Abstract
Fibrillar collagens are the principal structural molecules of connective tissues. The assembly of collagen fibrils is regulated by quantitatively minor fibrillar collagens, types V and XI. A unique amino-terminal propeptide domain of these collagens has been attributed this regulatory role. The structure of the amino terminal propeptide has yet to be determined. Low sequence similarity necessitated a secondary structure-based method to carry out homology modeling based upon the determined structure of LNS family members, named for a common structure in the laminin LG5 domain, the neurexin 1B domain and the sex hormone binding globulin. Distribution of amino acids within the model suggested glycosaminoglycan interaction and calcium binding. These activities were tested experimentally. Sequence analyses of existing genes for collagens indicate that 16 known collagen α chains may contain an LNS domain. A similar approach may prove useful for structure/function studies of similar domains in other collagens with similar domains. This will provide mechanistic details of the organization and assembly of the extracellular matrix and the underlying basis of structural integrity in connective tissues. The absolute requirement for collagen XI in skeletal growth is indicated by collagen XI deficiencies such as chondrodystrophies found in the cho/cho mouse and in humans with Stickler syndrome.
Keywords: type XI collagen, amino propeptide, LNS domain, homology modeling, β-sandwich, calcium, heparan sulfate
Collagens are a group of structurally similar proteins of the extracellular matrix. The primary structure of collagen is characterized by extensive Gly-X-Y repeats. Each collagen consists of three interacting α chains that form a triple helix (van der Rest and Garrone 1991). Tissue-specific extracellular matrix organization requires regulated assembly and growth. The absolute nature of the requirement for collagen XI in skeletal growth is indicated by the result of collagen XI deficiencies such as chondrodystrophies found in the cho/cho mouse and in humans with Stickler syndrome (Li et al. 1995).
Collagens are modular proteins, comprising distinct domains other than the triple helix, with characteristic features. To date, there are more than twenty-seven different collagens. Of these, sixteen share a noncollagenous domain originally identified as the Thsp1 or TSPN domain of thrombospondin (Bork 1992; Mayne and Brewton 1993; Bork et al. 1996). Similarity exists between this domain and a domain present in the proteins, laminin A, neurexin, and sex hormone binding globulin, abbreviated as the LNS domain. Sequence homology has been reported between TSPN and the noncollagenous domain of collagens discussed here. Three-dimensional structure information for TSPN does not exist in the Protein Data Bank (PDB). The LNS module, however, is represented by the laminin LG modules of the α2 chain (1dyk and 1quo) (Hohenester et al. 1999; Timpl et al. 2000; Tisi et al. 2000), neurexin-1 β (1c4r) (Rudenko et al. 1999), sex hormone-binding globulin (1d2s) (Grishkovskaya et al. 2000), and growth-arrest specific protein (1h3O) (Berman et al. 2000; Sasaki et al. 2002). Although sequence identity between the amino propeptide of collagen XI α1 chain [Npp α1(XI)] and LNS modules is relatively low (ranging from 21% to 25%), predicted secondary structure is high in similarity.
In the present work, we have derived a model for the structure of the Npp α1(XI) collagen using predicted secondary structural elements and molecular modeling techniques. Based upon predicted secondary structure, a tertiary structural template was identified using PSI-BLAST and FUGUE (Schaffer et al. 2001; Shi et al. 2001). Homology modeling was carried out using MODELLER (Sali and Blundell 1993) and the three-dimensional structure of laminin α2 LG domain (Tisi et al. 2000). Forty amino acids at the amino terminus of Npp α1(XI) collagen were modeled using an ab initio technique, and the predictions were merged to create one structural model. The model presented here includes 223 amino acids, beginning at the signal peptide processing site, includes the bone morphogenetic protein-1 (BMP-1) processing site (Medeck et al. 2003), and continues to the amino acid immediately preceding the variable region as illustrated in Figure 1 ▶. Sequence analysis and distribution of specific amino acid residues within the resulting structural model allowed for the prediction of divalent cation binding activity. In addition, analysis of the surface distribution of amino acids led to the prediction of a glycosaminoglycan binding site within this domain. Predicted binding functions were evaluated empirically, using fluorescence spectroscopy of tryptophan-128, and surface plasmon resonance to assess molecular interactions. Finally, sequence comparison among 16 collagen α chains that include this domain is presented. The LNS domains of collagens may be a key factor in the establishment of molecular interactions that play a role in cell–cell interactions, interactions between the cell and its surrounding extracellular matrix, as well as interactions between the molecular constituents of the extracellular matrix.
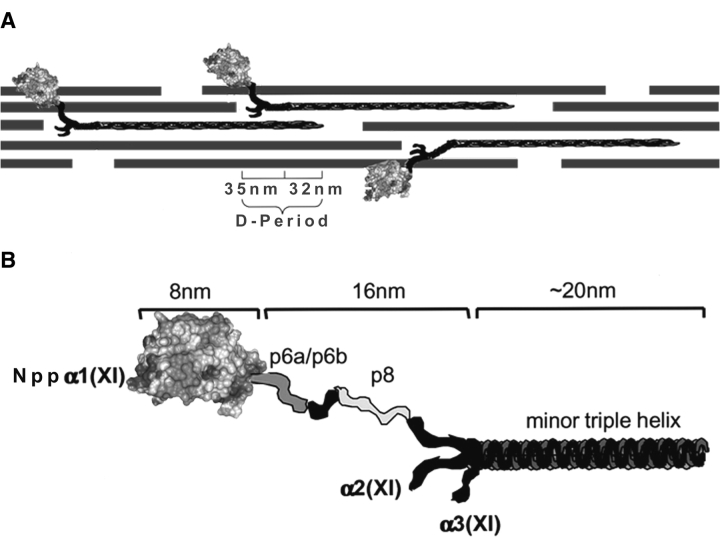
Figure 1.
Schematic of collagen fibril and Npp α1(XI) collagen. (A) Collagen fibril shown comprised of collagen types II, IX, and XI. The globular domain Nppα1(XI) collagen is shown to extend from the surface of the collagen fibril. The dimensions of the gap-overlap region of the D-period are indicated. (B) Schematic representation of the Npp α1(XI) and adjacent variable region and minor triple helix. The dimensions as determined (Gregory et al. 2000) by transmission electron microscopy are indicated. The position of the Npp α1(XI) domain is indicated as distal with respect to the adjacent p6a/p6b/p8 variable region and minor triple helix. The α2(XI) and α3(XI) chains are also indicated in the diagram.
Results
The Npp α1(XI) collagen domain represents a structure common to at least 16 collagen α chains. Although eventually removed by proteolysis in some cases, it can be relatively long-lived and found retained on the surface of collagen fibrils in embryonic tissues under steady state conditions (Thom and Morris 1991). Previous data suggest that this domain plays a major role in the mechanism of regulated fibrillar growth (Thom and Morris 1991; Keene et al. 1995). For the purposes of modeling the structure, the 223-residue amino propeptide, defined as that portion of the collagen XI α1 chain extending from the signal peptide cleavage site to the amino acid immediately preceding the variable region (p6a/p6b/p8 shown in Fig. 1 ▶), was divided into two regions; the amino-terminal 40 amino acids and the remaining 183 amino acid residues.
Template selection
Amino acid sequence from available species was obtained through GenBank (Berman et al. 2000). The amino acid sequence for Npp α1(XI) collagen is highly conserved among species. For example, 99% sequence identity exists between rat and mouse, 85% sequence identity exists between rat and human, 74% sequence identity exists between rat and chicken, and 70% identity is seen between rat and both bovine and red sea bream (data not shown).
The Npp α1(XI) collagen 223-amino-acid sequence of rat was used to query the PDB using both Gapped BLAST and FUGUE (Schaffer et al. 2001; Shi et al. 2001), and the results of this search are shown in Table 1. FUGUE analysis revealed members of the LNS family including laminin, neurexin, and sex hormone-binding globulin. Structurally similar growth-arrest-specific protein was also identified as a potential template. Gapped BLAST templates that overlapped with FUGUE results included laminin and sex hormone-binding globulin. The laminin α2 LG5 (LG5) domain contained the best alignment containing 184 residues, with 25% identity and 45% similarity. Consequently, the LG5 domain (residues 2934–3117 from the crystal structure of PBD:1dyk) (Tisi et al. 2000) was used as the template for modeling the Npp α1(XI) collagen.
Table 1.
Potential templates from the Protein Data Bank
| A. Gapped BLAST using BLOSUM62 Matrix | |||||||
| Accession no. | Description | Sequence length | Alignment length | E-value | Bit-score | Sequence identity (%) | Sequence similarity (%) |
| 1d2s_a | Sex hormone-binding globulin | 170 | 82 | 0.941166 | 26.7659 | 21 | 46 |
| 1qi7_a | N-glycosidase | 253 | 27 | 1.17302 | 26.4153 | 44 | 63 |
| 1dyk_a | Laminin α2 chain | 374 | 50 | 1.38631 | 26.0647 | 28 | 52 |
| 1quo_a | Laminin α2 chain | 181 | 69 | 3.14801 | 25.013 | 25 | 45 |
| 1ikp_a | Exotoxin A | 599 | 20 | 4.04452 | 24.6624 | 50 | 50 |
| 1aer_b | Exotoxin A | 195 | 20 | 5.07931 | 24.3119 | 50 | 50 |
| 1aer_a | Exotoxin A | 205 | 20 | 5.35662 | 24.3119 | 50 | 50 |
| 1wht_a | Serine carboxypeptidase II | 256 | 84 | 5.52182 | 24.3119 | 24 | 49 |
| 9gaf_c | Glycosylasparaginase | 292 | 62 | 6.88212 | 23.9613 | 24 | 45 |
| B. FUGUE Z-Score ≥6.0 (CERTAIN 99% confidence) | |||
| Accession no. | Description | Profile length | Z-score |
| (A) Gapped BLAST and (B) FUGUE were used to identify potential templates from PDB files available. The 223-amino-acid query sequence was used for Gapped BLAST using a BLOSUM62 Matrix. Potential templates are listed. Useful templates contain sequence that spans the 223-amino-acid query sequence and demonstrate maximum sequence identity. The FUGUE algorithm was used to detect templates not identified via Gapped BLAST. Z-scores ≥6.0 are marked as highly probable or “certain” templates. Adequate profile length while maximizing Z-score provides the criteria for selecting a template. Combining the two methods revealed the laminin α2 LG5 chain from 1dyk.pdb as the best template. | |||
| 1dyk | Laminin α2 chain | 213 | 17.08 |
| 1c4r | Neurexin-1 β | 180 | 15.09 |
| 1quo | Laminin α2 chain | 181 | 14.71 |
| 1d2s | Sex hormone-binding globulin | 170 | 10.02 |
| 1h30 | Growth-arrest-specific protein | 391 | 9.69 |
Sequence alignment
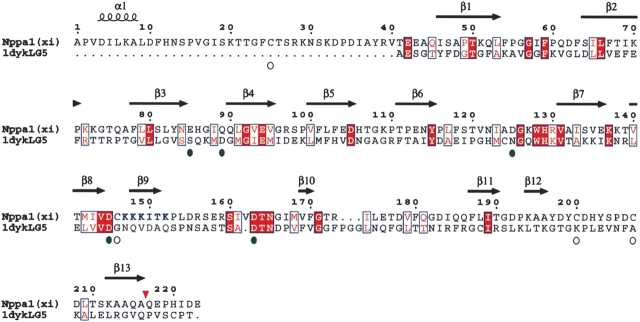
The amino acid sequence of Npp α1(XI) collagen was aligned with the sequence of laminin α2 LG domain, using the Align2D algorithm of MODELLER (Sali and Blundell 1993) and is shown in Figure 2 ▶. Manual inspection and modification of the alignment was performed after initial model generation and analysis of aligned secondary structural elements. Good sequence alignment was obtained for 183 amino acids of the Npp α1(XI) collagen sequence; however, the first 40 amino acids were not represented within the laminin template. Alignment of the predicted secondary structure with the known secondary structure of LG5 domain revealed higher sequence similarity in regions of β-strands than in regions predicted to be loops (11.4% identity and 30.7% similarity in β-strands versus 9.4% identity and 24.8% similarity in the loops).
Figure 2.
Alignment of 1dyk and Npp α1(XI) collagen. The laminin α2 LG chain was used as template for 183 amino acids of Npp α1(XI) collagen. Open circles indicate the position of four cysteine residues of Npp α1(XI) collagen; green circles indicate the position of residues of potential calcium binding sites. Conserved amino acid residues are shown as white letters on red background, while similar amino acid residues are shown as red letters on white background. Positions of predicted β-strands are indicated by arrows above the sequence. Position of predicted α-helix is indicated by a coil symbol above the sequence. Amino acids are numbered from 1 to 223, with 1 defined as the first amino acid after the signal peptide cleavage site. Bone morphogenetic protein-1 proteolytic processing site is indicated by an inverted red triangle. Putative heparin binding site is indicated by blue lettering at positions 147–152.
Topology of Npp α1(XI) collagen
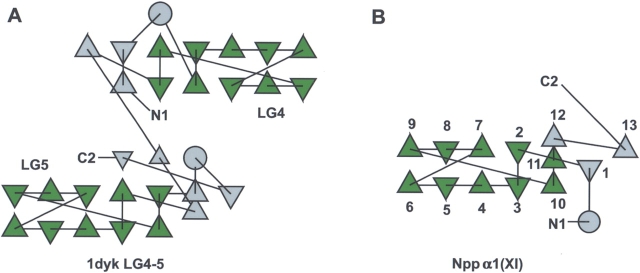
The structural topology was visualized for Npp α1(XI) collagen and compared to that of 1dyk (LG4-5) using tools for protein structural topology (TOPS) (Michalopoulos et al. 2004) in Figure 3 ▶. The secondary structural elements of 1dyk are preserved in Npp α1(XI) collagen (Fig. 3 ▶).
Figure 3.
Protein structural topology. (A) Topology of 1dyk LG4-5. The conserved motif is shown in green, while the outside regions are gray. β-Strands are shown as triangles and α-helices are shown as circles. (B) Topology of Npp α1(XI) collagen. The β-strands of 1dyk LG4-5 show identical topology with Npp α1(XI) collagen as well as other LNS family members (data not shown), and is characteristic of the LNS family of proteins. Although the directions of the triangles will vary depending on the total number of β-strands, the connectivity of the β-strands is constant.
Ab initio and homology model
Homology models were built based upon the alignment of sequence with 1dyk, LG5, shown in Figure 4A ▶ and by ab initio techniques. A structural model was generated for the region containing the first 40 amino acid residues using the HMMSTR/Rosetta Server (Bystroff et al. 2000; Bystroff and Shao 2002). This approach provided local structural motifs of the three-dimensional structure by fragment-insertion Monte Carlo. A model of the 40-amino-acid region was generated. The resulting model is shown in Figure 4B ▶, and predicts one α-helix, a loop region and a short β-strand.
Figure 4.
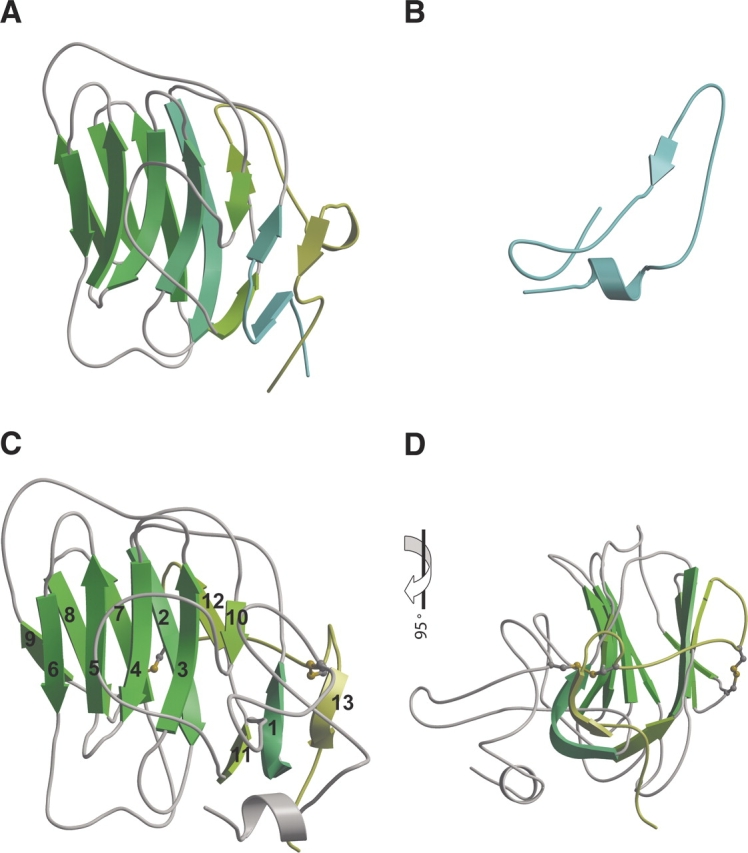
Ab initio and homology model. Three-dimensional structure of (A) 1dyk LG5, (B) the first 40 amino acids by ab initio modeling, (C) the model resulting from merger of ab initio and homology modeling, (D) model shown in 4-color after 90° rotation. Disulfide bonds are shown in panels C and D. β-Strands are numbered sequentially from the amino to carboxyl end of the protein.
Homology models were built based upon the alignment of sequence. Disulfide bonding within Npp α1(XI) collagen was specified as C25-C207 and C146-C200 as previously determined by liquid chromatography-tandem mass spectrometry for Npp α1(XI) collagen (Gregory et al. 2000). The models were subjected to evaluation and refinement using the program Verify3D (Eisenberg et al. 1997). Loops were refined using MODELLER (Sali et al. 1995) with default parameters and disulfide bonding specified. Between subsequent refinements, quality was ascertained to avoid buried polar atoms, buried hydrophilic groups, exposed hydrophobic atoms, holes, and close contacts. Maintenance of correct bond lengths, angles, chirality, and planarity of the peptide bond within the model were assessed as well. Residues falling within sterically forbidden regions of a Ramachandran plot were refined using loop optimization, rotamer libraries, and energy minimization. The complete model resulting fromthe combined ab initio/homology modeling approach is presented in Figure 4 ▶, C and D. The Npp α1(XI) collagen model is predicted to contain one α-helix and 13 β-strands, with intervening loop regions. The Npp α1(XI) collagen model superimposes onto LG5 with an RMSD of 1.31 Å using 135 residues (SIL to PKAA). The amino and carboxyl ends of the Npp α1(XI) collagen model are predicted to be in close proximity. An en face view with respect to the orientation of the concave surface of the β-sheets (Fig. 4C ▶) and the view upon a 90° rotation (Fig. 4D ▶) is shown. In our model, C146 and C200 form a disulfide bond. The amino and carboxyl termini are in close proximity, consistent with the previously determined C25–C207 disulfide bond.
Putative calcium binding activity
Two putative calcium binding sites were identified (Fig. 5 ▶). Analogy to the LG domain allowed the prediction that D163 and Q89 of Npp α1(XI) collagen may be involved in the partial coordination of a calcium ion (Wizemann et al. 2003). E85 of the Npp α1(XI) collagen model is in close proximity to the putative Ca2+ binding site and may contribute to the coordination of Ca2+. However, another site centered around D125 and D145 of Npp α1(XI) collagen may also coordinate calcium based upon sequence analysis carried out by Beckmann et al. (1998) (Fig. 5 ▶).
Figure 5.
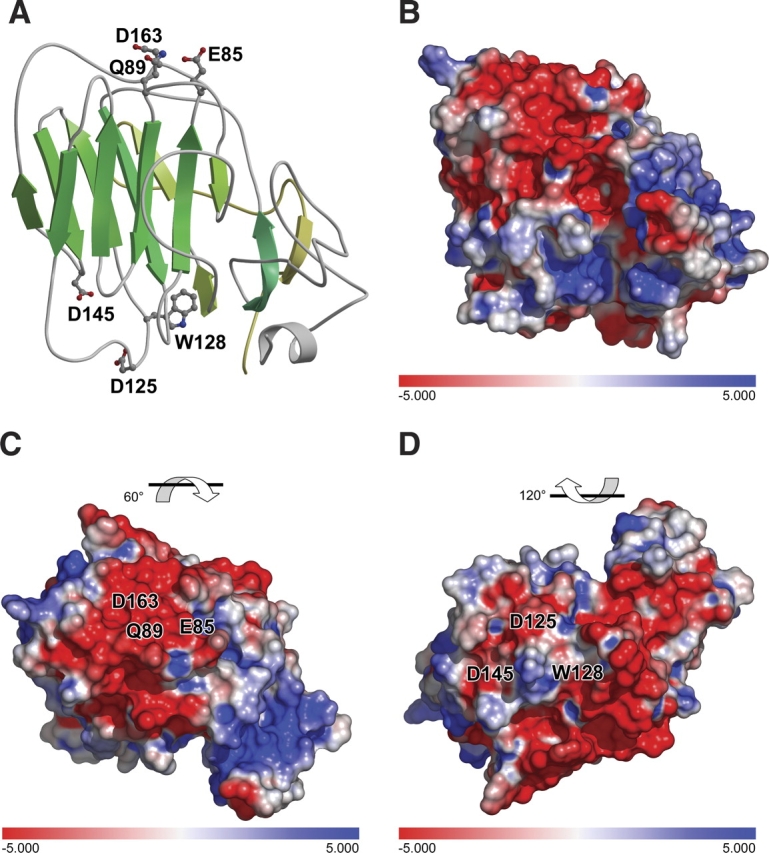
Putative calcium binding sites. Sequence homology with laminin α2 LG5 domain indicates the potential for similarity in calcium binding function, utilizing amino acid residues D163 and Q89. The amino acid side chain of E85 is in close proximity is also indicated. A second potential Ca2+ binding site is indicated near amino acid residues D125 and D145. (A) Ribbon diagram. (B) Electrostatic surface, identical orientation as that shown in panel A. (C) 60° rotation of model with respect to panel B to show Ca2+ binding site D163Q89. (D) 120° rotation of model with respect to panel B to show Ca2+ binding site D125D145.
Fluorescence spectroscopy
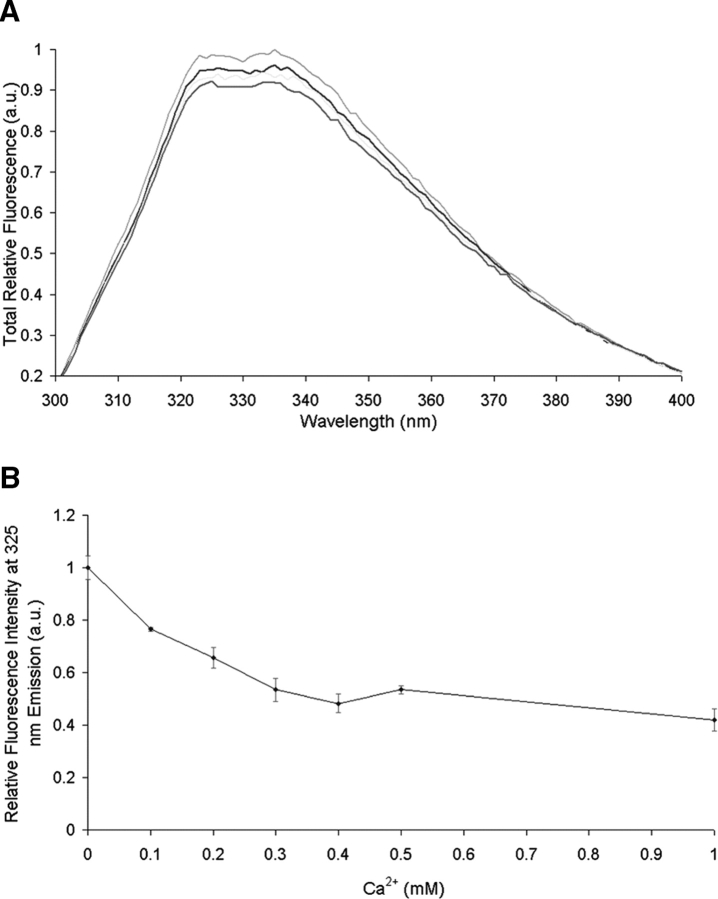
The effect of Ca2+ on the conformation of Npp α1(XI) collagen was studied by means of fluorescence spectroscopy. Using an excitation wavelength of 285 nm, the fluorescence emission of W128 was recorded from 300 to 400 nm as a function of Ca2+ concentration. The spectra revealed emission quenching as a result of the addition of Ca2+ (Fig. 6 ▶). Known fluorescence emission properties of isolated tryptophan residues suggest that changes in the environment of the tryptophan occur upon Ca2+ interaction, implying a conformational change in the protein structure. This change was both reversible and saturable. Similar results were observed for the divalent cations Mg2+ and Zn2+ (data not shown).
Figure 6.
Effect of divalent cation on the fluorescence emission spectrum of Npp α1(XI) collagen. Protein (40 μg/mL) in 5 mM MOPS buffer (pH 7.5), containing 150 mM NaCl was excited at 285 nm. (A) The emission spectra were recorded between 300 and 400 nm. (B) A decrease in fluorescence intensity was observed as a function of increasing Ca2+ concentration.
Putative heparin binding activity
Initial analysis of the surface distribution of amino acid side chains suggested a potential site for glycosaminoglycan interaction. The heparin binding site of the LG5 domain of laminin α2 was not conserved in the Npp α1(XI) collagen model, however, molecular modeling of an Npp-heparin interaction was performed using Auto-Dock 3.0 (Morris et al. 1998) and revealed an alternative putative binding site. For each ligand, one job of 100 docking runs was performed using a population of 200 individuals and an energy evaluation number of 2 × 106 employing the Lamarckian Genetic Algorithm (Morris et al. 1998). Two potential sites were identified, one with predicted higher affinity binding than the other. A further refined docking was centered at the putative higher affinity heparin-binding site on residues 147–152 with a grid spacing of 0.375 Å. The highly populated binding zone was studied with respect to residues involved in binding as well as final energy. The putative heparin-binding site comprised a positively charged area, and its location on the surface of the model is shown in Figure 7 ▶, comprised of 147-KKKITK-152. The putative heparinbinding site was consistent with motifs identified as heparin binding sites (XBBBXXB where B is a basic residue) (Cardin and Weintraub 1989). The amino acids of the putative heparin binding site form a tract of positively charged residues at physiological pH (Fig. 7 ▶). A similar arrangement of amino acid side chains forms the heparin-binding site in laminin α4 chain LG4 domain, which is a different site from that found for other laminin LG domains (Yamashita et al. 2004).
Figure 7.
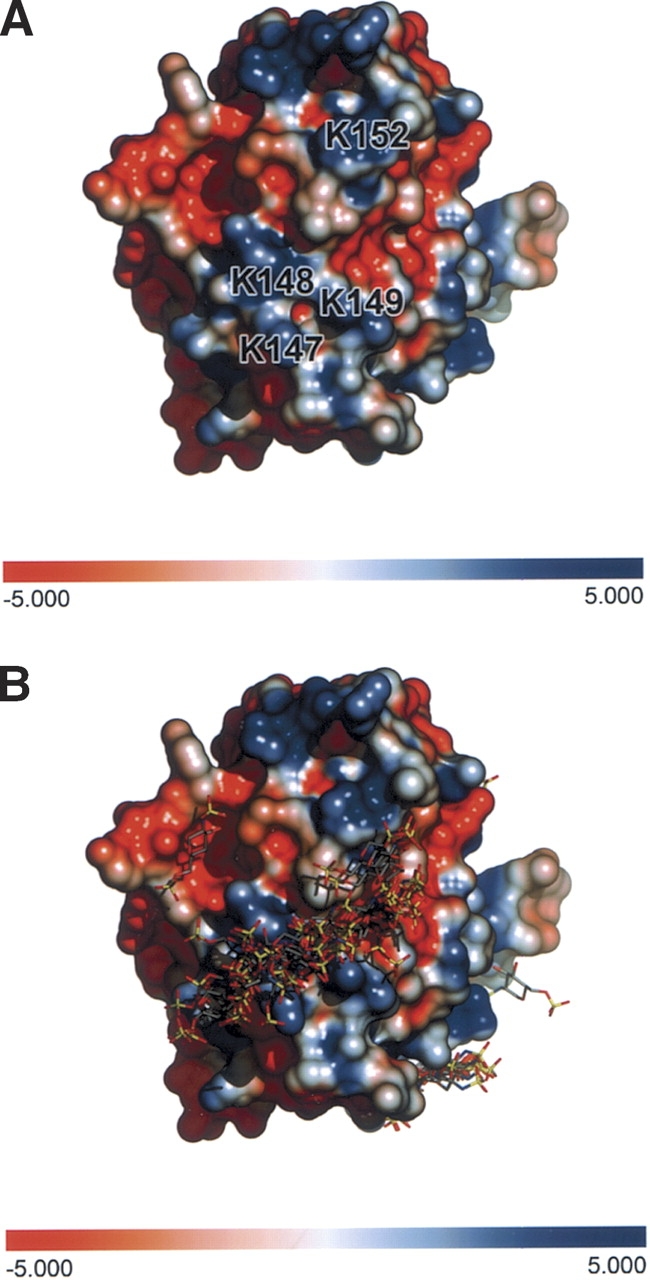
Putative heparin binding site. (A) Interactions between heparin and Npp α1(XI) collagen were predicted. For each heparin ligand, one job of 100 docking runs was performed using a population of 200 individuals and an energy evaluation number of 2 × 106 employing the Lamarckian Genetic Algorithm. A further refined docking was centered at the putative heparin binding site on residues 147–152 (KKKITK). (B) Amino acids 147–152 form a track of positively charged side chains on the surface of the protein.
Analysis of heparan sulfate binding to Npp α1(XI) collagen
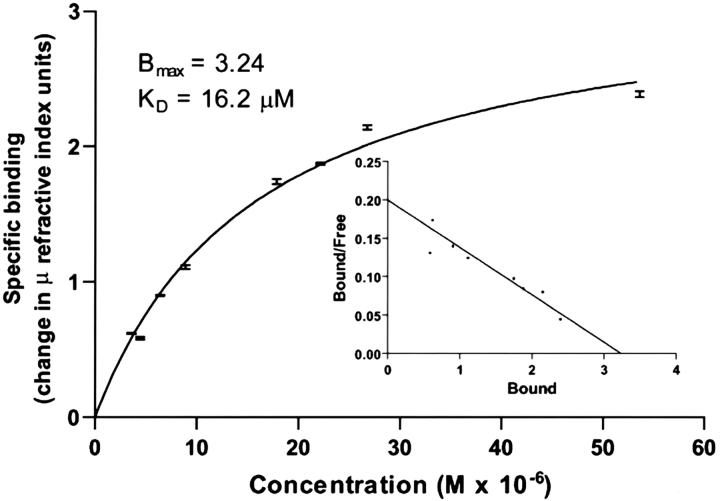
SPR measurements were performed in which Npp α1(XI) collagen was immobilized on the sensor chip and heparan sulfate was used as the analyte. Analysis of the data demonstrated that heparan sulfate bound to Npp α1(XI) collagen with an apparent Kd=16 μM (Fig. 8 ▶).
Figure 8.
Interaction between heparan sulfate and Npp α1(XI) collagen. Concentrations of heparan sulfate (0.625–0.75 mg/mL) were injected over immobilized Npp at a flow rate of 10 μL per min in phosphate buffered saline containing Tween-20 (0.05% v/v). An average of the response at equilibrium was determined for each concentration and the resulting equilibrium resonance units were plotted against concentration. The data were fit to a steady-state one-site affinity model to determine the equilibrium dissociation constant (Kd) of heparan sulfate for Npp α1(XI) collagen. Scatchard analysis was carried out and is presented in the inset.
Discussion
In this study, the three dimensional structure of the Npp α1(XI) collagen has been modeled as a member of the family of proteins containing an “LNS” domain, named for a common structure in the laminin, neurexin, and sex hormone binding globulin. On the basis of sequence characteristics, it is estimated that there are more than 400 LNS domains in proteins (Bateman et al. 2004). Sequence analyses of existing genes for collagens indicate that 16 known collagen α chains contain an LNS module, 10 of which are located at the extreme amino terminus of the protein (Fig. 9 ▶). Thus, the structural model of the Npp α1(XI) collagen may serve as a representative for homologous domains in other collagens and provide mechanistic details of the process of fibrillar assembly and extracellular matrix organization, as fibril-associated collagens with interrupted helices (FACIT) (Shaw and Olsen 2001) and the quantitatively minor fibrillar collagens types V and XI (Fichard et al. 1995) are predicted to contain an LNS domain. FACIT collagens are located on the surface of fibrillar collagen fibrils, and have been shown to mediate interactions between the collagen fibrils and other matrix constituents (Pihlajamaa et al. 2004). Likewise, the quantitatively minor collagens types V and XI are constituents of types I and II collagen fibrils, which serve a regulatory role in collagen fibril formation.
Figure 9.
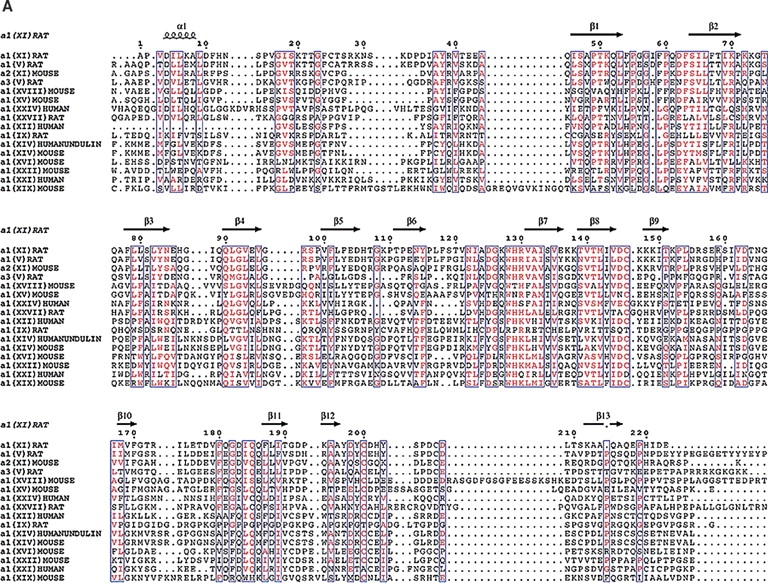
A comparison of collagens containing similar LNS domains. (A) Identical residues are red while similar residues are purple. The β-strands fall in highly conserved regions suggesting that our model may extended to other collagens that contain the LNS or thrombospondin-like domain. The β-strand which contains the putative heparin binding site is unique to Npp α1(XI) collagen and not conserved among the other collagens. The highly conserved cysteine residues suggest that they may be important in the structure of this domain. (B) Pfam schematic of LNS (TSPN) domain (shown as green box) in collagens. This domain is contained in a number of collagens but may vary in position.
Previously, Moradi-Améli et al. (1994) proposed that this domain within collagens adopted a structure containing nine β-strands within 110 amino acids, with these strands folded into two β-sheets. A disulfide bridge and specific hydrophobic residues were identified and proposed to stabilize the interaction between the β-sheets. The previous model was characterized as an immunoglobulin (Ig) domain, and proposed to act as a scaffold, which presented loops that can vary to present myriad functions and specificity of protein binding to specific ligands. Our work represents a refinement of the previous model and provides a structural basis on which to propose and evaluate putative binding activities.
We have demonstrated that activities shared among other LNS domains are also present within Npp α1(XI) collagen LNS domain, namely (1) glycosaminoglycan and (2) divalent cation binding. We have predicted which residues are involved in these interactions. Affinity for heparin has been documented for many LNS domains (Timpl et al. 2000), however, specific amino acids involved in the interaction are not strictly conserved, and the location of the binding site on the surface of the LNS domain has been shown to vary (Hohenester et al. 1999; Timpl et al. 2000; Yamashita et al. 2004). In the case of the Npp α1(XI) collagen LNS domain, docking analysis with heparin suggested that basic residues form a tract along the β-strand on the edge of the β-sandwich furthest from the amino and carboxyl termini. In contrast, the heparinbinding site of the LG5 domain of laminin α2 is located within loops along the “top” of the β-sandwich (as depicted in Fig. 4 ▶) opposite from the amino and carboxyl termini (Timpl et al. 2000).
The functional significance of the heparin-binding potential of the collagen LNS domains remains unknown. Other LNS domains found in collagens have been shown to interact with heparin, including the NC4 domain of collagen IX (Pihlajamaa et al. 2004), which is exposed at the surface of collagen fibrils. Unfortunately, no experimental data are available regarding the affinities of the LNS domains found in collagens and the heparan sulfate proteoglycans in situ. Such proteoglycans may fulfill a bridging function between adjacent collagen fibrils and/or between collagen fibrils and the cell surface. This would explain the apparent role of LNS-containing FACIT collagens types IX, XII, and XIV in the maintenance of mechanical integrity of the fibrillar network (Nishiyama et al. 1994; Olsen 1997), and the role of LNS-containing fibrillar collagens types V and XI in collagen fibrillogenesis. Another function for the LNS domains of collagens may be to interact with cell surface heparan sulfate proteoglycans to provide cell–extracellular matrix interactions. Extracellular matrix interactions often exhibit relatively low affinities for their ligands, with dissociation constants in the range of 10−6–10−8 M. Multiple weak interactions generated by the binding of many heparan sulfate groups to extracellular proteins may allow a sufficiently strong interaction. The significance of this dissociation constant may be that it facilitates remodeling of the tissue. Making and breaking specific contacts with the matrix may be facilitated if individual contacts are weak. It may be hypothesized that the macromolecular complex formation and cell-matrix interactions suitable for maintenance of tissue integrity are mediated by LNS domains of collagens. LNS domains may be central to tissue architecture, mediating both matrix–matrix interactions as well as the interactions between cells and their immediate environment.
The functional significance of calcium binding of LNS domains found in collagens remains unknown; however the possibility exists that calcium binding may facilitate interaction with heparan sulfate. Two potential sites for divalent cation coordination were identified. Future site-directed mutagenesis efforts will characterize the utilization of the two possible binding sites as well as the effect that calcium may have on the interaction between heparan sulfate and LNS domains of collagens.
Although Npp α1(XI) collagen is a relatively minor component in tissues, its ability to bind heparin and calcium is of particular interest. The regulation of fibrillogenesis by collagen type XI is based upon the hypothesis that the Npp α1(XI) collagen LNS domain plays a critical role. In this regulatory model, the triple helical domains of collagen XI are sequestered in the interior of the fibril, whereas the retained Npp α1(XI) LNS domains are excluded from the interior of the fibril, accumulating on the surface, and thus eventually sterically hinder the further deposition of type II collagen molecules onto the fibril surface. This model is supported by in vitro fibrillogenesis data with cartilage collagens that demonstrate uniformly thin collagen type II fibrils will assemble only when collagen XI is present (Blaschke et al. 2000). The LNS domain Npp α1(XI) collagen may play a more extensive role than the homologous domain of the α2(XI) chain because the Npp α2(XI) collagen is proteolytically processed much more rapidly than that of Npp α1(XI) collagen (Thom and Morris 1991). The result is that the LNS domain Npp α1(XI) collagen is retained at the surface of collagen fibrils for a longer period of time after incorporation into a collagen fibril than is the α2(XI) chain (Keene et al. 1995), and may mediate interaction between the surface of collagen fibrils and the surrounding extracellular matrix.
Since the Npp α1(XI) collagen LNS domain has been shown to be eventually cleaved proteolytically from the collagen triple helix in vivo and is not retained within the mature collagen fibril (Thom and Morris 1991; Keene et al. 1995), the heparan sulfate and divalent cation binding activity of Npp α1(XI) collagen may have biological function after release of the Npp α1(XI) collagen LNS domain from the triple helix of the α1(XI) collagen chain as well as while present on the surface of the collagen fibril.
The removal of the Npp α1(XI) collagen LNS domain from the surface of collagen fibrils is, in part, regulated by the adjacent variable region, the identity of which is determined by alternative splicing of exons that encode the variable region. The rate and extent of proteolytic processing by the enzyme BMP-1 was reduced for isoforms in which the Npp α1(XI) LNS domain was connected to the triple helix by a relatively short 31-amino-acid linker (Medeck et al. 2003).
In conclusion, we present a structural model for the organization of the Npp α1(XI) collagen LNS domain. The model predicts calcium and heparin binding activities. These activities are verified in this study. The regulation of fibrillogenesis is attributed to the Npp α1(XI) collagen domain which is retained on the surface of heterotypic collagen fibrils for an extended period of time. These findings suggest that interactions with divalent cation and glycosaminoglycan are crucial mechanistic details in the function of this LNS domain and perhaps LNS domains found in other collagens.
Materials and methods
Template selection
Structural templates for the Npp α1(XI) collagen from Rattus norvegicus (accession no.U20121) were obtained using sequence-structure based algorithms, PSI-BLAST (Schaffer et al. 2001) and FUGUE (Shi et al. 2001), a sequence–structure homology recognition method using environment-specific substitution tables and structure-dependent gap penalties. The best results were used as final templates for homology modeling based on E-Values, percent identity, and alignment length. The FUGUE results were analyzed with respect to Z-score and fold family.
Sequence alignment
Sequence alignment was performed using the Align2D algorithm employed in MODELLER (Sali and Blundell 1993; Sali et al. 1995). Default parameters were used with a Gonnet scoring matrix. Other models were created using the CLUSTALW (Thompson et al. 1994) multiple sequence alignment with default parameters using a Gonnet scoring matrix for both pairwise and multiple sequence alignments. Manual inspection and modification of the alignments were performed after initial model generation and analysis of alignment of secondary structures in the model.
Ab initio model generation
For the 40 amino acids at the amino terminus where no suitable template was available, ab initio models were generated using the HMMSTR/Rosetta Server (Bystroff et al. 2000; Bystroff and Shao 2002), which generates local structural motifs used by the Rosetta program to generate 3D structure by fragment-insertion Monte Carlo method. A model of the 40-amino-acid amino terminus was generated (Bystroff et al. 2000; Bystroff and Shao 2002).
Homology model generation
Homology models were built using MODELLER (Sali and Blundell 1993; Sali et al. 1995). Alignment parameters were used as discussed above and disulfide bonding was specified as C25-C207 and C146-C200 as determined by liquid chromatography-tandem mass spectrometry (Gregory et al. 2000). The structures of laminin and neurexin (PDB Idds: 1dyk and 1c4r, respectively) were used for homology modeling calculations. Only results using 1dyk are shown in the present work. All templates were typed with CHARMM (Brooks et al. 1983) (Chemistry at HARvard Molecular Mechanics) force-fields before they were submitted to MODELLER (Sali and Blundell 1993; Sali et al. 1995).
Model refinement and evaluation
Models were assessed using the program Verify3D (Eisenberg et al. 1997). Invalid regions were refined and evaluated using CHARMM energy minimization, rotamer libraries, and realignment of sequence. Loop refinement was done using MODELLER (Sali and Blundell 1993; Sali et al. 1995) with default parameters and disulfide bonding specified. The Ramachandran plot (Ramachandran and Sasiskharan 1968) was analyzed to find residues in forbidden regions. Such residues were refined using loop optimization, rotamer libraries, and energy minimization. Hydrogen bonding patterns and overall stability were taken into consideration with respect to putative structure/function relationships of the protein.
Electrostatic map
A DelPhi (Floratos et al. 2001) electrostatic potential map was generated within Insight II (Accelrys, San Diego, CA.) using default parameters with an ionic strength of zero. The map of the model was analyzed with respect to regions of substrate binding and surface charge.
Molecular modeling of Npp α1(XI)-heparin interactions
Docking of monomers was preformed using Auto-Dock 3.0 (Morris et al. 1998). For each heparin ligand, one job of 100 docking runs was performed using a population of 200 individuals and an energy evaluation number of 2 × 106 employing the Lamarckian Genetic Algorithm (Morris et al. 1998). The grid was housed around the entire protein for a “blind dock” using a grid spacing of 0.800 Å. A further refined docking was centered at the putative heparin binding site on residues 147–152 with a grid spacing of 0.375 Å. The highly populated binding zone was studied with respect to residues involved in binding as well as final energy.
Construction of the Npp α1(XI) collagen expression vector
The Npp α1(XI) collagen sequence was amplified and cloned into pET11a expression vector as previously described (Gregory et al. 2000). E. coli BL21(DE3)pLysS-competent cells were transformed and grown in LB medium.
Recombinant protein production and purification
Cells were grown to early log phase and recombinant protein expression was induced by the addition of IPTG. Cells were harvested by centrifugation and lysed by resuspension in a hypotonic detergent solution (B-PER, Pierce). Recombinant protein was collected by differential centrifugation to separate inclusion bodies from soluble proteins, followed by subsequent washes in phosphate buffered saline at 4°C until purity of the recombinant protein approached 70% as assessed by SDS-polyacrylamide gel electrophoresis. Inclusion body proteins were solubilized in 50 mM Tris-HCl (pH 7.5) containing 6 M guanidine hydrochloride and 150 mM NaCl, and clarified by centrifugation at 18,000g, 4°C for 30 min. Soluble protein was applied to a nickel-NTA affinity chromatography column at a flow rate of 1 mL/min. Protein was refolded while on the column. Absorbance at 280 nm and conductivity were monitored continuously. Bound protein was eluted using an imidazole concentration gradient from 5 mM to 250 mM. Protein in each fraction was assessed by SDS-polyacrylamide gel electrophoresis. Proper structure of folded protein was confirmed by circular dichroism and mass spectrometry as previously described (Gregory et al. 2000).
Fluorescence spectroscopy
Fluorescence measurements were made on a Varian Cary Eclipse with PCB 150 water Peltier system spectrofluorometer. Emission spectra were recorded from 300 to 400 nm using an excitation wavelength of 285 nm. Recombinant protein was prepared by dialysis against 5 mM MOPS buffer (pH 7.5), 150 mM NaCl and adjusted to a concentration of approximately 40 μg/mL. Aliquots of stock solutions of CaCl2, MgCl2, or ZnCl2 were added to the protein sample to assess the effect of changes in free Ca2+, Mg2+, and Zn2+.
Analysis of heparan sulfate binding to Npp α1(XI) collagen
All SPR measurements were performed at 20°C using a Reichert SR7000 instrument. Npp α1(XI) collagen was immobilized to a carboxyl mixed self-assembled monolayer surface on gold sensor chip by primary amine coupling. Briefly, the chip surface was activated with a 0.4-mM N-ethyl-N-(3-diethylaminopropyl) carbodiimide, 0.1-mM N-hydroxysuccinimide solution followed by the injection of Npp at a concentration of 20 μg/mL in 10 mM sodium acetate buffer (pH 4.5). When the desired level of binding was achieved, unreacted N-hydroxysuccinimide ester groups were blocked with 1 M ethanolamine hydrochloride. Samples of heparan sulfate were prepared in running buffer (phosphate buffered saline with 0.05% Tween-20), which was filtered through a 0.2-μm filter prior to use. To collect equilibrium binding data, various concentrations (0.0625–0.75 mg/mL) of the heparan sulfate analyte were injected in a volume of 200 μL over the Npp ligand at a flow rate of 10 μL/min. After 200 sec, the analyte solutions were replaced with running buffer for 200 sec. The surfaces were regenerated with a 100-sec injection of running buffer containing 1 M NaCl at a flow rate of 10 μL/min. The association, dissociation, and regeneration phases were followed in realtime by monitoring changes in signal expressed in resonance units and the data displayed as response units versus time. An average of the response at equilibrium was determined for each analyte concentration and the resulting equilibrium resonance units were plotted against concentration. Data were fit to a steady-state affinity model using GraphPad Prism (GraphPad Software) using a one-site association model.
Preparation of figures
Figures were generated using the PyMOL Molecular Graphics System (DeLano Scientific; DeLano 2002).
Acknowledgments
We thank Raquel Brown, Sorcha Cusack, Rohn McCune, and Noriko Hazeki-Taylor for technical assistance. This work was supported by grants from the Arthritis Foundation, NIH/NIAMS (RO1AR47985 and KO2AR48672), by a grant from NIH/NCRR (P20RR16454) and funding from the M.J. Murdock Foundation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051363105.
References
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., and Sonnhammer, E.L. et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32 138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann, G., Hanke, J., Bork, P., and Reich, J.G. 1998. Merging extracellular domains: Fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins. J. Mol. Biol. 275 725–730. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke, U.K., Eikenberry, E.F., Hulmes, D.J.S., Galla, H.-J., and Bruckner, P. 2000. Collagen XI nucleates self-assembly and limits lateral growth of cartilage fibrils. J. Biol. Chem. 275 10370–10378. [DOI] [PubMed] [Google Scholar]
- Bork, P. 1992. The modular architecture of vertebrate collagens. FEBS Lett. 307 49–54. [DOI] [PubMed] [Google Scholar]
- Bork, P., Downing, A.K., Kieffer, B., and Campbell, I.D. 1996. Structure and distribution of modules in extracellular proteins. Q. Rev. Biophys. 29 119–167. [DOI] [PubMed] [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4 187–217. [Google Scholar]
- Bystroff, C. and Shao, Y. 2002. Fully automated ab initio protein structure prediction using I-SITES, HMMSTR, and ROSETTA. Bioinformatics 18 Suppl 1: S54–S61. [DOI] [PubMed] [Google Scholar]
- Bystroff, C., Thorsson, V., and Baker, D. 2000. HMMSTR: A hidden Markov model for local sequence-structure correlations in proteins. J. Mol. Biol. 301 173–190. [DOI] [PubMed] [Google Scholar]
- Cardin, A. and Weintraub, H. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler. Thromb. Vasc. Biol. 9 21–32. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org.
- Eisenberg, D., Luthy, R., and Bowie, J.U. 1997. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 277 396–404. [DOI] [PubMed] [Google Scholar]
- Fichard, A., Kleman, J.P., and Ruggiero, F. 1995. Another look at collagen V and XI molecules. Matrix Biol. 14 515–531. [DOI] [PubMed] [Google Scholar]
- Floratos, A., Rigoutsos, I., Parida, L., and Gao, Y. 2001. DELPHI: A pattern-based method for detecting sequence similarity. IBM J. Res. & Dev. 45 455–473. [Google Scholar]
- Gregory, K.E., Oxford, J.T., Chen, Y., Gambee, J.E., Gygi, S.P., Aebersold, R., Neame, P.J., Mechling, D.E., Bachinger, H.P., and Morris, N.P. 2000. Structural organization of distinct domains within the non-collagenous N-terminal region of collagen type XI. J. Biol. Chem. 275 11498–11506. [DOI] [PubMed] [Google Scholar]
- Grishkovskaya, I., Avvakumov, G.V., Sklenar, G., Dales, D., Hammond, G.L., and Muller, Y.A. 2000. Crystal structure of human sex hormone-binding globulin: Steroid transport by a laminin G-like domain. EMBO. J. 19 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester, E., Tisi, D., Talts, J.F., and Timpl, R. 1999. The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin. Mol. Cell 4 783–792. [DOI] [PubMed] [Google Scholar]
- Keene, D.R., Oxford, J.T., and Morris, N.P. 1995. Ultrastructural localization of collagen types II, IX, and XI in the growth plate of human rib and fetal bovine epiphyseal cartilage: Type XI collagen is restricted to thin fibrils. J. Histochem. Cytochem. 43 967–979. [DOI] [PubMed] [Google Scholar]
- Li, Y., Lacerda, D.A., Warman, M.L., Beier, D.R., Yoshioka, H., Ninomiya, Y., Oxford, J.T., Morris, N.P., Andrikopoulos, K., and Ramirez, F. 1995. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell 80 423–430. [DOI] [PubMed] [Google Scholar]
- Mayne, R. and Brewton, R.G. 1993. New members of the collagen superfamily. Curr. Opin. Cell Biol. 5 883–890. [DOI] [PubMed] [Google Scholar]
- Medeck, R.J., Sosa, S., Morris, N., and Oxford, J.T. 2003. BMP-1-mediated proteolytic processing of alternatively spliced isoforms of collagen type XI. Biochem. J. 376 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos, I., Torrance, G.M., Gilbert, D.R., and Westhead, D.R. 2004. TOPS: An enhanced database of protein structural topology. Nucleic Acids Res. 32 D251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi-Améli, M., Deléage, G., Geourgjon, C., and van der Rest, M. 1994. Common topology within a non-collagenous domain of several different collagen types. Matrix Biol. 14 233–239. [DOI] [PubMed] [Google Scholar]
- Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E., Belew, R.K., and Olson, A.J. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19 1639–1662. [Google Scholar]
- Nishiyama, T., McDonough, A.M., Bruns, R.R., and Burgeson, R.E. 1994. Type XII and XIV collagens mediate interactions between banded collagen fibers in vitro and may modulate extracellular matrix deformability. J. Biol. Chem. 269 28193–28199. [PubMed] [Google Scholar]
- Olsen, B.R. 1997. Collagen IX. Int. J. Biochem. Cell Biol. 29 555–558. [DOI] [PubMed] [Google Scholar]
- Pihlajamaa, T., Lankinen, H., Ylostalo, J., Valmu, L., Jaalinoja, J., Zaucke, F., Spitznagel, L., Gosling, S., Puustinen, A., Morgelin, M., et al. 2004. Characterization of recombinant amino-terminal NC4 domain of human collagen IX: Interaction with glycosaminoglycans and cartilage oligomeric matrix protein. J. Biol. Chem. 279 24265–24273. [DOI] [PubMed] [Google Scholar]
- Ramachandran, G.N. and Sasiskharan, V. 1968. Conformation of polypeptides and proteins. Adv. Protein Chem. 23 283–437. [DOI] [PubMed] [Google Scholar]
- Rudenko, G., Nguyen, T., Chelliah, Y., Sudhof, T.C., and Deisenhofer, J. 1999. The structure of the ligand-binding domain of neurexin Iβ: Regulation of LNS domain function by alternative splicing. Cell 99 93–101. [DOI] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Sali, A., Potterton, L., Yuan, F., van Vlijmen, H., and Karplus, M. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23 318–326. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Knyazev, P.G., Cheburkin, Y., Gohring, W., Tisi, D., Ullrich, A., Timpl, R., and Hohenester, E. 2002. Crystal structure of a C-terminal fragment of growth arrest-specific protein Gas6. Receptor tyrosine kinase activation by laminin G-like domains. J. Biol. Chem. 277 44164–44170. [DOI] [PubMed] [Google Scholar]
- Schaffer, A.A., Aravind, L., Madden, T.L., Shavirin, S., Spouge, J.L., Wolf, Y.I., Koonin, E.V., and Altschul, S.F. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, L.M. and Olsen, B.R. 1991. FACIT collagens: Diverse molecular bridges in extracellular matrices. Trends Biochem. Sci. 16 191–194. [DOI] [PubMed] [Google Scholar]
- Shi, J., Blundell, T.L., and Mizuguchi, K. 2001. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310 243–257. [DOI] [PubMed] [Google Scholar]
- Thom, J.R. and Morris, N.P. 1991. Biosynthesis and proteolytic processing of type XI collagen in embryonic chick sterna. J. Biol. Chem. 266 7262–7269. [PubMed] [Google Scholar]
- Thompson, J., Higgins, D., and Gibson, T. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl, R., Tisi, D., Talts, J.F., Andac, Z., Sasaki, T., and Hohenester, E. 2000. Structure and function of laminin LG modules. Matrix Biol. 19 309–317. [DOI] [PubMed] [Google Scholar]
- Tisi, D., Talts, J.F., Timpl, R., and Hohenester, E. 2000. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 19 1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Rest, M. and Garrone, R. 1991. Collagen family of proteins. FASEB J. 5 2814–2823. [PubMed] [Google Scholar]
- Wizemann, H., Garbe, J.H.O., Friedrich, M.V.K., Timpl, R., Sasaki, T., and Hohenester, E. 2003. Distinct requirements for heparin and α-dystroglycan binding revealed by structure-based mutagenesis of the laminin α2 LG4-LG5 domain pair. J. Mol. Biol. 332 635–642. [DOI] [PubMed] [Google Scholar]
- Yamashita, H., Beck, K., and Kitagawa, Y. 2004. Heparin binds to the laminin α4 chain LG4 domain at a site different from that found for other laminins. J. Mol. Biol. 335 1145–1149. [DOI] [PubMed] [Google Scholar]