogen). Each sample was amplified using 40 cycles (20 sec 94°C, 20 sec 60°C, 20 sec 72°C) in a Bio-Rad iCycler iQ, and the data were analyzed using iCycler IQ version 3.0. The data were normalized using the reference genes IPP1, PDA1, and ACT1 (data shown for ACT1), and the signal was scaled to mRNA copies/cell according to a SAGE study (Velculescu et al. 1997), in which copies of mRNA/cell of all the reference genes had previously been determined.
cDNA array production
PCR products for the yeast ORFs were generated from yeast DNA using the ORF-specific primer set from Research Genetics and then reamplified with the Resgen universal forward and reverse primers (catalog no. 40612). PCR products were prepared by Randy Strich (Fox Chase Cancer Center) as part of a collaboration with the Wistar Institute. PCR products were spotted onto nylon filter arrays with a BioRobotics Microgrid TASII (Genomic Solutions). Each array contained 6319 individual yeast genes and 1169 gene repeats. The complete list of arrayed genes on array YA04 can be found at http://showelab.wistar.upenn.edu/Wistar_Showe_Lab_Gene_Lists.htm.
RNA amplification, hybridization, and scanning
Amplified RNA (aRNA) was prepared from 1 μg of total RNA (Fig. 1 ▶, samples 2–4, for each culture tested) using the RiboAmp kit (Arcturus). Labeled targets were prepared from 1.6 μg aRNA with Superscript II reverse transcriptase (Invitrogen), in the presence of 3000–5000 Ci/mM [α-33P]dCTP (Amersham Pharmacia Biotech), 1 mmol/L dATP, 1 mmol/L dTTP, 1 mmol/L dGTP, 67 ng/μg oligo-dT (Promega Bio-sciences), and 0.65× random decamer primers (Ambion). Labeled targets were hybridized to individual nylon arrays at 42°C for 18 h in 3 mL Microhyb buffer (Invitrogen). Arrays were washed twice in 2× standard saline citrate (SSC)/1% SDS solution for 30 min at 50°C, once in 0.5× SSC/1% SDS, and once in 0.1× SSC/0.5% SDS for 30 min at 55°C. The arrays were exposed to phosphor screens (Amersham Biosciences) for 6 d and scanned in a Storm 820 PhosphorImager (Molecular Dynamics). Quantitation of each spot was assessed by Imagene 5.0 software (http://www.biodiscovery.com) using manual spot alignment, measuring median pixel intensity for each spot, and subtracting the local background (http://www.wistar.upenn.edu/research_facilities/facilities/genomics/links.html).
Miniarray data analysis
Observed values <0.1 (11.6% [7698 values] of the 66,330 raw data values) were removed, in accordance with information from Wistar, the array manufacturer, who indicated that such values should not be regarded as meaningful measurements. An average was computed over spots with the same ORF, and data for each array were normalized by dividing by the median. After taking logarithms and performing scale normalization between the arrays, a linear model was fitted (Smyth 2004). In addition to the general level effect for each gene, the model contained one effect for the difference between 30°C (pH 5) and 35°C (pH 7) samples, one for the difference between 30°C (pH 5) and 35°C (pH 5) samples, and one for the difference between samples at time point 4 and samples at time points 2 or 3.
Empirical Bayes robustification of variance estimates was included, and t-tests for the difference between the 30°C (pH 5) and 35°C (pH 7) samples and the difference between the 30°C (pH 5) and 35°C (pH 5) samples were performed. All t-tests were jointly adjusted for multiple testing using the false discovery rate method (disregarding correlations between genes). Essentially, then p-values were adjusted, so that when selecting all genes with p-values less than a threshold q, a proportion of q false positives would be expected amongst these genes. The estimated effects are shown as factors in Table 3, as the linear modeling was performed on a log scale. The p-values in Table 3 are the adjusted values, so that selecting genes with p-values <0.05 gives an expected false discovery rate of 5%.
Acknowledgments
This work was supported by the European Commission via contracts LSHG-CT-2004-504601 (E-MeP) to R.M.B., and QLG2-CT-2002-00988 (SPINE) to L.G. and R.M.B. We also acknowledge Chalmers Bioscience Initiative and the Wallenberg Foundation through their support of the Membrane Protein Centre, Lundberg Laboratory, Göteborg, Sweden. The miniarray data was performed in collaboration with Dr. L. Showe, W. Horng, and L.Gudipati of theWistar Institute, Philadelphia, PA.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051435705.
References
- Arechaga, I., Miroux, B., Runswick, M.J., and Walker, J.E. 2003. Overexpression of Escherichia coli F1F0-ATPase subunit a is inhibited by instability of the uncB gene transcript. FEBS Lett. 547 97–100. [DOI] [PubMed] [Google Scholar]
- Avaro, S., Belgareh-Touze, N., Sibella-Arguelles, C., Volland, C., and Haguenauer-Tsapis, R. 2002. Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast 19 351–371. [DOI] [PubMed] [Google Scholar]
- Bill, R.M. 2001. Yeast—A panacea for the structure-function analysis of membrane proteins? Curr. Genet. 40 157–171. [DOI] [PubMed] [Google Scholar]
- Bill, R.M., Hedfalk, K., Karlgren, S., Mullins, J.G., Rydström, J., and Hohmann, S. 2001. Analysis of the pore of the unusual major intrinsic protein channel, yeast Fps1p. J. Biol. Chem. 276 36543–36549. [DOI] [PubMed] [Google Scholar]
- Borkovich, K.A., Farrelly, F.W., Finkelstein, D.B., Taulien, J., and Lindquist, S. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9 3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman, G.J., van Oostrum, J., Breikers, G., Bovee-Geurts, P.H., Klaassen, C.H., and DeGrip, W.J. 2003. Functional expression of His-tagged rhodopsin in Sf9 insect cells. Methods Mol. Biol. 228 73–86. [DOI] [PubMed] [Google Scholar]
- Butz, J.A., Niebauer, R.T., and Robinson, A.S. 2003. Co-expression of molecular chaperones does not improve the heterologous expression of mammalian G-protein coupled receptor expression in yeast. Biotechnol. Bioeng. 84 292–304. [DOI] [PubMed] [Google Scholar]
- Caplan, S., Green, R., Rocco, J., and Kurjan, J. 1991. Glycosylation and structure of the yeast MF α 1 α-factor precursor is important for efficient transport through the secretory pathway. J. Bacteriol. 173 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keyzer, J., Van Der Does, C., and Driessen, A.J. 2002. Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J. Biol. Chem. 277 46059–46065. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. and Schekman, R. 1989. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J. Cell. Biol. 109 2653–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijken, J.P.V., Bauer, J., Brambilla, L., Duboc, P., Francois, J.M., Gancedo, C., Giuseppin, M.L.F., Heijnen, J.J., Hoare, M., Lange, H.C., et al. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enz. Microb. Technol. 26 706–714. [DOI] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, D.A., Delipala, C., Leadsham, J., Jarvis, S.M., and Oesterhelt, D. 2003. A novel yeast expression system for the overproduction of qualitycontrolled membrane proteins. FEBS Lett. 553 45–50. [DOI] [PubMed] [Google Scholar]
- Grisshammer, R. and Tate, C. 1995. Overexpression of integral membrane proteins for structural studies. Q. Rev. Biophys. 3 315–422. [DOI] [PubMed] [Google Scholar]
- Hedfalk, K., Bill, R.M., Hohmann, S., and Rydström, J. 2000. Overexpression and purification of the glycerol transport facilitators, Fps1p and GlpF in S. cerevisiae and E. coli. In Molecular biology and physiology of water and solute transport: The MIP family of channel proteins (eds. S. Hohmann and S. Nielsen), pp. 29–34. Kluwer/Plenum, New York.
- Hedfalk, K., Bill, R.M., Mullins, J.G., Karlgren, S., Filipsson, C., Bergstrom, J., Tamas, M.J., Rydstrom, J., and Hohmann, S. 2004. A regulatory domain in the C-terminal extension of the yeast glycerol channel Fps1p. J. Biol. Chem. 279 14954–14960. [DOI] [PubMed] [Google Scholar]
- Huth, J., Werner, S., and Muller, H.-G. 1990. The proton extrusion of growing yeast cultures as an on-line parameter in fermentation processes: Ammonium assimilation and proton extrusion are correlated by 1:1 stoichiometry in nitrogen-limited fed-batch fermentations. J. Basic Microbiol. 30 561–567. [Google Scholar]
- Joseph-Liauzun, E., Farges, R., Le Fur, G., Ferrara, P., and Loison, G. 1995. High-level production of a human membrane protein in yeast: The peripheral-type benzodiazepine receptor. Gene 155 195–199. [DOI] [PubMed] [Google Scholar]
- Kennedy, M.A. and Bard, M. 2001. Positive and negative regulation of squalene synthase (ERG9), an ergosterol biosynthetic gene, in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1517 177–189. [DOI] [PubMed] [Google Scholar]
- Kjeldsen, T. 2000. Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol. 54 277–286. [DOI] [PubMed] [Google Scholar]
- Krebs, M.P., Noorwez, S.M., Malhotra, R., and Kaushal, S. 2004. Quality control of integral membrane proteins. Trends Biochem. Sci. 29 648–655. [DOI] [PubMed] [Google Scholar]
- Kunji, E.R., Slotboom, D.J., and Poolman, B. 2003. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta 1610 97–108. [DOI] [PubMed] [Google Scholar]
- Larsson, C. and Gustafsson, L. 1993. The role of physiological state in osmotolerance of the salt-tolerant yeast Debaryomyces hansenii. Can. J. Microbiol. 39 603–609. [Google Scholar]
- Lundstrom, K. 2003. Semliki Forest virus vectors for rapid and high-level expression of integral membrane proteins. Biochim. Biophys. Acta 1610 90–96. [DOI] [PubMed] [Google Scholar]
- Miroux, B. and Walker, J.E. 1996. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260 289–298. [DOI] [PubMed] [Google Scholar]
- Mizuta, K. and Warner, J.R. 1994. Continued functioning of the secretory pathway is essential for ribosome synthesis.Mol. Cell. Biol. 14 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebauer, R.T., Wedekind, A., and Robinson, A.S. 2004. Decreases in yeast expression yields of the human adenosine A2a receptor are a result of translational or post-translational events. Protein Expr. Purif. 37 134–143. [DOI] [PubMed] [Google Scholar]
- Ogg, S.C. and Walter, P. 1995. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81 1075–1084. [DOI] [PubMed] [Google Scholar]
- Oh, M.K. and Liao, J.C. 2000. DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab. Eng. 2 201–209. [DOI] [PubMed] [Google Scholar]
- Pool, M.R. 2003. Getting to the membrane: How is co-translational protein targeting to the endoplasmic reticulum regulated? Biochem. Soc. Trans. 31 1232–1237. [DOI] [PubMed] [Google Scholar]
- Reilander, H. and Weiss, H.M. 1998. Production of G-protein-coupled receptors in yeast. Curr. Opin. Biotechnol. 9 510–517. [DOI] [PubMed] [Google Scholar]
- Rothblatt, J.A., Deshaies, R.J., Sanders, S.L., Daum, G., and Schekman, R. 1989. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell. Biol. 109 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarramegna, V., Talmont, F., Demange, P., and Milon, A. 2003. Heterologous expression of G-protein-coupled receptors: Comparison of expression systems from the standpoint of large-scale production and purification. Cell. Mol. Life Sci. 60 1529–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T., Chen, P.S., and Pellegrini, M. 1985. The induction of ribosome biosynthesis in a nonmitotic secretory tissue. J. Biol. Chem. 260 7645–7650. [PubMed] [Google Scholar]
- Schwartz, T. and Blobel, G. 2003. Structural basis for the function of the β subunit of the eukaryotic signal recognition particle receptor. Cell 112 793–803. [DOI] [PubMed] [Google Scholar]
- Seidel, J. and Tanner, W. 1997. Characterization of two new genes downregulated by α-factor. Yeast 13 809–817. [DOI] [PubMed] [Google Scholar]
- Serrano, R., Ruiz, A., Bernal, D., Chambers, J.R., and Arino, J. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: Evidence for calcium-mediated signalling. Mol. Microbiol. 46 1319–1333. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article 3. [DOI] [PubMed] [Google Scholar]
- Stirling, C.J., Rothblatt, J., Hosobuchi, M., Deshaies, R., and Schekman, R. 1992. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas, M.J., Karlgren, S., Bill, R.M., Hedfalk, K., Allegri, L., Ferreira, M., Thevelein, J.M., Rydstrom, J., Mullins, J.G., and Hohmann, S. 2003. A short regulatory domain restricts glycerol transport through yeast Fps1p. J. Biol. Chem. 278 6337–6345. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Zhou, W., Vogelstein, J., Basrai, M.A., Bassett Jr., D.E., Hieter, P., Vogelstein, B., and Kinzler, K.W. 1997. Characterization of the yeast transcriptome. Cell 88 243–251. [DOI] [PubMed] [Google Scholar]
- Wang, A., Clapper, J., Guderian, J.A., Foy, T.M., Fanger, G.R., Retter, M.W., and Skeiky, Y.A. 2003. A novel method for increasing the expression level of recombinant proteins. Protein Expr. Purif. 30 124–133. [DOI] [PubMed] [Google Scholar]
- Ward, A., Sanderson, N.M., O’Reilly, J., Rutherford, N.G., Poolman, B., and Henderson, P.J.F. 2000. In Membrane transport, a practical approach (ed. S.A. Baldwin), pp. 141–166. Oxford University Press, New York.
- Westermann, B. and Neupert, W. 1997. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J. Mol. Biol. 272 477–483. [DOI] [PubMed] [Google Scholar]
- Youker, R.T., Walsh, P., Beilharz, T., Lithgow, T., and Brodsky, J.L. 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15 4787–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]