Abstract
YggX is a highly conserved protein found only in eubacteria and is proposed to be involved in the bacterial response to oxidative stress. Here we report the solution structure of YggX from Escherichia coli determined by nuclear magnetic resonance spectroscopy. The structure of YggX displays a fold consisting of two N-terminal antiparallel β-sheets and three α-helices, which shares significant structural similarity to the crystal structure of a hypothetical protein PA5148 from Pseudomonas aeruginosa. Previous studies propose YggX as an iron binding protein that is involved in cellular iron trafficking. Our data indicate that the protein alone does not bind iron in vitro, suggesting other cofactors or different conditions may be necessary for metal binding.
Keywords: NMR spectroscopy, protein structure, YggX, E. coli, iron-mediated oxidative damage
The YggX protein is a protein highly conserved across 37 bacterial species but lacks homology to proteins of known function. Mutations in the yggX gene were originally isolated during a genetic screen for thi+ revertants from a thi− strain of Salmonella enterica (Gralnick and Downs 2001). Mutants that overproduce the YggX protein show the suppression of several phenotypes related to oxidative damage, including deficiency in aconitase activity, elevated spontaneous mutation rate, and sensitivity to superoxide stress (Gralnick and Downs 2001). Purified YggX decreases chelatable iron in solution and protects DNA from iron-mediated oxidative damage, both in vivo and in vitro (Gralnick and Downs 2003). The protection of DNA and FeS clusters by YggX, coupled with its apparent iron-binding abilities, leads to a model for YggX function. YggX is proposed as a mediator in iron transactions between iron acquisition and iron-requiring processes, such as synthesis and/or repair of FeS clusters in biosynthetic enzymes. In this model, the absence of YggX produces an increase in intracellular iron that results in enhanced damage to DNA and proteins. Conversely, overproduction of YggX decreases damage to DNA by diminishing the intracellular concentration of free iron (Gralnick and Downs 2003).
Recent studies have shown that the yggX of Escherichia coli is part of the SoxRS regulon, a group of coregulated genes that code for antioxidant defenses. The regulon is under transcriptional control of the SoxR and SoxS proteins, a sensor for oxidative stress and a transcriptional regulator, respectively (Pomposiello and Demple 2001). Transcriptional profiling studies showed that exposure to superoxide-producing agents or the ectopic expression of the transcriptional regulator SoxS in the absence of stress stimulated the transcription of yggX (Pomposiello et al. 2001). Promoter deletion fusions to a reporter lacZ gene, and gel mobility shift assays with purified SoxS showed that SoxS binds to an unusual site at the yggX promoter. Northern blot analysis of yggX transcription confirmed the observations from the gene array experiments. The yggX is transcribed in two different forms (Pomposiello et al. 2003), a monocystronic transcript containing the yggX gene, and a dicystronic transcript containing yggX and the downstream gene, mltC, which codes for a membrane-bound peptydoglycan hydrolase. The transcriptional regulation of yggX is consistent with the proposed function of YggX as a mediator in intracellular iron transactions.
Results and Discussion
Resonance assignments
Genomic sequencing identified the open reading frame for the yggX gene in E. coli (Wasinger and Humphery-Smith 1998) as an 11-kDa protein, and expression studies demonstrated that the gene codes for a small and abundant protein (Link et al. 1997). We expressed and purified recombinant YggX from E. coli for structure determination by NMR spectroscopy. 1H-15N HSQC and translational diffusion NMR spectra indicated YggX is a globular, folded, monomeric protein and soluble at ~1 mM. Complete 1H, 15N, and 13C assignments were made for the polypeptide backbone of the recombinant YggX sequence except for the N-terminal methionine and the NH of G2. In addition some NH resonances for the C-terminal 6xHis tag were visible (H93–H96) but exhibited no NOE connectivity. Side-chain assignments for the 13C, 15N, and nonlabile 1H resonances are complete for the recombinant YggX sequence with the exception of M1; Cζ/Hζ of F7, F10, F20, F74, and F76; Cɛ1/Hɛ1 of H45 and H82; and Hγ of L67. The high percentage of assignments was important for the success of the automated NOE assignment procedure in CYANA (Guntert et al. 1997). Chemical shifts have been deposited at BMRB (accession code 6453).
Solution structure of YggX
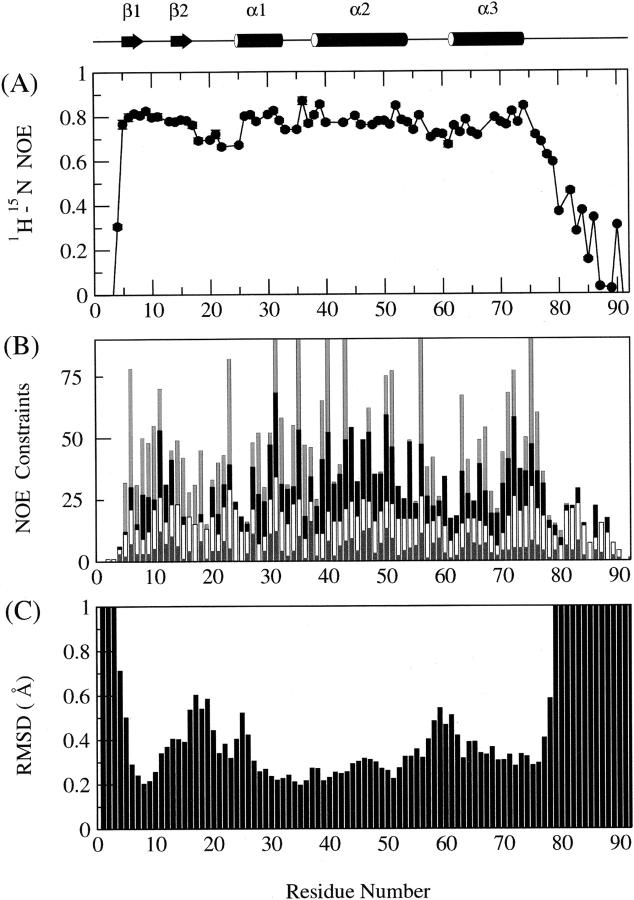
YggX is a single domain protein (Fig. 1 ▶) consisting of two small N-terminal antiparallel β-sheets (T5-F7, E14-E16); a core body comprising three helices: helix α1 (G25-E34), helix α2 (K37-K54), helix α3 (A61-F74); and an unstructured C-terminal extension (E77-H96, plus the C-terminal 6xHis tag). Hydrophobic contacts at the core of YggX are crucial for determining its global fold. In particular, residues from the three helices are at the interior of the protein and make stabilizing contacts (L27, G28, I31, A39, W40, W43, Q47, L50, I51, L67, M71, V72, and L75). In addition, residues in the small loop connecting helices α1 and α2 (I35, S36) are completely protected from the solvent by residues in and adjacent to the β-sheets. This region (T5-Q18) also appears to be important for stabilizing residues at the C termini of helices α1 and α3 and the N terminus of helix α2, with residues C8, G17 and Q18 exhibiting low solvent accessibility.
Figure 1.
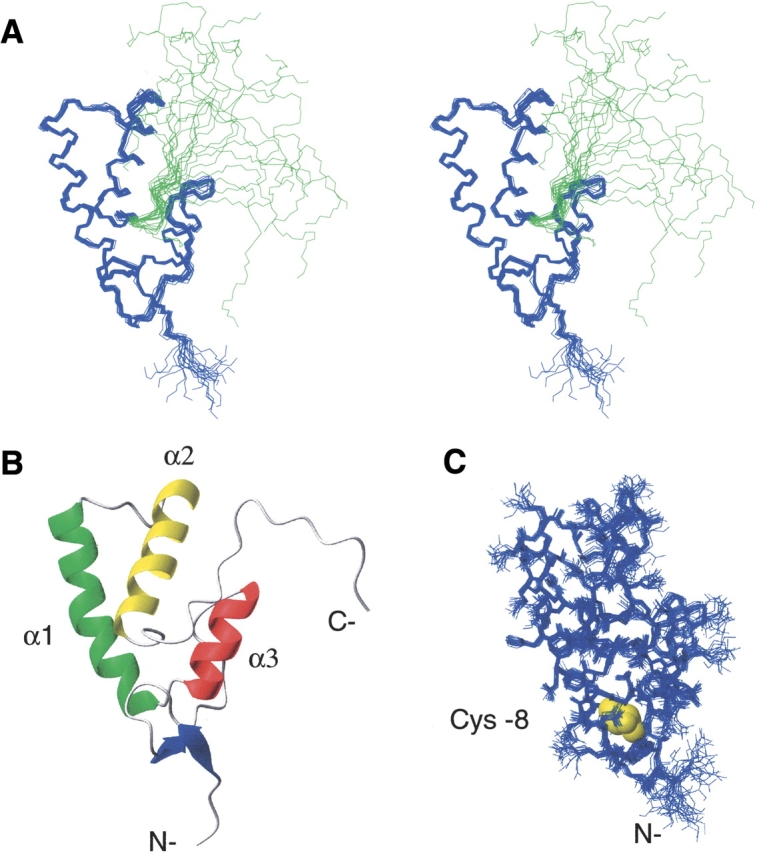
The NMR structure of recombinant YggX (residues 1– 96). (A) Cross-eyed stereo diagram of superimposed backbone atoms from the 20 lowest energy solution structures of YggX. Residues 1–76 are shown in blue and C-terminal tail in green. (B) Ribbon representation of the lowest energy YggX conformer. (C) Solution structure of YggX showing all nonhydrogen atoms of the core fold. Conserved cysteine-8, highlighted in yellow, is shown buried in the core of YggX.
The fold of the core residues (4–76) of YggX is well defined, exhibiting RMSDs of 0.35 Å and 0.78 Å for backbone and all nonhydrogen atoms, respectively (Table 1). The heteronuclear NOE data (Fig. 2A ▶) for this region indicate structural rigidity, although, as expected, the larger exposed loop regions exhibit slightly lower NOE values. The tight compact structure of the core residues is also evidenced by retarded NH exchange, which was not complete after 1 mo with residues in the three helices and the loop connecting helices α1 and α2 being the most protected. Heteronuclear NOE data confirm the C-terminal tail, spanning residues E77-K92 and the 6xHis-tag, to be ill-defined and flexible on the nanosecond time scale, consistent with the large RMSDs exhibited in the structural ensemble (Fig. 2C ▶). Although residues within this region exhibit some short-range NOE connectivity (Fig. 2B ▶), there appears to be no preference for any secondary structural elements.
Table 1.
NMR constraints used for structure calculation and structural statistics for YggX
| Constraints used for structure calculation (residues 2–92) | |
| Total NOEs | 2059 |
| Intraresidue NOEs (n = 0) | 440 |
| Sequential NOEs (n = 1) | 512 |
| Medium range NOEs (n = 2,3,4) | 550 |
| Long range NOEs (n > 4) | 557 |
| Dihedral angle constraints | 72 |
| 15N-1H residual dipolar couplings | 42 |
| Total number of constraints | 2173 |
| Average RMSD to mean structure (Å) residues 4–76 | |
| Backbone atoms | 0.350 ± 0.057 |
| All heavy (nonhydrogen atoms) | 0.783 ± 0.058 |
| Average energy values (kcal/mol−1) | |
| Etotal | −461.19 ± 7.46 |
| Ebond | 5.56 ± 0.48 |
| Eangle | 57.38 ± 1.52 |
| Eimproper | 6.41 ± 0.56 |
| EVdW | −590.19 ± 6.73 |
| ENOE | 10.70 ± 1.21 |
| Edihedral | 0.26 ± 0.21 |
| Esani | 22.10 ± 3.72 |
| Deviation from idealized covalent geometry | |
| Etotal | 0.0019 ± 0.0001 |
| Ebond | 0.3590 ± 0.0032 |
| Eangle | 0.2210 ± 0.0050 |
| RMSD from experimental data | |
| Distance restraints (Å) | 0.0430 ± 0.0005 |
| Dihedral angle restraints (Å) | 0.0980 ± 0.0180 |
| Average Ramachandran statistics for 20 lowest energy structures (residues 1–78) | |
| Residues in most favored regions | 82.7% |
| Residues in additional allowed regions | 17.3% |
| Residues in generously allowed regions | 0.0% |
| Residues in disallowed regions | 0.0% |
| Analysis of residual dipolar coupling | |
| RMSD (Hz) | 1.624 ± 0.105 |
| Q-factor | 0.147 ± 0.011 |
| Correlation coefficient | 0.988 |
Figure 2.
Plots of 1H-15N heteronuclear NOE, NOE constraints, and RMSD statistics. Shown above is a cartoon diagram representing the secondary structure of YggX. (A) 1H-15N heteronuclear NOE data acquired at 600 MHz. (B) Summary of all assigned unambiguous NOE constraints: intraresidue, sequential, medium, and long-range NOEs are shown as dark gray, white, black, and medium gray bars, respectively. (C) Backbone RMSDs calculated for the 20 lowest energy YggX structures based on the superposition of residues 1–78.
Sequence analysis and structural comparisons of YggX
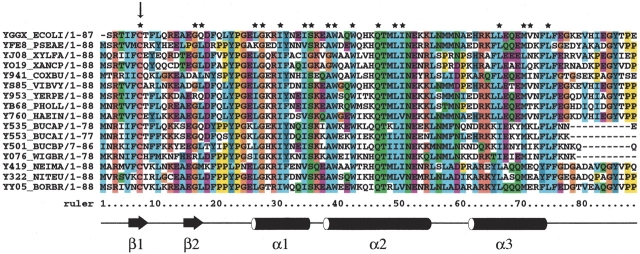
YggX is found throughout Gram-negative bacteria, which could point to its biological relevance in the response to oxidative stress. Primary sequence comparison of YggX homologs from other Gram-negative bacterial species shows a remarkably high degree of sequence conservation (Fig. 3 ▶). However, in some species, there is slight sequence variation at the C-terminal end, which corresponds to the unstructured region of the protein. This variability might reflect differences in target specificity within this protein family. Overall, the generally high sequence similarity, particularly of residues at the interior of the protein, implies that its structure is also well conserved throughout bacterial species. This is supported when comparing the crystal structure of the hypothetical protein PA5148 from Pseudomonas aeruginosa (RCSB code 1T07) to the solution structure of YggX from E. coli. Their primary sequence share 51% sequence identity and structures only deviate by 2.16 Å over their backbone atoms of the core residues (4–78 in E. coli and 3–77 P. aeruginosa), indicating that this hypothetical protein is an YggX homolog.
Figure 3.
Alignment of primary sequences for YggX related proteins (Pfam 04362.5). Sequence relatives for YggX was identified by Pfam (Bateman et al. 2002). First two sequences correspond to YggX from E. coli and the hypothetical PA5148 protein from P. aeruginosa. The black arrow shows the lone conserved cysteine residue found in all YggX relatives. The stars denote residues exhibiting an average of <5% of the accessible surface area. These residues form stabilizing hydrophobic contacts, which are essential for determining its global fold. Shown below is a cartoon representation of the secondary structure of YggX.
YggX as an iron binding protein?
The exact function of YggX remains elusive. It was reported that YggX decreases chelatable iron in solution both in vivo and in vitro, and a model was proposed by which YggX can bind iron (Gralnick and Downs 2003). A DALI server search (Holm and Sander 1996) for structural homologs of YggX shows a number of hits with a Z-score above 3, with the hypothetical protein PA5148 from P. aeruginosa giving the highest Z-score of 8.4. However, within the other DALI hits, no iron binding proteins were identified, and none of the other proteins could shed light on the role for YggX in trafficking iron.
Under the conditions used for NMR structure determination, we were unable to detect iron binding by YggX using an assay as previously reported by Gralnick and Downs (2003). In addition, titration of YggX with both Fe(II) and Fe(III) exhibited no perturbations to the signature 1H-15N HSQC spectrum (data not shown). Thus, under our conditions YggX alone can not bind iron. A critical role for the highly conserved and solitary cysteine residue Cys-7 (Cys-8 in our numbering) was previously proposed (Gralnick and Downs 2003), suggesting it may act as a ligand for iron binding. Our solution structure shows cysteine (Fig. 1C ▶) to be buried (0% solvent accessibility) and unavailable for binding to iron. Similarly, this cysteine residue in the structural homolog from P. aeruginosa is also completely buried. This does not exclude a role for Cys-8 in iron binding, which could occur via local unfolding induced by interactions with other protein cofactors or under other solution conditions. Alternatively, loss of iron chelating upon mutation of this cysteine may arise from loss of structure of this highly buried residue. Clearly, more detailed experiments are required to address this issue.
Conclusions
The solution structure of YggX has been determined by NMR, showing a well-folded protein comprising three helices, a small N-terminal β-sheet, and an unstructured C-terminal tail. Under our conditions, YggX alone was unable to bind iron; indicating that either other cofactors or proteins are necessary or other specific conditions are needed for iron recognition.
Materials and methods
Cloning and expression of YggX
The yggX gene was amplified by PCR, the fragment was cloned into the NcoI–XhoI sites of pET28b+(Kanr) vector (Novagen), and proteins were expressed in BL21 Gold-DE3 E. coli strain (Stratagene). For unlabeled and isotopically enriched protein sample preparations, recombinant YggX with a C-terminal 6x-histidine tag was expressed in either Luria broth or M9 media containing 15NH4Cl (Isotech, Inc.) or 15NH4Cl and D-(13C6) glucose (Cambridge Isotope Laboratory). All cultures were grown at 37°C until an OD600 of ~0.8. The temperature was then reduced to 30°C, and 1 mM isopropyl-1-thio-β- D-galactopyranoside was added to the culture and shaken for 3 h to induce expression of the YggX His-tag fusion protein.
Purification and characterization of YggX
Harvested cells were resuspended in lysis buffer (5mM imidazole, 50 mM HEPES, 500 mM NaCl, 5% glycerol, 100 μg/mL lysozyme, 1 mM of the protease inhibitor PMSF at pH 7.5) and disrupted by sonication. Total lysate was centrifuged and supernatant collected for subsequent purification steps. Histidine-tagged YggX was isolated by standard protocols using affinity chromatography with Ni2+-loaded Sepharose and further purified by HPLC gel filtration (HiLoad 16/60 Superdex 75, Amersham Biosciences). Sequence composition and characterization of YggX was completed by DNA sequencing and ESI mass spectrometry. Two cloning artifacts were present in the recombinant YggX protein. At the N terminus, a glycine residue was inserted after the initial methionine residue, and at the C terminus, an eight-residue Leu-Glu-His6 affinity-tag was added.
NMR spectroscopy
For NMR analysis, purified recombinant YggX was exchanged by gel-filtration into buffer containing 50 mM sodium phosphate, 150 mM NaCl, and 1 mM NaN3, and 10% D2O (pH 6.3). All NMR spectra were recorded at 303 K on a Bruker Avance DRX 600-MHz spectrometer equipped with a triple-resonance CryoProbe and processed with NMRPipe (Delaglio et al. 1995). Backbone 1H, 13C, and 15N assignments were completed from CBCA(CO)NH, HNCACB, and HBHA(CBCACO)NH spectra. 1H, 13C, and 15N side-chain assignments were obtained by analysis of the H(CC)(CO)NH, C(C)(CO)NH, and HCCH-TOCSY experiments. Aromatic 1H/13C ring assignments were obtained from constant time two-dimensional HBCBCGCDHD and HBCBCGCDCEHE spectra. All NMR data were analyzed by using NMRView (Johnson and Blevins 1994) and in-house scripts. 1H, 13C, and 15N chemical shifts were referenced to DSS according to the IUPAC recommendation (Markely et al. 1998). Distance constraints were obtained from a simultaneous three-dimensional 13C/15N-edited NOESY experiment (τm = 120 msec) in 90% H2O/10% D2O, and 13C-edited NOESY (τm = 100 msec) and 13C-edited NOESY (aromatic region) (τm = 100 msec) experiments acquired in 99.9% D2O. A four-dimensional 13C-13C NOESY experiment with a mixing time of 80 msec was also recorded to confirm a number of long range constraints but was not used to derive distance restraints. Additional restraints used in structure calculations were dihedral restraints, derived from 3JHN-Cα coupling constants obtained from the HNHA experiment (Kuboniwa et al. 1994), and 1H-15N residual dipolar couplings extracted from comparison of IPAP-HSQC experiments recorded on YggX with and without 8 mg/mL Pf1 phage (Ottiger and Bax 1998).
NMR structure calculation
A set of unambiguous NOE constraints were extracted from the NOESY spectra and used in conjunction with dihedral angle restraints to generate a preliminary fold of YggX using CNS 1.1 (Brunger et al. 1998). The resulting structures were used as model templates for automated assignment of NOESY cross peaks (Herrmann et al. 2002) and structure calculation with torsion angle dynamics using CYANA 2.0 (Guntert et al. 1997). The standard CYANA protocol of seven iterative cycles of NOE assignment and structure calculation, followed by a final structure calculation, was applied. The resulting structures were refined with dihedral angle and residual dipolar coupling restraints in addition to 2059 NOE restraints obtained from CYANA using the standard CNS refinement protocol. Initial axial and rhombic components of the alignment tensor were obtained from the histogram method (Clore et al. 1998a) and optimized by a grid search (Clore et al. 1998b) and determined to be Da = −12.5 and R = 0.3. The 20 lowest energy structures with the fewest violations were selected to represent the YggX structure. No NOE violations >0.2 Å were observed. Structural statistics for this ensemble as calculated by CNS (Brunger et al. 1998), PROCHECK (Laskowski et al. 1996), and SSIA (Zweckstetter and Bax 2000) are summarized in Table 1. The coordinates have been deposited in the RCSB under PDB code 1YHD.
Titration of YggX with iron and in vitro binding assay
For the following experiments, purified YggX was exchanged into the NMR assay buffer 50 mM MES, 150 mM NaCl, 1 mM NaN3, and 10% D2O (pH 6.3) using a PD-10 gel-filtration column. Iron titration experiments were carried out on a 0.5 mM 15N-labeled YggX sample by addition of up to a three-fold excess of Fe(II) (FeSO4) or Fe(III) (FeCl3) and monitoring of the perturbations in chemical shifts of amide signals from the 15N-1H HSQC spectrum. Minimal changes in pH and volume were ensured throughout. All spectra were acquired on a Bruker 600-MHz AVANCE spectrometer at 303 K. Detection of chelatable Fe(II) by purified YggX in vitro were carried out similarly to a previously described assay (Gralnick and Downs 2003) in our solution conditions. Briefly, varying concentrations of protein (a final of 0–200 μM) were distributed in 1-mL aliquots containing our NMR assay buffer and freshly prepared FeSO4 at a final concentration of 100 μM. Chelatable Fe(II) was detected by adding 1, 10-phenanthroline (a final of 500 μM) to each cuvette, incubated for 30 min at 25°C, and then measured at an absorbance of 510 nm using a Varian Cary 1 Bio UV-VIS spectrophotometer.
Acknowledgments
We thank Demetra Elias for her assistance and helpful discussions. The Montreal-Kingston Bacterial Structural Genomics Initiative supported this work under a Canadian Institute of Health Research (CIHR) grant GSP-48370. N.S. is a recipient of the McGill Faculty of Medicine Internal Fellowship. The senior authors (P.P. and K.G.) dedicate this article to Dr. Robert A. Bender, advisor and mentor.
Note added in proof
During the submission of this manuscript, the solution structure of YggX from S. enterica was released (PDB 1XS8). YggX from both S. enterica and E. coli are nearly identical, having 99% sequence similarity (88% identity), a DALI Z-score of 8.8 and an RMSD of 2.23 Å over their backbone atom (4–78 for E. coli and 3–77 for S. enterica). We also note that similar to the E. coli and P. arguinosa structures, the single cysteine residue in S. enterica YggX is also completely buried.
Abbreviations
HSQC, heteronuclear single quantum coherence
NMR, nuclear magnetic resonance
NOE, nuclear Overhauser effect
RCD, residual dipolar coupling
RMSD, root mean square deviation
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051358105.
References
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., and Nilges, M., Pannu, N.S. et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt 5) 905–921. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., and Bax, A. 1998a. A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J. Magn. Reson. 133 216–221. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., and Tjandra, N. 1998b. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 131 159–162. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Gralnick, J. and Downs, D. 2001. Protection from superoxide damage associated with an increased level of the YggX protein in Salmonella enterica. Proc. Natl. Acad. Sci. 98 8030–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. The YggX protein of Salmonella enterica is involved in Fe(II) trafficking and minimizes the DNA damage caused by hydroxyl radicals: Residue CYS-7 is essential for YggX function. J. Biol. Chem. 278 20708–20715. [DOI] [PubMed] [Google Scholar]
- Guntert, P., Mumenthaler, C., and Wuthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Herrmann, T., Guntert, P., and Wuthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319 209–227. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1996. Mapping the protein universe. Science 273 595–603. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Kuboniwa, H., Grzesiek, S., Delaglio, F., and Bax, A. 1994. Measurement of HN-H alpha J couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR 4 871–878. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Link, A.J., Robison, K., and Church, G.M. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18 1259–1313. [DOI] [PubMed] [Google Scholar]
- Markely, J.L., Bax, A., Arata, Y., Hilbers, C.W., Kaptein, R., Sykes, B.D., Wright, P.E., and Wutrich, K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl. Chem. 70 117–142. [DOI] [PubMed] [Google Scholar]
- Ottiger, M. and Bax, A. 1998. Characterization of magnetically oriented phospholipid micelles for measurement of dipolar couplings in macromolecules. J. Biomol. NMR 12 361–372. [DOI] [PubMed] [Google Scholar]
- Pomposiello, P.J. and Demple, B. 2001. Redox-operated genetic switches: The SoxR and OxyR transcription factors. Trends Biotechnol. 19 109–114. [DOI] [PubMed] [Google Scholar]
- Pomposiello, P.J., Bennik, M.H., and Demple, B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183 3890–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello, P.J., Koutsolioutsou, A., Carrasco, D., and Demple, B. 2003. SoxRS-regulated expression and genetic analysis of the yggX gene of Escherichia coli. J. Bacteriol. 185 6624–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasinger, V.C. and Humphery-Smith, I. 1998. Small genes/gene-products in Escherichia coli K-12. FEMS Microbiol. Lett. 169 375–382. [DOI] [PubMed] [Google Scholar]
- Zweckstetter, M. and Bax, A. 2000. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J. Am. Chem. Soc. 122 3791–2792. [Google Scholar]