Abstract
Grb7 is a member of the Grb7 family of proteins, which also includes Grb10 and Grb14. All three proteins have been found to be overexpressed in certain cancers and cancer cell lines. In particular, Grb7 (along with the receptor tyrosine kinase erbB2) is overexpressed in 20%–30% of breast cancers. Grb7 binds to erbB2 and may be involved in cell signaling pathways that promote the formation of metastases and inflammatory responses. In a prior study, we reported the solution structure of the Grb7-SH2/erbB2 peptide complex. In this study, T1, T2, and steady-state NOE measurements were performed on the Grb7-SH2 domain, and the backbone relaxation behavior of the domain is discussed with respect to the potential function of an insert region present in all three members of this protein family. Isothermal titration calorimetry (ITC) studies were completed measuring the thermodynamic parameters of the binding of a 10-residue phosphorylated peptide representative of erbB2 to the SH2 domain. These measurements are compared to calorimetric studies performed on other SH2 domain/phosphorylated peptide complexes available in the literature.
Receptor tyrosine kinases (RTKs) play a major role in intracellular communication and cell signaling. The general mechanism of action involves binding of a ligand to the RTK, RTK dimerization and autophosphorylation, and initiation of a signaling cascade within the cell. The growth factor receptor-bound (Grb) family of proteins binds to the epidermal growth factor receptor (EGFR, erbB1) via their SH2 domains. A subclass of Grb proteins was identified based on similar domain architecture (Margolis 1994). The members of the Grb7 family of proteins, Grb7, Grb10, and Grb14, have an N-terminal proline-rich domain, followed by a Ras associating-like (RA) domain identified via sequence homology searches (Wojcik et al. 1999), a pleckstrin homology (PH) domain, a short phosphotyrosine interaction region (PIR, BPS), and a C-terminal Src Homology 2 (SH2) domain.
The members of the Grb7 family may carry out a more specialized signaling pathway than other more ubiquitously expressed SH2 domain-containing proteins, as their expression is considerably more tissue-specific. In particular, Grb7 is expressed in the liver, kidney, and gonads (Margolis et al. 1992), and correlations are seen between overexpression and cancer. Grb7 is co-overexpressed with erbB2 in 20%–30% of all breast cancers (Stein et al. 1994). A potential role for Grb7 in enhanced FAK-mediated cell migration (Han and Guan 1999; Han et al. 2000; Shen and Guan 2001) was postulated, bringing up the speculation that cancers overexpressing Grb7 may be particularly prone to metastasis. In addition, research describing an interaction between erbB2, Grb7, and NIK (Chen et al. 2003) provided a link between erbB2/Grb7 overexpression and downstream inflammation that promotes an environment favorable for the initiation and progression of cancer. The causal relationship between chronic inflammation and cancer is now widely accepted (for review, see Coussens and Werb 2002).
The SH2 domains of Grb7, Grb10, and Grb14 share 68%–72% sequence identity, and they each preferentially bind to different receptor tyrosine kinases. Grb7 binds strongly to the erbB2 receptor via its SH2 domain in coimmunoprecipitation experiments (Stein et al. 1994), while Grb10 and 14 only weakly associate with erbB2 (Janes et al. 1997). The hGrb7-SH2 domain binds to phosphorylated tyrosine 1139 on the erbB2 receptor and prefers an asparagine in the +2 position, while the Grb14-SH2 domain binds to phosphorylated tyrosine 766 on the fibroblast growth factor receptor (FGFR) and prefers a hydrophobic residue in the +3 position (Daly et al. 1996). Throughout this manuscript, peptide residue positions subsequent (C-terminal) to the phosphotyrosine residue are referred to as +1 (the first residue after the pTyr), +2 (the second residue after the pTyr), etc. Residue positions prior (N-terminal) to the phosphotyrosine are referred to as −1 (the residue immediately preceding the pTyr), and so forth.
The SH2 domain of Grb10 was shown to interact with many different proteins, including the insulin and IGF1 receptors (He et al. 1998), platelet-derived growth factor (PDGF) receptor-β (Wang et al. 1999), Ret (Pandey et al. 1995), Kit (Jahn et al. 2002), Raf1 and MEK1 (Nantel et al. 1998), and Nedd4 (Morrione et al. 1999). Despite this abundance of binding partners and a recently solved Grb10-SH2 domain crystal structure (Stein et al. 2003), a consensus binding sequence for Grb10 has not been determined.
SH2 domains bind their phosphorylated tyrosine ligands in an extended conformation, with the exception of the Grb2-SH2 and Grb7-SH2 domains, which bind their ligands in a β-turn conformation (Rahuel et al. 1998; Ogura et al. 1999; Ivancic et al. 2003). In the Grb2-SH2 domain, a bulky tryptophan residue (Trp 66) within the EF-loop region occludes the hydrophobic pocket normally occupied in the phosphorylated peptide complexed state by a hydrophobic residue in the +3 position (i.e., the +3 binding pocket). In a similar manner, a four-residue insertion in the EF loop of the Grb7- SH2 domain (Met 83–Gly 86) occludes the +3 binding pocket. This insert is present in all of the Grb7 protein family SH2 domains. As with the Grb7-SH2 domain, the preferred consensus-binding site of the Grb2-SH2 domain contains an asparagine in the +2 position relative to the phosphorylated tyrosine. In the Grb2-SH2 domain, this asparagine has been postulated to provide binding-site recognition through favorable enthalpic interactions with a β-strand lysine residue (βD6, Lys 54) (McNemar et al. 1997).
In a prior study, the solution structure of the human Grb7-SH2 domain in complex with a phosphorylated peptide representative of the erbB2 receptor was presented (Ivancic et al. 2003). In the present study we present a biophysical characterization of Grb7 binding to erbB2, through analytical ultracentrifugation methods, nuclear magnetic resonance (NMR) relaxation measurements, and ITC. These studies are presented in support of a hypothesis that the dynamic nature of a region in these SH2 domains (including the insertion region unique to this protein family) may play a role in the binding specificity differences observed between the Grb7 protein family members.
Nuclear relaxation studies by NMR spectroscopy are a useful method for studying protein dynamics, as they are sensitive to a wide range of internal molecular motions (from the millisecond to picosecond timescale). Both global and local bond motions affect the relaxation of the observed nuclei, and thus the linewidths of the individual NMR resonances. Through measurement of the longitudinal relaxation time (T1), transverse relaxation time (T2), and heteronuclear steady-state nuclear Overhauser effect (NOE), information describing the timescale and type of motion a nucleus is experiencing can be monitored. This approach has been utilized to good effect in many protein dynamics studies (Kay et al. 1989; Clore et al. 1990; Barbato et al. 1992; Schneider et al. 1992; Stone et al. 1992; Li and Montelione 1995; Nicholson et al. 1995; Loh et al. 1999; Pawley et al. 2002; and others).
Certain per residue combinations of T2 and NOE values can be used to determine the existence or non-existence of mobile sites in a biomolecule, without the full Lipari and Szabo (1982a,b) model-free analysis needed to resolve the order parameters (S2) and effective correlation times (τe). For instance, a residue that possesses a T2 value substantially lower than the average T2 value for the bulk of the biomolecule, coupled with a higher than average NOE value, is an indication of decreased internal motion for that residue. Analogously, a residue that possesses a T2 value substantially higher than the average T2 value for the bulk of the biomolecule, coupled with a lower than average NOE value, is an indication of increased internal motion for that residue (Loh et al. 1999). In the nuclear relaxation studies of the human Grb7-SH2 domain presented here such an approach is taken, with the goal of locating regions of the protein domain displaying increased and/or decreased mobility with respect to the unliganded and liganded forms of the SH2 domain. A complete model-free analysis of the nuclear relaxation parameters of the human Grb7-SH2 domain has not been possible due to SH2 domain dimerization processes (addressed in the Results and Discussion sections).
Materials and methods
Sample preparation
Expression and purification of the human Grb7-SH2/pY1139 complex were described in detail previously (Brescia et al. 2002). For both the free Grb7-SH2 domain and Grb7-SH2/pY1139 complex, sample conditions for NMR spectroscopy were 0.6–1.0 mM protein, 0.6–1.0 mM pY1139 peptide ligand, 50 mM sodium acetate, 100 mM NaCl, 5 mM DTT, 1 mM NaN3, and 1 mM EDTA, at pH 6.6. Sample conditions for the analytical ultracentrifugation and ITC studies were the same as that for the NMR studies, in terms of buffer conditions and pH. The concentration of the Grb7-SH2 domain solution for ultracentrifugation was 51 μM. Sample conditions for the ITC studies were 30–50 μM Grb7-SH2 domain and 300–500 μM pY1139 peptide ligand, with omission of the DTT and EDTA.
Analytical ultracentrifugation
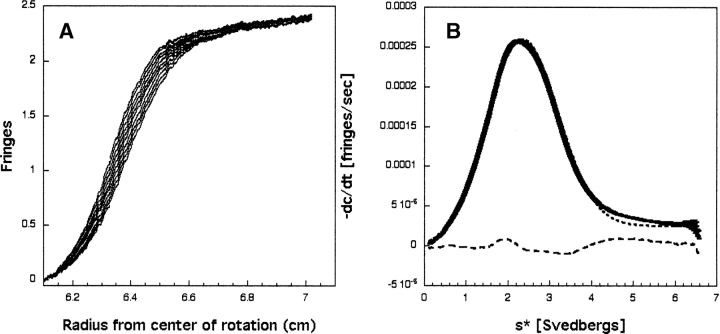
Sedimentation velocity data were obtained on the unliganded human Grb7-SH2 domain using a Beckman XLI analytical ultracentrifuge fitted with an An50-Ti rotor. Data were collected using a two-sector centerpiece and centrifuged at 50,000 rpm at 20°C. Figure 1A ▶ shows a representative set of interference scans from the sedimentation velocity analysis of the Grb7- SH2 domain.
Figure 1.
(A) Representative interference scans for the Grb7-SH2 domain. Data were acquired at 1-min intervals. For clarity, every third scan is shown from a sedimentation velocity run at 50,000 rpm and 20°C, and starting with a run time of 2 h. (B) Shown are dc/dt data for the Grb7-SH2 domain from scans taken 1 min apart. The solid line (showing error bars) represents the average dc/dt. The dotted line represents the best-fit to a single species, and the dashed line indicates the residuals of the fit. The rms deviation for this fit is 5.85 × 10−6.
Backbone nuclear relaxation measurements
All NMR spectra of both the hGrb7 SH2/erbB2 pY1139 peptide complex and free Grb7-SH2 domain were obtained at a temperature of 298 K on a Varian INOVA 500 MHz spectrometer. The spectrometer was equipped with a 1H/13C/15N resonance pulsed-field gradient probe. Spectra were recorded in the States-TPPI mode for quadrature detection with carrier frequencies for 1H and 15N at 4.73 ppm and 120.0 ppm, respectively. Sensitivity-enhanced pulse sequences used to obtain the 15N T1 and T2 values and the steady-state NOE values were obtained from the Lewis E. Kay laboratory and are described elsewhere (Kay et al. 1992; Farrow et al. 1994). 15N T1 values were measured from spectra recorded with eight different durations of the T delay: 11, 67, 111, 278, 444, 611, 888, and 1332 msec. 15N T2 values were measured from spectra recorded with nine different durations of the T delay: 0, 16, 31, 47, 63, 78, 94, 110 and 126 msec. For estimation of noise levels, duplicate spectra were collected for T=67 and 444 msec (T1 data) and for T=49 msec (T2 data). Relaxation delays of 3 sec were employed in the measurement of 15N T1 and T2 values; 512 real data points were collected in the direct dimension, and 256 real data points were collected in the indirect dimension with 32 scans per point. Spectral widths of 2000 Hz and 8000 Hz were employed in F1 and F2, respectively. To determine the NOE values, spectra were recorded in the presence and absence of a proton presaturation period of 3 sec. For the NOE spectra, a relaxation delay of 5 sec prior to a 3-sec proton presaturation period was used, and a net relaxation delay of 8 sec was used for the NOE spectra without a proton presaturation period. Again, 512 real data points were collected in the direct dimension, and 128 real data points were collected in the indirect dimension with 64 scans per point. The same spectral widths were used as in the T1 and T2 experiments.
Data were processed using the Azara 2.6 software package (Wayne Boucher, University of Cambridge) on a Silicon Graphics workstation. All spectra were processed identically with sinebell squared apodization functions and zero filling to the next power of 2 in both dimensions and a polynomial baseline correction in the direct dimension. The T1 and T2 values were determined by fitting the measured peak volumes to a two-parameter function: I(t)=I0 exp(−t/T1,2), where I0 is the intensity at time t=0 and I(t) is the intensity after a delay of time t. The software program DASHA was used in calculating the T1 and T2 values for each residue in both the free and complexed hGrb7-SH2 domain (Orekhov et al. 1995). The steady-state NOE values were determined from the ratios of the average volumes of the peaks with and without proton saturation. The standard deviation of the NOE values was determined as described in Farrow et al. (1994).
ITC studies
Titrations were performed using a VP-ITC ultrasensitive isothermal titration calorimeter (MicroCal) (Wiseman et al. 1989). Grb7-SH2 was dialyzed with three exchanges of buffer (1/1000 v/v). To reduce errors arising from heats of dilution due to buffer differences between samples in the syringe and the reaction vessel, the pY1139 peptide ligand was dialyzed concurrently with the Grb7-SH2 sample. Both peptide and protein concentrations were determined using measured A280 values and were degassed prior to loading into the syringe and reaction vessel. For each titration, 10 μL of peptide ligand (300–500 μM) was delivered into the protein sample (30–50 μM) over a 20-sec duration with an adequate interval (250 sec) allowing complete equilibration. Binding curves involved the addition of 30 injections that enabled 50% saturation to occur by the fifteenth injection. The heats of dilution, obtained by titrating the identical peptide solution into the reaction cell containing only the sample buffer, were subtracted prior to analysis. Data were recorded at a baseline of 20 μcal/sec and at a high gain mode, thus providing the fastest re-equilibration between injections. The syringe mixing speed was 300 rpm in order to ensure sufficient mixing while keeping baseline noise at a minimum. The temperature-dependent binding studies were conducted at 15°, 25°, and 37°C (±0.2°) after the instrument had been equilibrated for 0.5 to 1 h at the required temperature. The data were collected automatically, and analyzed with a one-site model via the Origin software (V7.0) provided from MicroCal. Origin utilizes a nonlinear least-squares algorithm (minimization of χ2), the concentration of the titrant and sample, and an equilibrium equation to provide best-fit values of the binding curve thus providing stoichiometry (n), change in enthalpy (ΔH), and binding constant (K) (Zhang et al. 2000).
Results
Ultracentrifugation studies
The analytical ultracentrifugation dc/dt data were analyzed using the program DCDT+ (Philo 2000). The partial specific volume of the protein was estimated according to the method of Cohn and Edsall (1943). The solution density was estimated according to the method of Laue et al. (1992). Figure 1B ▶ shows the resultant dc/dt versus s* curve for the Grb7-SH2 domain at a loading concentration of 51 μM. The data were fit to various models to determine the weight-average sedimentation coefficient (s*). The data were best fit to a single species, with the corresponding residuals of the fit also shown in Figure 1B ▶. Fitting the dc/dt data to s/D, which returns molecular weight, returns an s20,w=2.18+0.01 and a molecular weight of 20.02+0.02 kilodalton. Inclusion of additional species did not improve the fit, as judged by the rms deviations of the fits (data not shown).
Nuclear relaxation data
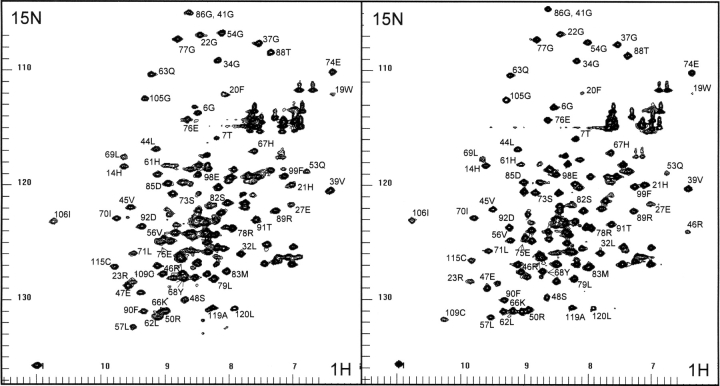
1H-15N correlation HSQC spectra are shown of the human Grb7-SH2 domain and the human Grb7-SH2/erbB2 peptide (pY1139) complex in Figure 2 ▶. The functional regions of the Grb7-SH2 domain include the phosphate binding residues Arg 26, Arg 46, and Ser 48; residues Val 56 and Leu 69 that are in direct contact with the erbB2 peptide phosphotyrosine aromatic ring; and residues Tyr 68 and Ser 82 that are in direct contact with the +1 Val and +2 Asn residues of the pY1139 peptide. These contacts and the Grb7-SH2/pY1139 complex structure were previously described in detail (Ivancic et al. 2003).
Figure 2.
The proton-nitrogen (1H-15N) correlation spectra of the unliganded Grb7-SH2 domain (left panel) and the Grb7- SH2/pY1139 complex (right panel) are shown. The F2 (X-axis) labels represent amide and Gln/Asn side chain proton resonances, and F1 (Y-axis) labels represent the attached nitrogen resonances.
Sequence assignments of the human Grb7-SH2/pY1139 complex have been published (Brescia at al. 2002). Of the 130 residues composing the Grb7-SH2/pY1139 complex, the two N-terminal residues of the SH2 domain and the two N-terminal residues of the pY1139 peptide remain unassigned. The backbone assignments of the free Grb7- SH2 domain were established through analysis of three-dimensional HNCA, HN(CO)CA, CBCA(CO)NH, and HNCACB triple-resonance spectral data sets. Of the 120 residues composing the free Grb7-SH2 domain, 20 remain unassigned, primarily due to the poor quality of NMR spectral data acquired.
Values for the 1H-15N backbone relaxation parameters T1, T2, and the steady-state NOE of the free Grb7-SH2 domain and the Grb7-SH2/pY1139 complex were obtained from exponential decay fits of the spectral volumes for each assigned resonance. The same relaxation parameters measured from exponential fits of the resonance spectral intensities gave essentially identical results. Reliable relaxation parameters were not obtained in some cases due to severe resonance overlap or because the resonance signals were too weak for analysis at the latter time points.
The backbone T1, T2, and steady-state NOE relaxation parameters for the human Grb7-SH2 domain free and in complex with the erbB2 peptide pY1139 are presented in Figure 3 ▶, A and B. The T1 (or T2) relaxation data are plotted as the variation of T1 (or T2) per residue from the average T1 (or T2) value for the SH2 domain determined from the central highly structured region (residues 20– 70). For this region the average T1 values are 0.973 and 0.683 sec (free vs. complex) and the average T2 values are 0.0449 and 0.0511 sec (free vs. complex). The average NOE values are 0.671 and 0.706 (free vs. complex).
Figure 3.

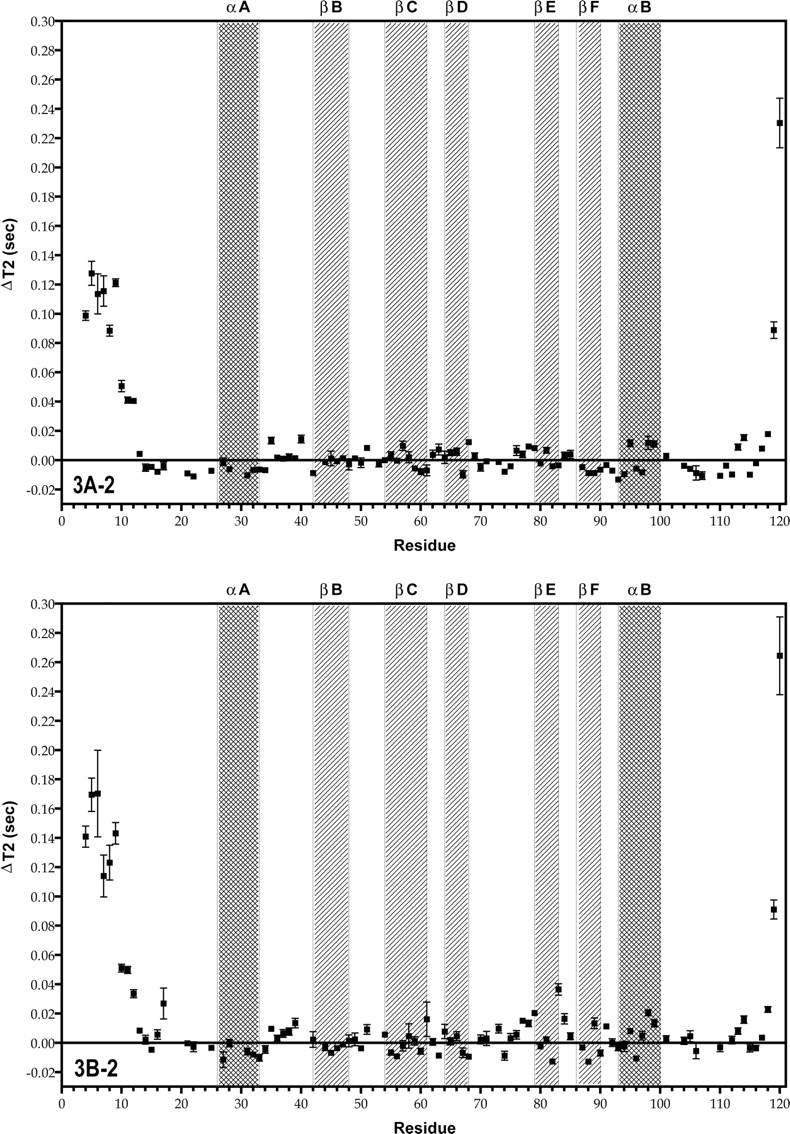
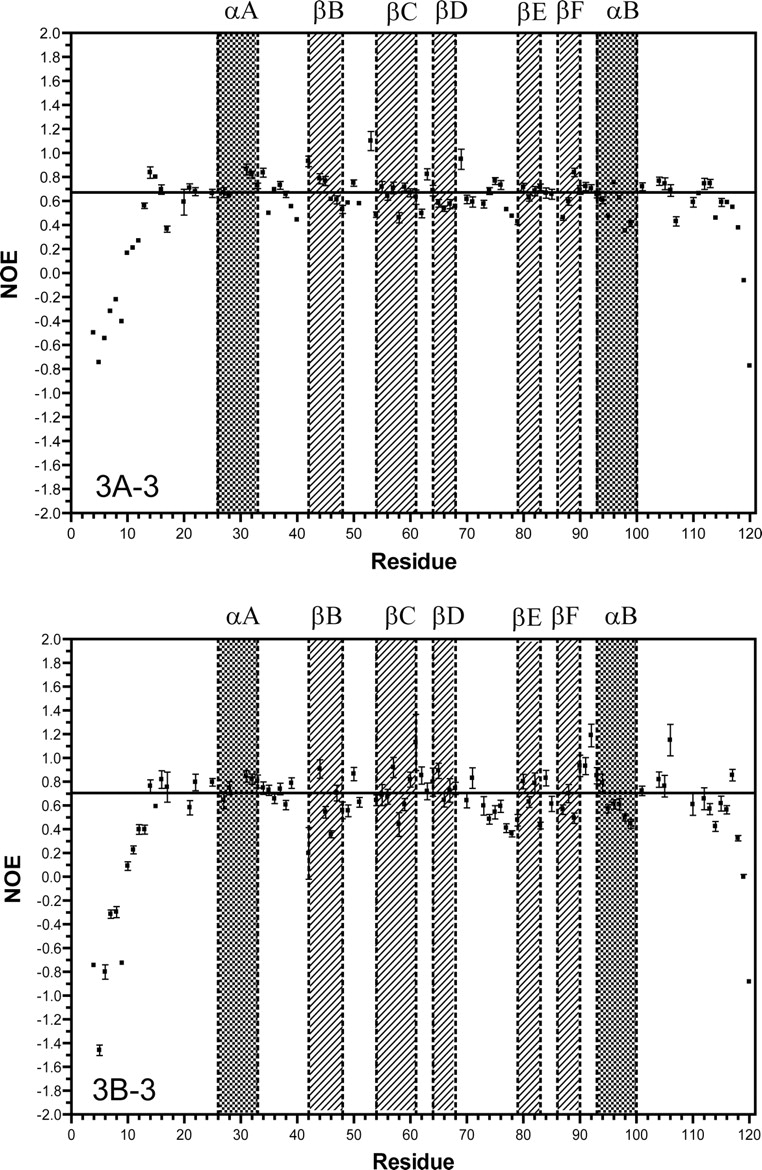
Plots of T1, T2, and NOE as a function of residue number for the free human Grb7-SH2 domain (A1–3), and the Grb7-SH2/pY1139 peptide complex (B1–3), measured at 500 MHz. For the T1 and T2 measurements, the data are represented as the difference (Δ) in the T1 or T2 value from the average T1 or T2 value for the protein domain (described in the text). The average values for the T1, T2, and NOE are represented in the plots by a solid horizontal line. The secondary structural regions of the Grb7-SH2 domain are indicated by filled squares and labeled accordingly.
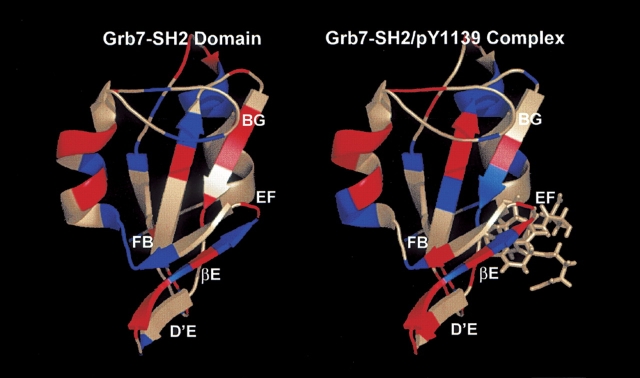
For the free Grb7-SH2 domain, the backbone T1, T2, and NOE values vary little from the average values except at the N-terminal and C-terminal ends of the domain, where the T1 values drop, the T2 values rise, and the NOE values drop. This is expected for unstructured random coil regions. A higher concentration of residues with lower than average T2 values and higher than average NOE values is found in the D′ E loop, βE strand, and FB loop regions in comparison to the nuclear relaxation parameters for the same regions in the Grb7- SH2/pY1139 complex (Fig. 3A-2, B-2, A-3, B-3 ▶; Fig. 4 ▶).
Figure 4.
Backbone nuclear relaxation results for the free Grb7-SH2 domain (left panel) and Grb7-SH2/pY1139 peptide complex (right panel). Residues highlighted in blue represent those that exhibit lower than average T2 values and higher than average NOE values. Residues highlighted in red represent those that exhibit higher than average T2 values and lower than average NOE values. Residues shown in light gray either exhibit more complex relaxation behavior (e.g., higher than average T2 values and higher than average NOE values) or relaxation parameters were not obtained. The structured residues of the pY1139 peptide are shown in a stick format. The labeling of secondary structural elements follows the nomenclature of Eck et al. (1993).
In the Grb7-SH2/pY1139 complex, the same pattern is observed for the T1, T2, and NOE relaxation parameters at the unstructured N-terminal and C-terminal ends of the SH2 domain. In addition, noticeable changes in these relaxation parameters are observed for many residues either in direct contact with the erbB2 peptide or in some way affected by ligand binding. For clarity, the raw T2 values, 15N NOE values, and the change in these two parameters from the average values are reported in Tables 1A and 1B for the residues highlighted in the following discussion.
Table 1A.
T2 and15N NOE relaxation parameters for the Grb7-SH2 domain (uncomplexed) residues
| Residue | 2°Region | T2 (sec) | ΔT2 (sec) | 15N NOE | Δ15N NOE |
| 46 | βB | 0.0441 | −0.0008 | 0.614 | −0.057 |
| 56 | βC | 0.0445 | −0.0004 | 0.640 | −0.031 |
| 57 | βC | 0.0546 | +0.0097 | 0.708 | +0.037 |
| 67 | βD | 0.0352 | −0.0097 | 0.574 | −0.097 |
| 68 | βD | 0.0572 | +0.0123 | 0.556 | −0.115 |
| 73 | D′E | 0.0436 | −0.0013 | 0.575 | −0.096 |
| 75 | D′E | 0.0407 | −0.0042 | 0.766 | +0.095 |
| 76 | D′E | 0.0514 | +0.0065 | 0.733 | +0.062 |
| 77 | D′E | 0.0488 | +0.0039 | 0.529 | −0.142 |
| 78 | D′E | 0.0542 | +0.0093 | 0.474 | −0.197 |
| 79 | βE | 0.0531 | +0.0082 | 0.427 | −0.244 |
| 82 | βE | 0.0407 | −0.0042 | 0.682 | +0.011 |
| 83 | βE | 0.0413 | −0.0036 | 0.714 | +0.043 |
| 85 | EF | 0.0488 | +0.0039 | 0.656 | −0.015 |
| 89 | βF | 0.0360 | −0.0089 | 0.838 | +0.167 |
| 91 | FB | 0.0415 | −0.0034 | 0.721 | +0.050 |
| 98 | αB | 0.0567 | +0.0118 | 0.351 | −0.320 |
The residue number, location of the residue by secondary structural region, raw T2 value, change in T2 from the average T2 value, raw 15N NOE value, and change in the 15N NOE value from the average 15N NOE value are reported.
Table 1B.
T2 and15N NOE relaxation parameters for the Grb7-SH2 domain (complexed with pY1139) residues
| Residue | 2° Region | T2 (sec) | ΔT2 (sec) | 15N NOE | Δ15N NOE |
| 46 | βB | 0.0474 | −0.0037 | 0.358 | −0.348 |
| 56 | βC | 0.0419 | −0.0092 | 0.673 | −0.033 |
| 57 | βC | 0.0491 | −0.0020 | 0.920 | +0.214 |
| 67 | βD | 0.0443 | −0.0068 | 0.727 | +0.021 |
| 68 | βD | 0.0417 | −0.0094 | 0.744 | +0.038 |
| 73 | D′E | 0.0608 | +0.0097 | 0.597 | −0.109 |
| 75 | D′E | 0.0542 | +0.0031 | 0.543 | −0.163 |
| 76 | D′E | 0.0566 | +0.0055 | 0.594 | −0.112 |
| 77 | D′E | 0.0662 | +0.0151 | 0.410 | −0.296 |
| 78 | D′E | 0.0643 | +0.0132 | 0.362 | −0.344 |
| 79 | βE | 0.0714 | +0.0203 | 0.477 | −0.229 |
| 82 | βE | 0.0383 | −0.0128 | 0.787 | +0.081 |
| 83 | βE | 0.0876 | +0.0365 | 0.431 | −0.275 |
| 85 | EF | 0.0556 | +0.0045 | 0.613 | −0.093 |
| 89 | βF | 0.0644 | +0.0178 | 0.492 | −0.214 |
| 91 | FB | 0.0624 | +0.0113 | 0.931 | +0.225 |
| 98 | αB | 0.0716 | +0.0205 | 0.498 | −0.208 |
The residue number, location of the residue by secondary structural region, raw T2 value, change in T2 from the average T2 value, raw 15N NOE value, and change in the 15N NOE value from the average 15N NOE value are reported.
A group of residues, Glu 75 through Leu 79, and especially Met 83 and Asp 85, experience higher than average T2 values with lower than average heteronuclear steady-state NOEs (Fig. 3B-2, B-3 ▶). Several residues (Leu 57, His 67, Tyr 68, and Ser 82) have lower than average T2 relaxation values with higher than average heteronuclear NOE measurements. A stretch of residues, 31–45, displays lower than average T1 values in the Grb7-SH2/pY1139 complex (Fig. 3B-1 ▶). Precise measurements of T2 and the steady-state NOE are the most useful for determining the widest range of internal molecular motion parameters (i.e., S2 and τe) (Jin et al. 1998). As a full model-free analysis is not possible here, the T1 results will not be discussed further except with respect to determination of the molecular correlation time τm (see below).
In general, the largest variations in the relaxation parameters in the Grb7-SH2/pY1139 complex are concentrated either in residues in direct contact with the pY1139 peptide or in the C-terminal one-third of the SH2 domain (Figs. 3 ▶, 4 ▶).
Molecular correlation time
For both the free Grb7-SH2 domain and the Grb7-SH2/pY1139 complex, the approximate molecular correlation time (τm) was calculated from the measured T1/T2 ratios. The ratio of T1 over T2 may be used to estimate τm if τe (the effective correlation time)<100 psec, τm>1 nsec, and T2 is not decreased by chemical exchange processes (Kay et al. 1989; Clore et al. 1990; Farrow et al. 1994). In this case, the ratio of T1/T2 is effectively independent of both the order parameter S2 and τe. To ensure that the T1 and T2 measurements met these requirements, T1/T2 ratios with one standard deviation (SD) greater than, or one SD less than, the mean were excluded from the calculation of τm. For T1/T2 ratios greater than one SD from the mean, exchange processes may contribute to a reduction in the T2 value; while for those with values less than one SD from the mean, the system may not be modeled by a simple single-time- scale spectral density function (Farrow et al. 1994).
Using the above-described prerequisites, the free human Grb7-SH2 domain has a τm of 21.5 nsec based on the T1/T2 ratios of the 73 residues that met the one-SD variance criterion. For the Grb7-SH2/pY1139 peptide complex, the τm was calculated to be 11.7 nsec based on the T1/T2 ratios of the 64 residues that met the one-SD variance criterion.
Binding thermodynamics of the erbB2 pY1139 ligand
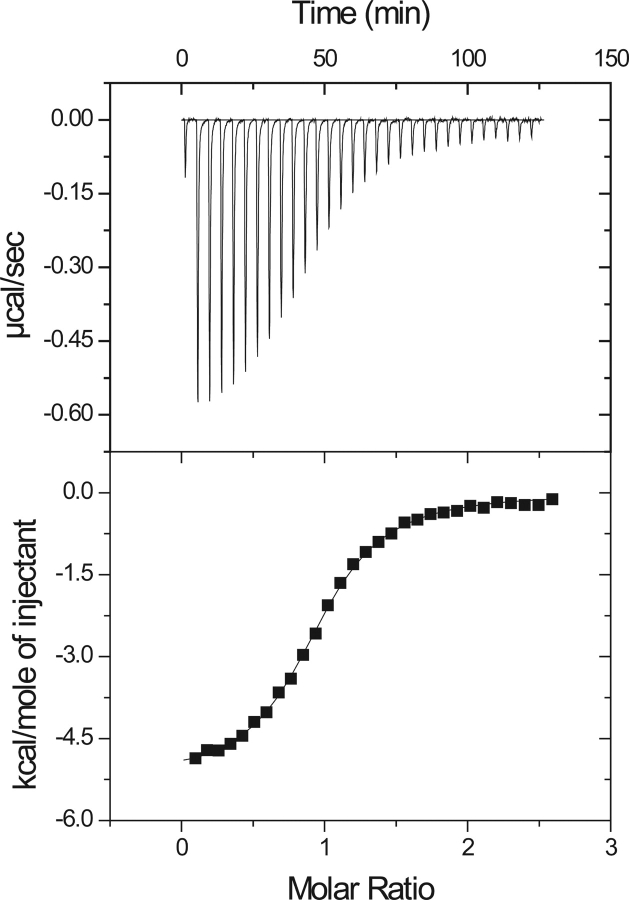
A thermodynamic description for the binding of the pY1139 phosphotyrosine decapeptide (NH2- PQPEpYVNQPD-COOH) to the Grb7-SH2 domain was determined using ITC. Figure 5 ▶ (top) shows the heat of reaction for 30 titrations of pY1139 into a solution of Grb7-SH2. The binding reaction is exothermic, and the integrated heats of reaction for the data in Figure 5 ▶ (top) are shown as a binding curve in Figure 5 ▶ (bottom). A nonlinear least squares fit of the binding curve provides values for the stoichiometry of binding (n, ligand:protein), equilibrium binding constant (Kb), and change in enthalpy (ΔH). A thermodynamic description of binding is obtained from these parameters including the free energy of binding, ΔG, and change in entropy of reaction, ΔS:
Figure 5.
(Top) Exothermic heats of reaction (μcal/sec) measured for 30 injections of the pY1139 peptide into a Grb7-SH2 sample. (Bottom) The solid line is the best fit to the data for a single binding site model using a nonlinear least squares fit to solve for n (number of binding sites), Kb (equilibrium binding constant), and ΔH (change in enthalpy of reaction), and reported in Table 2.
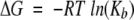 |
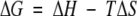 |
where R is the gas constant and T is the temperature in Kelvin. The solid line shown in Figure 5 ▶ (bottom) is the best fit for one of the reactions performed at 25°C, and a summary of the average binding parameters for five reactions, including the dissociation constant (Kd=1/Kb), is presented in Table 2. These data show that the free energy of binding (ΔG=−7.70 kcal mol−1) is favored by both enthalpic (ΔH=−4.66 kcal mol−1) and entropic contributions (−TΔS=−3.04 kcal mol−1).
Table 2.
Thermodynamic parameters for the binding of erbB2 peptide pY1139 to Grb7-SH2 at 25°C
| Kd (μM) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | −TΔS (kcal mol−1) |
| 2.28 ± 0.15 | −7.70 ± 0.04 | −4.66 ± 0.05 | −3.04 ± 0.09 |
The reported values are the average of five experiments. The stoichiometry, n, was also fit and had values between 0.94 and 1.06 for each analysis.
Temperature dependence of ligand binding: ΔCp of binding
Titrations were also performed between 15° and 37°C to determine the change in heat capacity, ΔCp, upon ligand binding. ΔCp was calculated from the dependence of ΔHreact on temperature (Fig. 6 ▶) and based on the slope has a value of −99 cal mol−1 K−1. The size and sign of ΔCp are consistent with a binding interaction that involves a net increase in buried apolar surface area (Connelly 1997).
Figure 6.
Temperature-dependent ΔH energetics of binding for Grb7- SH2 and pY1139: The heat capacity of binding (ΔCp), determined from the slope for ΔH, has a value of ′99 cal mol′1 K′1. ITC binding reactions were performed at 15°, 25°, and 37°C under the solution conditions described in the text (Materials and Methods).
Discussion
Ultracentrifugation studies (Fig. 1 ▶) of the Grb7-SH2 domain demonstrate a larger than expected molecular weight, at 20 kDa, than the actual molecular weight (13.8 kDa). Since the maximum sedimentation coefficient (s*) value for a spherical molecule of molecular weight 13.8 kDa would be 1.858, and the measured s* is 2.18, we can conclude that the quaternary state is not monomeric (Philo 2000). The dimerization behavior of the SH2 domains of this protein family is also reflected in the measured molecular correlation time (τm) of the ligand-free Grb7-SH2 domain (21.5 nsec) and the Grb7-SH2/pY1139 complex (11.7 nsec). Since τm is proportional to the volume for an isotropically tumbling spherical macromolecule (Cantor and Schimmel 1980; Venable and Pastor 1988), it should roughly increase in proportion to molecular weight. The fact that the τm of the Grb7-SH2 domain in its ligand-free form is almost twice that of the τm of the bound form is a strong indication that higher molecular weight forms exist for the free SH2 domain.
Human Grb7-SH2 domain backbone nuclear relaxation
We have shown that there are regions of the Grb7-SH2 domain and Grb7-SH2/pY1139 peptide complex that have backbone nuclear relaxation characteristics different from those of the bulk of the SH2 domain; these are residues within the D′ E loop, βE strand, EF loop, FB loop, and many of the SH2 domain residues in direct contact with the erbB2 pY1139 peptide.
The T2 difference plots of the Grb7-SH2 domain free and in complex with the erbB2-derived pY1139 peptide demonstrate substantial changes in the relaxation behavior of many residues upon ligand binding. Of immediate note is that in the complex, several residues in direct contact with the pY1139 peptide experience large decreases in the backbone T2 relaxation rate with respect to the average backbone T2 relaxation rate of the SH2 domain (Fig. 3B-2 ▶). These residues are Arg 46, Val 56, Tyr 68, and Ser 82. While Tyr 68 and Ser 82 concurrently show increased heteronuclear 15N NOE values, Arg 46 and Val 56 experience average or lower heteronuclear 15N NOE values (Fig. 3B-3 ▶). For Tyr 68 and Ser 82, the logical interpretation of these decreased T2 relaxation rates with increased NOE values is that these residues experience decreased motion upon ligand binding and/or increased chemical exchange with the pY1139 peptide. This is also consistent with an increased spin density surrounding these residues in the Grb7-SH2/pY1139 complex. The more complex relaxation behavior of Arg 46 and Val 56 cannot be interpreted in terms of the simple determination of mobile sites and will require further study for complete interpretation. For two other residues in contact with the pY1139 peptide, Arg 26 and Leu 69, the complete set of relaxation parameters was not available.
Of additional interest is that residues Ser 73, Glu 75 through Leu 79, Met 83, Asp 85, and Arg 89 (D′ E loop, βE strand, EF loop, and FB loop) all have higher than average T2 relaxation times with lower than average 15N heteronuclear NOE values in the Grb7-SH2/pY1139 complex (Fig. 3B-2, B-3 ▶). These data suggest that the residues are flexible and somewhat disordered in the liganded form of the SH2 domain. Some of these same residues in the free Grb7-SH2 domain display similar relaxation behavior (Gly 77–Leu 79, and Asp 85); however, Glu 75, Met 83, and Arg 89 have decreased T2 values with increased NOE measurements, suggesting decreased mobility (Fig. 3A-2, A-3 ▶). Ser 73 and Glu 76 have both average T2 values and heteronuclear 15N NOE values in the unliganded SH2 domain. Interestingly, Glu 75 through Leu 79 corresponds to many of the same residues that have missing electron density in the X-ray diffraction structure of the human Grb10- SH2 domain (Stein et al. 2003), further corroborating the highly dynamic nature of this SH2 domain region in the Grb7 protein family. Increased backbone T2 relaxation times are also seen for a substantial Number of residues found at or near the dimerization interface recorded by Stein et al. (2003) (e.g., Arg 89, Thr 91, and Glu 98). These higher T2 values may be due to increased motion for these residues in the SH2 domain complex resulting from the breakup of the dimerization interface upon ligand binding. This conclusion is further supported by the fact that NMR spectral data recorded on the free Grb7-SH2 domain are of poorer quality, with broader resonances and lower signal-to-noise ratios, than data acquired on the Grb7-SH2/pY1139 complex (B. Lyons, unpubl.).
A full model-free Lipari and Szabo treatment of the Grb7-SH2 domain has not been possible due to exchange broadening effects observed both visually in the recorded NMR data sets and in the measured τm, and demonstrated by the sedimentation velocity ultra-centrifugation study herein. A similar dimerization equilibrium exists in the study of Troponin C (Mercier et al. 2001). In that study, the authors were able to establish a lower concentration limit where Troponin C exists in greater than 90% monomeric form, thus allowing the determination of S2 and τe values. In the Grb7-SH2 domain system, monomeric solution conditions have eluded characterization.
Potential role of the flexible D′E, EF, and FB loop regions in the Grb7 protein family
Our study of the backbone relaxation behavior of the human Grb7-SH2 domain has provided clues to a potential functional difference related to the conformation of the D′E, EF, and FB loop regions in this family of proteins. The conformation of these regions may provide a “bind or not-bind” switch in the SH2 domains of these proteins. The demonstrated mobile nature of this region in the Grb7-SH2 domain may allow sampling of multiple conformations until stabilizing interactions with the +2 Asn residue and/or the +1 residue of the erbB2 peptide lock the loop into a conformation upon complex formation. This conformation blocks the +3 binding pocket (“closed”), but is characterized by increased motion in residues near to, but not in direct contact with, the ligand. Such energetic characteristics may partially compensate for the unfavorable decrease in entropy associated with ligand binding.
If the mobile nature of the D′E, EF, and FB loops is a general feature of the Grb7 protein family, we can infer function for these regions in the Grb14 and Grb10 SH2 domains. In the Grb14-SH2 domain, the same dynamic movement of the D′E, EF, and FB loops can allow sampling of conformations. When a phosphopeptide ligand with a hydrophobic residue (e.g., Leu, such as in the Grb14 binding site of the fibroblast growth factor receptor) in the +3 position makes stabilizing contacts with the SH2 domain EF loop and βD strand, the EF loop may be locked into a conformation that preserves the +3 binding pocket observed in “standard” SH2 domains (“open”). Though the +3 binding pocket is occupied by the ligand, it may still be possible for nearby residues to show increased motion, again compensating for the unfavorable entropy of ligand binding. There are subtle sequence differences in the βE strands and EF loops of these two SH2 domains that may affect the type of stabilizing interactions formed upon ligand binding, thus directing specificity. A thorough investigation of the effect of mutations in the βE strand and EF loop, specifically residues Ser 82 and Met 83 in the Grb7- SH2 domain and Thr 79 and Leu 80 of the Grb14-SH2 domain, is the next step in confirming this hypothesis.
The dynamic nature of the D′E, EF, and FB loop regions in the Grb7 protein family could explain why a consensus binding sequence for the Grb10-SH2 domain has not been determined (He et al. 1998). An ability to sample many conformations and adapt to different sequences in phosphopeptides could result in Grb10 binding a broad spectrum of proteins. This may be evolutionarily advantageous in signaling pathways involving the Grb10 protein.
We have hypothesized that increased mobility in the D′E, EF, and FB loop regions in the Grb7 protein family SH2 domains allows the selection of either a “closed” conformation (where the +3 binding pocket is occluded and the ligand binds in a turn conformation) or an “open” conformation (in which the +3 binding pocket is available and the ligand binds in an extended conformation) upon binding. Evidence for mobility in the D′E region has been observed in at least one other SH2 domain. The order parameter values (S2) for the D′E transition and loop region of the phospholipase C-γ1 C-terminal SH2 domain display the lowest stretch of values for the entire molecule in both the unliganded and liganded states (Farrow et al. 1994). There is also a high degree of correlation between the location of mobile sites in the Grb7-SH2 domain identified through the T2 and 15N NOE analysis in this study and exchanging regions of the N-terminal SH2 domain of PI3-kinase identified through relaxation dispersion measurements (Mittag et al. 2003). Specifically, exchanging residues in the BC loop, βD strand, EF loop, and BG loop of the N-terminal PI3-Kinase SH2 domain correspond roughly to the location of mobile regions in the Grb7-SH2 domain. Amplified exchange and/or mobility in this region of SH2 domains could be a phenomenon related to their function in general.
Increased flexibility and dynamics have been shown to be important components of binding in many other protein interaction systems. For example, increased dynamics are observed in three regions of Cdc42Hs important for function (Loh et al. 1999, 2001). Other examples of a link between increased dynamics and binding site location or function include HIV-1 protease (Nicholson et al. 1995), Borrelia OspA (Pawley et al. 2002), and CheW (Griswold and Dahlquist 2002), among others.
ITC studies
A thermodynamic analysis for the binding of a phosphotyrosine peptide from the receptor tyrosine kinase erbB2 (pY1139) to the SH2 domain of Grb7 was conducted using ITC. The results from ITC experiments indicate that the binding reaction occurs with a stoichiometry of one pY peptide to one SH2 domain with ΔG=−7.70 kcal mol−1 at 25°C. The interaction is both enthalpically (ΔH=−4.66 kcal mol−1) and entropically (−TΔS=−3.04 kcal mol−1) favored. As with all SH2 domains for which the temperature dependence upon ligand binding is available, the ΔCp for the Grb7-SH2 domain binding the erbB2 peptide is small and negative (−99 cal/mol) (Mandiyan et al. 1996; McNemar et al. 1997; Bradshaw and Waksman 1999). This value compares favorably to a ΔCp of −146 cal/mol for the Grb2- SH2 domain binding the Shc peptide (McNemar et al. 1997). Positive values of ΔCp are interpreted as evidence for the release of tightly bound water molecules upon binding, while negative ΔCp values are interpreted as increased removal of apolar groups from the solvent (Dill 1990; Privalov et al. 1990; Connelly 1997).
In a calorimetric study of an Shc peptide binding to the Grb2-SH2 domain (McNemar et al. 1997), the authors suggested that hydrogen bond formation between Leu 65 and Lys 54 of the SH2 domain and the +2 Asn of the peptide may be reflected in the measured enthalpy term. Replacement of the +2 Asn residue of the Shc peptide with an Ala resulted in a disfavorable 5.6 kcal/mol change in the enthalpy of binding. In Grb2-SH2/phosphopeptide complex structures, a hydrogen bond is formed between the carbonyl oxygen of Leu 65 and the peptide +2 asparagine side chain amide proton (Rahuel et al. 1998; Ogura et al. 1999). This orientation places the asparagine side chain carbonyl in a favorable position to form a hydrogen bond with the amide hydrogen of the βD6 residue (Lys 54) in the Grb2-SH2 domain.
A similar argument does not hold for the Grb7-SH2 domain. The residue corresponding to Leu 65 in Grb2- SH2 is Ser 82 in Grb7-SH2, and a hydrogen bond is formed between the Ser 82 side chain hydroxyl proton and the ligand +2 asparagine side chain carbonyl (Ivancic et al. 2003). This orientation of the +2 Asn 1141 residue in the Grb7-SH2/pY1139 complex makes the formation of hydrogen bonds between the SH2 domain βD6 residue Leu 69 and Asn 1141 less likely, since greater distances exist between the potentially interacting functional groups. This may be reflected in the measured enthalpy of binding for the pY1139 peptide. The Grb2-SH2 domain interaction with Shc peptide is dominated by enthalpic contributions (87% of the free energy of binding). For the Grb7-SH2 domain, the enthalpic contributions to the free energy of binding are less dominating (61% of the free energy of binding). The lower favorable ΔH value for the pY1139 peptide may result from the peptide’s ability to only form hydrogen bonds to Ser 82, and not additionally to Leu 69.
Amino group-containing residues at the βD6 position in SH2 domains are involved in enthalpically stabilizing interactions with the ligand phosphotyrosine aromatic ring (Waksman et al. 1993; Pascal et al. 1994; Ogura et al. 1999). The introduction of a leucine at the βD6 position (as in the Grb7-SH2 domain) removes this amino-aromatic interaction to the RTK phosphotyrosine, and thus may also explain the lower favorable ΔH value for the Grb7-SH2 domain binding the erbB2 peptide. The only other SH2 domain that does not have a Q/K/R at the βD6 position is the p85α subunit C-terminal SH2 domain of PI3-kinase, which also has a hydrophobic residue (a valine) at this position. This SH2 domain binds its ligands in an extended conformation, and its free energy of binding is dominated by a large favorable enthalpy term (O’Brien et al. 2000). For this SH2 domain, the large enthalpy term is presumably at least partially due to a significant number of noncovalent bonds associated with SH2 domain/ligand +3 binding pocket interactions. Since the Grb7-SH2 domain does not bind its ligand in an extended conformation, and the +3 binding pocket is occluded, a further direct comparison of these two SH2 domains may not be warranted.
Yet another argument exists for the smaller observed ΔH for Grb7-SH2 binding of the erbB2 peptide ligand. It is assumed that the formation of hydrogen bonds between the SH2 domain pTyr binding pocket and the phosphate moiety of the ligand dominates the ΔH contribution to the free energy of binding. It has been suggested that the BC loop in the Grb7 protein family SH2 domains may be defective in coordinating phosphate groups due to the presence of a non-glycl residue at position BC5 (Stein et al. 2003). This defective phosphate coordination may be reflected in the smaller enthalpic contribution to binding observed for the Grb7-SH2 domain.
Whatever the reasons for the smaller relative enthalpic contribution to the free energy change of binding, it appears the loss is partially recouped through favorable entropic interactions. The entropic term contributes 39% of the total free energy of binding for the Grb7- SH2/erbB2 peptide complex (−TΔS=−3.04 kcal/mol). For ITC studies of SH2 domains available in the literature (Table 3), −TΔS values range from +1.3 kcal/mol for the p85α subunit C-terminal SH2 domain of PI3- kinase (O’Brien et al. 2000) to −1.7 kcal/mol for the SH2 domain of p56lck (Lemmon and Ladbury 1994). The unfavorable negative entropy contributions have been interpreted as an energetic penalty incurred through restriction of side chain vibrational modes in the bound state relative to the unbound state (Ladbury et al. 1996). This entropy penalty may be particularly large for SH2 domains that bind their ligands in an extended conformation and for which +3 ligand binding is tight. Extrapolation of this view would imply that greater favorable entropic contributions to the binding free energy would be required in general for SH2 domains that bind their ligands in a turn conformation, as compensation for the loss of SH2 domain/ligand interactions within the +3 binding pocket. This is certainly borne out for the observed thermodynamic parameters of the Grb7 and Grb2 SH2 domains. It should be noted that this interpretation of Grb7 binding thermodynamics may be affected by the postulated monomer/dimer Grb7-SH2 domain transition. It is currently not obvious how this possible additional effect on the thermodynamics can be readily separated from the global thermodynamic parameters measured.
Table 3.
Thermodynamic binding parameters for representative SH2 domains
| SH2 domain | Peptide | ΔHa | −TΔSa | KD (μM) | Techniqueb | Reference |
| Grb2 | Shc | −7.9 | −1.2 | 0.20 | ITC | McNemar et al. 1997 |
| Grb2 | erbB1 | N/A | N/A | 0.004 | SPR | Suenaga et al. 2003 |
| erbB4 | N/A | N/A | 0.19 | SPR | ||
| Grb2 | Shc | N/A | N/A | 0.38 | SPR | deMol et al. 2004 |
| Src | pYEEI | −8.4 | −0.12 | 0.55 | ITC | Ladbury et al. 1995 |
| Src | pYEEI | N/A | N/A | 0.67 | SPR | Ladbury et al. 1995 |
| Src | pYEEI | −7.3 | 0.60 | ITC | Bradshaw and Waksman 1998 | |
| Src | pYEEI | −9.3 | −0.33 | 0.10 | ITC | Chung et al. 1998 |
| Lck | EGFR (erbB1) | −1.1 to −1.5 | 7.1 to 3.3 | ITC | Lemmon and Ladbury 1994 | |
| Lck | Lck(pY505) | −8.4 | 1.1 | 4.2 | ITC | Ladbury et al. 1995 |
| p85α N-term | PDGFR | −9.4 | −0.73 | 0.47 | ITC | Ladbury et al. 1995 |
| p85α N-term | PDGFR | N/A | N/A | 0.24 | SPR | Ladbury et al. 1995 |
| p85α C-term | PDGFR | −11.2 | 1.3 | 0.048 | ITC | O’Brien et al. 2000 |
| p85α C-term | PDGFR | N/A | N/A | 0.064 | SPR | Panayotou et al. 1993 |
a Kcal/mol.
b ITC, isothermal titration calorimetry; SPR, surface plasmon resonance.
It is intriguing to consider the relatively large favorable TΔS contribution to Grb7-SH2 binding in light of the backbone nuclear relaxation studies presented here. Positive changes in entropy with binding imply an increase in disorder. Although intuitively this is offensive to the thought that protein complexes are more stable and thus less disordered than the free proteins, this result correlates positively with the observed increase in the total number of mobile sites in the Grb7-SH2/pY1139 complex relative to the uncomplexed Grb7-SH2 domain.
The Grb2-SH2 domain binding an Shc-derived peptide is also entropically favored. However, the entropic contribution to the free energy of binding is only 13%. There is some evidence that the Grb2-SH2 domain may undergo a conformational change upon ligand binding, based on comparisons of the structures of the free and bound states (Nioche et al. 2002) and kinetic analysis of SPR data (de Mol et al. 2004). Unfortunately the backbone nuclear relaxation characteristics of the Grb2-SH2 domain have not yet been reported, making a direct comparison with the Grb7-SH2 domain nuclear relaxation behavior impossible.
The findings presented here and the literature of biophysical characterization of protein-protein interactions as a whole point out the continuing complexity of understanding biomolecular energetics, and the wisdom of approaching each interaction as unique.
Acknowledgments
This work was supported by U.S. Department of Energy Grant DE-FG02-00er45828 and a grant from the Lake Champlain Cancer Research Organization.
Abbreviations
BPS, between Plekstrin and Src
EGFR, epidermal growth factor receptor
erbB2 (aka HER2, EGFR2), erythroblastosis B
FGFR, fibroblast growth factor receptor
Grb, growth factor receptor bound
HSQC, heteronuclear single-quantum coherence
J(ω), spectral density function
IR, insulin receptor
ITC, isothermal titration calorimetry
NMR, nuclear magnetic resonance
NOE, nuclear Overhauser effect
NOESY, NOE spectroscopy
PDGFR, platelet derived growth factor receptor
T1, longitudinal relaxation time (aka spin-lattice)
T2, transverse relaxation time (aka spin–spin)
τm, molecular correlation time
τe, effective correlation time
RMSD, root-mean-square deviation
RTK, receptor tyrosine kinase
S2, order parameter
SH2, Src homology 2.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041102305.
References
- Barbato, G., Ikura, M., Kay, L.E., Pastor, R.W., and Bax, A. 1992. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31 5269–5278. [DOI] [PubMed] [Google Scholar]
- Bradshaw, J.M. and Waksman, G. 1998. Calorimetric investigation of proton linkage by monitoring both the enthalpy and association constant of binding: Application to the interaction of the Src SH2 domain with a high-affinity tyrosyl phosphopeptide. Biochemistry 37 15400–15407. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Calorimetric examination of high-affinity Src SH2 domain-tyrosyl phosphopeptide binding: Dissection of the phosphopeptide sequence specificity and coupling energetics. Biochemistry 38 5147–5154. [DOI] [PubMed] [Google Scholar]
- Brescia, P.J., Ivancic, M., and Lyons, B.A. 2002. Assignment of backbone 1H, 13C, and 15N resonances of human Grb7-SH2 domain in complex with a phosphorylated peptide ligand. J. Biomol. NMR 23 77–78. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Techniques for the study of biological structure and function. In Biophysical chemistry. Freeman, San Francisco.
- Chen, D., Xu, L.G., Chen, L., Li, L., Zhai, Z., and Shu, H.B. 2003. NIK is a component of the EGF/heregulin receptor signaling complexes. Oncogene 22 4348–4355. [DOI] [PubMed] [Google Scholar]
- Chung, E., Henriques, D., Renzoni, D., Zvelebil, M., Bradshaw, J.M., Waksman, G., Robinson, C.V., and Ladbury, J.E. 1998. Mass spectrometric and thermodynamic studies reveal the role of water molecules in complexes formed between SH2 domains and tyrosyl phosphopeptides. Structure 6 1141–1151. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Driscoll, P.C., Wingfield, P.T., and Gronenborn, A.M. 1990. Analysis of the backbone dynamics of interleukin-1 β using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry 29 7387–7401. [DOI] [PubMed] [Google Scholar]
- Cohn, E.J. and Edsall, J.T. 1943. Proteins, amino acids and peptides as ions and dipolar ions. Reinhold, New York.
- Connelly, P.R. 1997. The cost of releasing site-specific, bound water molecules from proteins: Toward a quantitative guide for structure-based drug design. In Structure-based drug design: Thermodynamics, modeling, and strategy (eds. J.E. Ladbury and P.R. Connelly), pp. 143–157. Springer-Verlag, Berlin.
- Coussens, L.M. and Werb, Z. 2002. Inflammation and cancer. Nature 420 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, R.J., Sanderson, G.M., Janes, P.W., and Sutherland, R.L. 1996. Cloning and characterization of GRB14, a novel member of the GRB7 gene family. J. Biol. Chem. 271 12502–12510. [DOI] [PubMed] [Google Scholar]
- de Mol, N.J., Catalina, M.I., Fischer, M.J., Broutin, I., Maier, C.S., and Heck, A.J. 2004. Changes in structural dynamics of the Grb2 adaptor protein upon binding of phosphotyrosine ligand to its SH2 domain. Biochim. Biophys. Acta 1700 53–64. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. 1990. The meaning of hydrophobicity. Science 250 297–298. [DOI] [PubMed] [Google Scholar]
- Eck, M.J., Shoelson, S.E., and Harrison, S.C. 1993. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362 87–91. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15NNMR relaxation. Biochemistry 33 5984–6003. [DOI] [PubMed] [Google Scholar]
- Griswold, I.J. and Dahlquist, F.W. 2002. The dynamic behavior of CheW from Thermotoga maritima in solution, as determined by nuclear magnetic resonance: Implications for potential protein–protein interaction sites. Biophys. Chem. 101–102 359–373. [DOI] [PubMed] [Google Scholar]
- Han, D.C. and Guan, J.L. 1999. Association of focal adhesion kinase with Grb7 and its role in cell migration. J. Biol. Chem. 274 24425–24430. [DOI] [PubMed] [Google Scholar]
- Han, D.C., Shen, T.L., and Guan, J.L. 2000. Role of Grb7 targeting to focal contacts and its phosphorylation by focal adhesion kinase in regulation of cell migration. J. Biol. Chem. 275 28911–28917. [DOI] [PubMed] [Google Scholar]
- He, W., Rose, D.W., Olefsky, J.M., and Gustafson, T.A. 1998. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J. Biol. Chem. 273 6860–6867. [DOI] [PubMed] [Google Scholar]
- Ivancic, M., Daly, R.J., and Lyons, B.A. 2003. Solution structure of the human Grb7-SH2 domain/erbB2 peptide complex and structural basis for Grb7 binding to ErbB2. J. Biomol. NMR 27 205–219. [DOI] [PubMed] [Google Scholar]
- Jahn, T., Seipel, P., Urschel, S., Peschel, C., and Duyster, J. 2002. Role for the adaptor protein Grb10 in the activation of Akt. Mol. Cell Biol. 22 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes, P.W., Lackmann, M., Church, W.B., Sanderson, G.M., Sutherland, R.L., and Daly, R.J. 1997. Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J. Biol. Chem. 272 8490–8497. [DOI] [PubMed] [Google Scholar]
- Jin, D., Andrec, M., Montelione, G.T., and Levy, R.M. 1998. Propagation of experimental uncertainties using the Lipari-Szabo model-free analysis of protein dynamics. J. Biomol. NMR 12 471–492. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Torchia, D.A., and Bax, A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28 8972–8979. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Nicholson, L.K., Delaglio, F., Bax, A., and Torchia, D.A. 1992. Pulse sequences for removal of the effects of cross-correlation between dipolar and chemical-shift anisotropy relaxation mechanism on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 97 359–375. [Google Scholar]
- Ladbury, J.E., Lemmon, M.A., Zhou, M., Green, J., Botfield, M.C., and Schlessinger, J. 1995. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: A reappraisal. Proc. Natl. Acad. Sci. 92 3199–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury, J.E., Hensmann, M., Panayotou, G., and Campbell, I.D. 1996. Alternative modes of tyrosyl phosphopeptide binding to a Src family SH2 domain: Implications for regulation of tyrosine kinase activity. Biochemistry 35 11062–11069. [DOI] [PubMed] [Google Scholar]
- Laue, T.M., Shaf, B.S., Ridgeway, T.M., and Pelletier, S.L. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical ultracentrifugation in biochemistry and polymer science, pp. 90–125. Royal Society of Chemistry, Cambridge, UK.
- Lemmon, M.A. and Ladbury, J.E. 1994. Thermodynamic studies of tyrosyl-phosphopeptide binding to the SH2 domain of p56lck. Biochemistry 33 5070–5076. [DOI] [PubMed] [Google Scholar]
- Li, Y.C. and Montelione, G.T. 1995. Human type-α transforming growth factor undergoes slow conformational exchange between multiple backbone conformations as characterized by nitrogen-15 relaxation measurements. Biochemistry 34 2408–2423. [DOI] [PubMed] [Google Scholar]
- Lipari, G. and Szabo, A. 1982a. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104 4546–4559. [Google Scholar]
- ———. 1982b. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104 4559–4570. [Google Scholar]
- Loh, A.P., Guo, W., Nicholson, L.K., and Oswald, R.E. 1999. Backbone dynamics of inactive, active, and effector-bound Cdc42Hs from measurements of (15)N relaxation parameters at multiple field strengths. Biochemistry 38 12547–12557. [DOI] [PubMed] [Google Scholar]
- Loh, A.P., Pawley, N., Nicholson, L.K., and Oswald, R.E. 2001. An increase in side chain entropy facilitates effector binding: NMR characterization of the side chain methyl group dynamics in Cdc42Hs. Biochemistry 40 4590–4600. [DOI] [PubMed] [Google Scholar]
- Mandiyan, V., O’Brien, R., Zhou, M., Margolis, B., Lemmon, M.A., Sturtevant, J.M., and Schlessinger, J. 1996. Thermodynamic studies of SHC phosphotyrosine interaction domain recognition of the NPXpY motif. J. Biol. Chem. 271 4770–4775. [DOI] [PubMed] [Google Scholar]
- Margolis, B. 1994. The GRB family of SH2 domain proteins. Prog. Biophys. Mol. Biol. 62 223–244. [DOI] [PubMed] [Google Scholar]
- Margolis, B., Silvennoinen, O., Comoglio, F., Roonprapunt, C., Skolnik, E., Ullrich, A., and Schlessinger, J. 1992. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc. Natl. Acad. Sci. 89 8894–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNemar, C., Snow, M.E., Windsor, W.T., Prongay, A., Mui, P., Zhang, R., Durkin, J., Le, H.V., and Weber, P.C. 1997. Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2- SH2. Biochemistry 36 10006–10014. [DOI] [PubMed] [Google Scholar]
- Mercier, P., Spyracopoulos, L., and Sykes, B.D. 2001. Structure, dynamics, and thermodynamics of the structural domain of troponin C in complex with the regulatory peptide 1–40 of troponin I. Biochemistry 40 10063–10077. [DOI] [PubMed] [Google Scholar]
- Mittag, T., Schaffhausen, B., and Gunther, U.L. 2003. Direct observation of protein-ligand interaction kinetics. Biochemistry 42 11128–11136. [DOI] [PubMed] [Google Scholar]
- Morrione, A., Plant, P., Valentinis, B., Staub, O., Ku, S., Rotin, D., and Baserga, R. 1999. mGrb10 interacts with Nedd4. J. Biol. Chem. 274 24094–24099. [DOI] [PubMed] [Google Scholar]
- Nantel, A., Mohammad-Ali, K., Sherk, J., Posner, B.I., and Thomas, D.Y. 1998. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J. Biol. Chem. 273 10475–10484. [DOI] [PubMed] [Google Scholar]
- Nicholson, L.K., Yamazaki, T., Torchia, D.A., Grzesiek, S., Bax, A., Stahl, S.J., Kaufman, J.D., Wingfield, P.T., Lam, P.Y., Jadhav, P.K., et al. 1995. Flexibility and function in HIV-1 protease. Nat. Struct. Biol. 2 274–280. [DOI] [PubMed] [Google Scholar]
- Nioche, P., Liu, W.Q., Broutin, I., Charbonnier, F., Latreille, M.T., Vidal, M., Roques, B., Garbay, C., and Ducruix, A. 2002. Crystal structures of the SH2 domain of Grb2: Highlight on the binding of a new high-affinity inhibitor. J. Mol. Biol. 315 1167–1177. [DOI] [PubMed] [Google Scholar]
- O’Brien, R., Rugman, P., Renzoni, D., Layton, M., Handa, R., Hilyard, K., Waterfield, M.D., Driscoll, P.C., and Ladbury, J.E. 2000. Alternative modes of binding of proteins with tandem SH2 domains. Protein Sci. 9 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, K., Tsuchiya, S., Terasawa, H., Yuzawa, S., Hatanaka, H., Mandiyan, V., Schlessinger, J., and Inagaki, F. 1999. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J. Mol. Biol. 289 439–445. [DOI] [PubMed] [Google Scholar]
- Orekhov, V.Y., Nolde, D.E., Golovanov, A.P., Korzhnev, D.M., and Arseniev, A.S. 1995. Processing of heteronuclear NMR relaxation with the new software DASHA. Appl. Magn. Reson. 9 581–588. [Google Scholar]
- Panayotou, G., Gish, G., End, P., Truong, O., Gout, I., Dhand, R., Fry, M.J., Hiles, I., Pawson, T., and Waterfield, M.D. 1993. Interactions between SH2 domains and tyrosine-phosphorylated platelet-derived growth factor β-receptor sequences: Analysis of kinetic parameters by a novel biosensor-based approach. Mol. Cell Biol. 13 3567–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A., Duan, H., Di Fiore, P.P., and Dixit, V.M. 1995. The Ret receptor protein tyrosine kinase associates with the SH2-containing adapter protein Grb10. J. Biol. Chem. 270 21461–21463. [DOI] [PubMed] [Google Scholar]
- Pascal, S.M., Singer, A.U., Gish, G., Yamazaki, T., Shoelson, S.E., Pawson, T., Kay, L.E., and Forman-Kay, J.D. 1994. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-γ 1 complexed with a high affinity binding peptide. Cell 77 461–472. [DOI] [PubMed] [Google Scholar]
- Pawley, N.H., Koide, S., and Nicholson, L.K. 2002. Backbone dynamics and thermodynamics of Borrelia outer surface protein A. J. Mol. Biol. 324 991–1002. [DOI] [PubMed] [Google Scholar]
- Philo, J.S. 2000. A method for directly fitting the timer derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279 151–163. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L., Gill, S.J., and Murphy, K.P. 1990. The meaning of hydrophobicity: Response to Dill. Science 250 297–298. [DOI] [PubMed] [Google Scholar]
- Rahuel, J., Garcia-Echeverria, C., Furet, P., Strauss, A., Caravatti, G., Fretz, H., Schoepfer, J., and Gay, B. 1998. Structural basis for the high affinity of amino-aromatic SH2 phosphopeptide ligands. J. Mol. Biol. 279 1013–1022. [DOI] [PubMed] [Google Scholar]
- Schneider, D.M., Dellwo, M.J., and Wand, A.J. 1992. Fast internal mainchain dynamics of human ubiquitin. Biochemistry 31 3645–3652. [DOI] [PubMed] [Google Scholar]
- Shen, T.L. and Guan, J.L. 2001. Differential regulation of cell migration and cell cycle progression by FAK complexes with Src, PI3K, Grb7 and Grb2 in focal contacts. FEBS Lett. 499 176–181. [DOI] [PubMed] [Google Scholar]
- Stein, D., Wu, J., Fuqua, S.A., Roonprapunt, C., Yajnik, V., D’Eustachio, P., Moskow, J.J., Buchberg, A.M., Osborne, C.K., and Margolis, B. 1994. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 13 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, E.G., Ghirlando, R., and Hubbard, S.R. 2003. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J. Biol. Chem. 278 13257–13264. [DOI] [PubMed] [Google Scholar]
- Stone, M.J., Fairbrother, W.J., Palmer 3rd, A.G., Reizer, J., Saier Jr., M.H., and Wright, P.E. 1992. Backbone dynamics of the Bacillus subtilis glucose permease IIA domain determined from 15NNMR relaxation measurements. Biochemistry 31 4394–4406. [DOI] [PubMed] [Google Scholar]
- Suenaga, A., Hatakeyama, M., Ichikawa, M., Yu, X., Futatsugi, N., Narumi, T., Fukui, K., Terada, T., Taiji, M., Shirouzu, M., et al. 2003. Molecular dynamics, free energy, and SPR analyses of the interactions between the SH2 domain of Grb2 and ErbB phosphotyrosyl peptides. Biochemistry 42 5195–5200. [DOI] [PubMed] [Google Scholar]
- Venable, R.M. and Pastor, R.W. 1988. Frictional models for stochastic simulations of proteins. Biopolymers 27 1001–1014. [DOI] [PubMed] [Google Scholar]
- Waksman, G., Shoelson, S.E., Pant, N., Cowburn, D., and Kuriyan, J. 1993. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell 72 779–790. [DOI] [PubMed] [Google Scholar]
- Wang, J., Dai, H., Yousaf, N., Moussaif, M., Deng, Y., Boufelliga, A., Swamy, O.R., Leone, M.E., and Riedel, H. 1999. Grb10, a positive, stimulatory signaling adapter in platelet-derived growth factor BB-, insulin-like growth factor I-, and insulin-mediated mitogenesis. Mol. Cell Biol. 19 6217–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman, T., Williston, S., Brandts, J.F., and Lin, L.N. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179 131–137. [DOI] [PubMed] [Google Scholar]
- Wojcik, J., Girault, J.A., Labesse, G., Chomilier, J., Mornon, J.P., and Callebaut, I. 1999. Sequence analysis identifies a ras-associating (RA)- like domain in the N-termini of band 4.1/JEF domains and in the Grb7/10/14 adapter family. Biochem. Biophys. Res. Commun. 259 113–120. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Akilesh, S., and Wilcox, D.E. 2000. Isothermal titration calorimetry measurements of Ni(II) and Cu(II) binding to His, GlyGlyHis, HisGlyHis, and bovine serum albumin: A critical evaluation. Inorg. Chem. 39 3057–3064. [DOI] [PubMed] [Google Scholar]