Abstract
Recently, the solution structure of the hypothetical protein HI1450 from Haemophilus influenzae was solved as part of a structure-based effort to understand function. The distribution of its many negatively charged residues and weak structure and sequence homology to uracil DNA glycosylase inhibitor (Ugi) suggested that HI1450 may act as a double-stranded DNA (dsDNA) mimic. We present supporting evidence here and show that HI1450 interacts with the dsDNA-binding protein HU-α. The interaction between HI1450 and HU-α from H. influenzae is characterized using calorimetry and NMR spectroscopy. HU-α binds to HI1450 with a Kd of 3.0 ± 0.2 μM, which is similar in affinity to its interaction with dsDNA. Chemical shift perturbation data indicate that the β1-strand of HI1450 and neighboring regions are most directly involved in interactions with HU-α. These results show that HI1450 and its structural homolog, Ugi, use similar parts of their structures to recognize DNA-binding proteins.
Keywords: double-stranded DNA mimic, HU-α, structural genomics
HI1450 from Haemophilus influenzae was originally annotated as a hypothetical protein of unknown function with sequence homologs in a number of species including Yersinia pestis, Vibrio cholerae, and Escherichia coli (see http://s2f.umbi.umd.edu). The solution structure of HI1450 was solved recently as part of a structural genomics project in an effort to determine the function of the protein (Parsons et al. 2004). HI1450 has weak structure and sequence homology to uracil DNA glycosylase inhibitor (Ugi), which inhibits the DNA repair enzyme uracil DNA glycosylase (UDG) by mimicking double-stranded DNA (dsDNA). In addition, HI1450 has many negatively charged residues, comprising 29% of its sequence, which are distributed in a pattern that resembles the phosphodiester backbone of dsDNA. Thus, HI1450 was identified as a putative dsDNA mimic.
During the preparation of HI1450 for structural studies, it was observed that several proteins copurified with HI1450 that were not seen in the preparation of other proteins by the same method. Here, we describe the separation and mass spectrometric analysis of these proteins. One of these was identified unambiguously as HU-α (heat-unstable protein). HU-α is a small histone-like protein that binds sequence-independently to dsDNA and is thought to function in gene regulation and DNA compaction due to its ability to bend and stabilize DNA (Dorman and Deighan 2003; van Noort et al. 2004). It is also involved in DNA repair and is one of the most abundant proteins in the cell (Ali Azam et al. 1999). Interactions between HI1450 and HU-α from H. influenzae were characterized quantitatively using calorimetry and NMR spectroscopy.
Results and Discussion
Identification of the DNA-binding protein HU-α in HI1450 preparations
In the process of purifying HI1450 for structure determination, it was noted that several proteins copurified with His6-tagged HI1450. The number of contaminants was more than normally seen in other similarly purified proteins. In addition, several of these contaminants ran at a lower molecular weight on SDS-PAGE than was observed in other protein purifications. Removal of the His6-tag and a second run over the Ni-NTA column did not remove the contaminants, indicating that they were interacting with HI1450 and not the affinity column. The contaminants could be removed with a high salt (1M NaCl) wash. A mass spectrum of the total salt wash could not be interpreted unambiguously so the contaminants were purified using SDS-PAGE and a mass spectrum of each was taken. Samples were recovered for proteins with molecular ions at 9544, 10,657, and 11,189 Da. Before purification there were also proteins at about 26.0 and 66.0 kDa but very little remained after purification with no dominant peaks detectable by mass spectrometry.
A search of all the proteins in E. coli found only one, HU-α (MW9534.96), which has a molecular weight within 10 Da of the 9544-Da peak. HU-α, like other dsDNA binding proteins, has a pI close to 10. No peaks were observed in the mass spectrum for the other form of HU, HU-β (MW 9193.49). E. coli proteins with molecular weights close to the 10,657 Da peak are Integration Host Factor (IHF, β-subunit, MW10, 651.11), which is another dsDNA binding protein with a pI of 9.89, and a hypothetical protein, b3427 (MW 10,659.08). It is also interesting to note that HU and IHF have sequence homology (30%–40%) and a common global fold (Rice et al. 1996; White et al. 1999). Proteins with molecular weights close to the 11,189 Da peak are ribosomal protein L23 (MW 11,199.09) and a hypothetical protein, b1044 (MW 11,193.97), and both have a pI >9. None of the hypothetical proteins have close homologs in H. influenzae but this does not mean the E. coli versions do not interact with HI1450. SinceHU-α was the only protein identified unambiguously and is a known dsDNA binding protein, the H. influenzae version was cloned and purified for further biophysical characterization of its interaction with HI1450. H. influenzae HU-α has 78.9% sequence identity to E. coli HU-α and only the α-form of HU exists in H. influenzae.
Characterization of binding between HU-α and HI1450
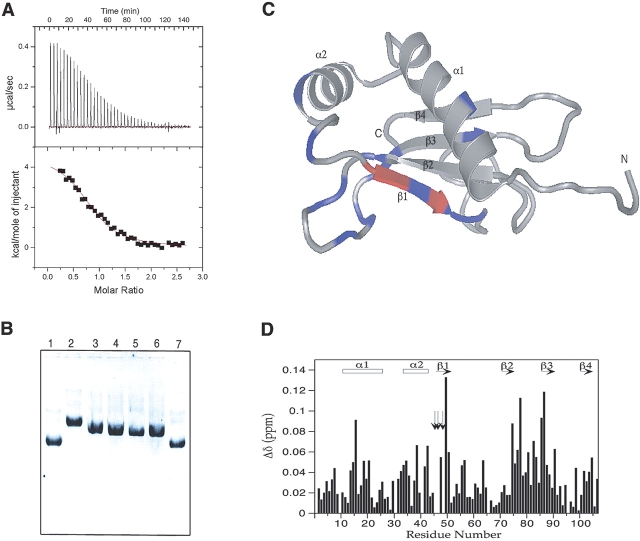
The calorimetric titration of HI1450 into a solution of H. influenzae HU-α shows that the proteins bind with a Kd of 3.0 ± 0.2 μM (Fig. 1A ▶) in 200 mM NaCl. This is close to the 0.45 μM, 1.3 μM, and 2.5 μM values reported for HU binding to supercoiled DNA, linear DNA, and bulk RNA, respectively, as determined by gel mobility shift assays and measured under similar stringent conditions (Balandina et al. 2002). Thus, the interaction with HU-α may be biologically relevant. Alternatively, the target protein for HI1450 may bind more tightly but be produced at much lower levels in the cell. DNA gel shift analysis showed that HU–DNA complexes could be partially disrupted by the addition of HI1450, demonstrating that the HI1450 and dsDNA binding sites on HU-α overlap. It was noted that high concentrations of HI1450 (up to 68 times greater than the concentration of HU-α) did not completely return the dsDNA to its original unbound form. The reasons for this are not understood at present.
Figure 1.
(A) Binding curve from calorimetry for the interaction between HU-α and HI1450. (B) DNA gel showing results from the competitive binding assay. Lane 1, plasmid DNA; lane 2, plasmid DNA plus HU-α; lane 3, HU-α /DNA mixture plus HI1450 (4 μL, 23-fold excess over HU-α); lane 4, HU-α /DNA mixture plus HI1450 (6 μL, 34-fold excess over HU-α); lane 5, HU-α /DNA mixture plus HI1450 (9 μL, 51-fold excess over HU-α); lane 6, HU-α /DNA mixture plus HI1450 (12 μL, 68-fold excess over HU-α); lane 7, plasmid DNA plus HI1450. (C) Chemical shift changes from a 1:3 (HU-α : HI1450) mixture mapped onto the 3D structure of HI1450 (red, exchange broadened; blue, Δtot>0.05 ppm). (D) Histogram plot of residue number vs. chemical shift changes for the above mixture. The Δδ values were determined using Equation 1. Residues with exchange-broadened peaks are indicated with vertical arrows.
The binding of H. influenzae HU-α to HI1450 was also monitored by NMR spectroscopy. Chemical shift changes were measured for HI1450 amide protons and nitrogens in a 2D 15N HSQC spectrum in the absence and presence of HU-α. Figure 1C ▶ maps the shift changes onto the three-dimensional structure of HI1450 to highlight the residues most affected by the addition of HU-α. In particular, the signals due to residues 46, 47, and 49 in the β1-strand are highly perturbed in the presence of approximately 0.3-mol equivalents of HU-α, disappearing due to exchange broadening effects. Peaks due to nearby residues in the β2-β3 loop are also perturbed indicating that the β1-strand and neighboring regions are most directly involved in interactions with HU-α. In the HU-dsDNA structure, a β-hairpin from each HU subunit binds to the minor groove of dsDNA (Swinger et al. 2003). The alignment of HI1450 with dsDNA indicates that the β1-strand mimics part of the minor groove (Parsons et al. 2004). Other shifts in the β2-β3 loop are also consistent with HU-α interacting with a pseudo-minor groove in HI1450.
The HSQC spectrum of more even ratio mixtures of HU-α and HI1450 was difficult to interpret due to line broadening effects, consistent with intermediate exchange between free and bound forms of HI1450 on the chemical shift timescale. The addition of an excess of HU-α (3.0 equivalents) to HI1450 does not lead to a reappearance of the exchange broadened peaks. It is noteworthy that the β1-strand of the structural homolog, Ugi, plays a key role at the binding interface of the Ugi-UDG complex (Putnam et al. 1999). Superposition of the β1-strands from HI1450 and Ugi reveals a pattern and distribution of negatively charged residues very similar to that found in the phosphodiester backbone of dsDNA (Fig. 4d in Parsons et al. 2004).
In summary, the results presented here identify the DNA-binding protein HU as a potential binding partner for HI1450 in vivo and provide additional support to the hypothesis that HI1450 functions as a dsDNA mimic, analogous to Ugi in its interactions with UDG. Further, NMR experiments show that HI1450 and Ugi use similar parts of their structures to recognize DNA-binding proteins.
Materials and methods
HU-α identification and purification
E. coli BL21(DE3) cells were transformed with a pet15b vector containing the HI1450 gene and grown to a density of 1 O.D. in 3 L of minimal media with 50 mg/L each of carbenicillin and ampicillin. Protein expression was induced with 1 mM IPTG and the cells were grown a further 3 h at 37°C. The culture was centrifuged for 10 min at 7000 g, resuspended in binding buffer (50 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole [pH 8.0]), and sonicated for a total of 6 min. The lysate was centrifuged for 25 min at 25,000g. The cell supernatant was loaded onto a Ni-NTA column (Qiagen) and washed with 50 mM sodium phosphate, 300 mM sodium chloride, 20 mM imidazole (pH 8.0). Proteins with a suspected affinity for HI1450 were removed with a salt gradient of 0.3 to 1.5 M NaCl. These proteins were then concentrated with a 5000 MWCO Centricon filter (Amicon) and dialyzed into 25 mM Tris (pH 7.6). Mass spectrometric analysis showed many peaks with molecular weights from 5600 to 26,000 Da. To simplify the spectra, individual protein bands were cut from a large (20×34 cm) SDS-PAGE gel run under denaturing conditions and stained with cupric sulfate (Lee et al. 1987). A total of seven bands were cut out. One was later identified as belonging to HI1450. The gel bands were destained with 0.25 mM EDTA and 0.25 mM Tris-HCl (pH 9.0), and the protein extracted using a modified version of the syringe maceration extraction method (Scheer and Ryan 2001). Specifically, the gels were crushed by passing them through a 5-mL syringe with no needle five times. Water was added so that the crushed gel was covered by at least half its volume. After a strong shaking, the tubes were placed at 4°C overnight. The water was removed by placing the crushed gel into PD10 columns. After collecting the water, 2–3 mL of fresh water was added to the columns. After sitting for 30 min at room temperature, the water was again removed and collected. Individual protein samples were dialyzed into 20 mM Tris (pH 7.6). The molecular weights of the resulting protein samples were measured by mass spectrometry. Spectra were collected on a MALDI-TOF Voyager-DE Biospectrometry Workstation (PerSeptive Biosystems) equipped with a nitrogen laser (337 nm, 3-nsec pulse). The samples were mixed with equal volumes of sinapinic acid solution (10 mg/mL in 50% acetonitrile, 0.3% TFA).
Preparation of HU-α from H. influenzae
The gene for HU-α was cloned from H. influenzae genomic DNA and inserted into a pet28 vector containing a His6-GST tag with a Tev cleavage site. The protein was produced in the same way as HI1450 except it was grown in LB with kanamyacin as the antibiotic. It was purified in the same way as HI1450, including a 1-M NaCl wash and eluted with binding buffer containing 250 mM imidazole. The protein was concentrated, dialyzed into Tev cleavage buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 1 mM dTT [pH 8.0]), and incubated with Tev that had a permanent His6-tag on its N terminus. The cleaved GST and Tev were removed by passing the protein back over the Ni-NTA column. The reported concentrations of HU-α are for monomer.
Purification of HI1450 for binding studies
Since the salt wash did not remove all the contaminants from HI1450, it was further purified over an HQ50 anion exchange column (Applied Biosystems) in 25 mM MOPS (pH 7.5) using a 1-M NaCl gradient. HI1450 purified in this manner was greater than 95% pure based on SDS-PAGE gels and was used in all binding experiments with HU-α.
Calorimetry
The binding affinity of HI1450 to HU-α was determined using a Microcal VP Titration Calorimeter. The proteins were both dialyzed into 20 mM Tris-HCl, 200 mM NaCl (pH 8.0). Five microliters (5 μL) of 0.5-mM HI1450 were injected into 0.03 mM of HU-α every 4 min until HU-α was saturated.
Competitive binding assay
For each sample, 4 μL of 22-μM HU-α was mixed with 1 μL of 20-μM pet28a plasmid DNA in 20 mM Tris-HCl, 200 mM NaCl (pH 8.0). Varying amounts of 0.5-mM HI1450 (0, 4, 6, 9, and 12 μL) were added to these different samples, so that a 0–68-fold range of HI1450 to HU-α concentration ratios was established. As a control, 12 μL of HI1450 was added to 1 μL of plasmid DNA without HU-α. The samples were run on a 0.8% agarose gel and stained with ethidium bromide.
NMR spectroscopy
15NHSQC spectra were collected at 297Kon a BrukerDRX-600 equipped with 3-axis gradient probes and recorded in States-TPPI mode. Data were processed on a Linux workstation using nmrPipe/nmrDraw (Delaglio et al. 1995) and analyzed using Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco). For the binding experiment, a solution of unlabeled HU-α was added to 15N-labeled HI1450 (0.9 mM) in steps of 0.33 equivalents up to 1mol equivalent and a 15NHSQC spectrum was recorded after each addition. The buffer conditions used were 50 mM sodium phosphate, 200 mM sodium chloride (pH7.0).
The total chemical shift change after addition of HU was calculated using Equation 1,
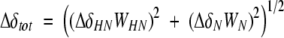 |
(1) |
with weighting factors WHN=1 and WN=0.154 (Ayed et al. 2001).
Acknowledgments
This work was supported by NIH grants GM57890 and 1S10RR15744, and the W.M. Keck Foundation.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041275705.
References
- Ali Azam, T., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed, A., Mulder, F.A., Yi, G.S., Lu, Y., Kay, L.E., and Arrowsmith, C.H. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8 756–760. [DOI] [PubMed] [Google Scholar]
- Balandina, A., Kamashev, D., and Rouviere-Yaniv, J. 2002. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277 27622–27628. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. and Deighan, P. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13 179–184. [DOI] [PubMed] [Google Scholar]
- Lee, C., Levin, A., and Branton, D. 1987. Copper staining: A five-minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 166 308–312. [DOI] [PubMed] [Google Scholar]
- Parsons, L.M., Yeh, D.C., and Orban, J. 2004. Solution structure of the highly acidic protein HI1450 from Haemophilus influenzae, a putative double-stranded DNA mimic. Proteins 54 375–383. [DOI] [PubMed] [Google Scholar]
- Putnam, C.D., Shroyer, M.J., Lundquist, A.J., Mol, C.D., Arvai, A.S., Mosbaugh, D.W., and Tainer, J.A. 1999. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 287 331–346. [DOI] [PubMed] [Google Scholar]
- Rice, P.A., Yang, S., Mizuuchi, K., and Nash, H.A. 1996. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell 87 1295–1306. [DOI] [PubMed] [Google Scholar]
- Scheer, J.M. and Ryan, C.A. 2001. A method for the quantitative recovery of proteins from polyacrylamide gels. Anal. Biochem. 298 130–132. [DOI] [PubMed] [Google Scholar]
- Swinger, K. K., Lemberg, K. M., Zhang, Y., and Rice, P.A. 2003. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 22 3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort, J., Verbrugge, S., Goosen, N., Dekker, C., and Dame, R.T. 2004. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. 101 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S.W., Wilson, K.S., Appelt, K., and Tanaka, I. 1999. The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta. Crystallogr. D Biol. Crystallogr. 55(Pt. 4) 801–809. [DOI] [PubMed] [Google Scholar]