Abstract
The effects of spectral magnitude on the calculated secondary structures derived from circular dichroism (CD) spectra were examined for a number of the most commonly used algorithms and reference databases. Proteins with different secondary structures, ranging from mostly helical to mostly β-sheet, but which were not components of existing reference databases, were used as test systems. These proteins had known crystal structures, so it was possible to ascertain the effects of magnitude on both the accuracy of determining the secondary structure and the goodness-of-fit of the calculated structures to the experimental data. It was found that most algorithms are highly sensitive to spectral magnitude, and that the goodness-of-fit parameter may be a useful tool in assessing the correct scaling of the data. This means that parameters that affect magnitude, including calibration of the instrument, the spectral cell pathlength, and the protein concentration, must be accurately determined to obtain correct secondary structural analyses of proteins from CD data using empirical methods.
Keywords: circular dichroism (CD) spectroscopy, calibration, secondary structure analyses, synchrotron radiation circular dichroism (SRCD)
Circular dichroism (CD) spectroscopy can be a valuable method for determining the secondary structures of proteins. However, an accurate analysis relies on (among other things) having the correct magnitude for the spectrum. The determination of the magnitude of a circular dichroism spectrum can be adversely affected by a number of factors including errors in instrument calibration, cell pathlength, and protein concentration. Often these factors are not carefully considered when spectra are reported in the literature or analyzed by empirical secondary structure calculation methods.
A number of years ago we examined the effects of magnitude on constrained, unconstrained, and normalized least-squares methods of analyses of protein secondary structures from CD data (Wallace and Teeters 1987). We demonstrated that all except the normalized methods were highly influenced by the magnitudes of the spectra. Since that time, much more sophisticated algorithms have been developed for such empirical analyses, including variable selection, neural network, and principal component methods (van Stokkum et al. 1990; Andrade et al. 1993; Sreerama and Woody 2000). We therefore felt it was important to revisit this issue and examine the magnitude effects on these types of analyses, expanding our study to β-sheet and mixed proteins as well as mostly helical proteins. This study was facilitated by the availability of the Web server DICHROWEB (Lobley et al. 2002; Whitmore and Wallace 2004), which enables calculations using five different algorithms and seven different reference databases which can be used in various combinations, and for which it is possible to use an optional scale factor feature to facilely alter the magnitude of a spectrum to be analyzed.
Results
Test proteins
Several examples of each of the classes of all β, all α, and mixed secondary structures were examined using a wide range of algorithms, reference databases, and scaling factors. Each of the proteins reported herein were chosen for the study based on the following criteria: (1) the protein spectrum was not already in any of the existing reference databases, (2) the availability of highly purified protein, (3) the availability of an X-ray structure, and (4) the classification of the protein in the CATH database (Orengo et al. 1997) as a mainly α, mainly β, or a mixed α– β protein. Ceruloplasmin (CATH = 2.60.40.420) contains 34% β-sheet and 12% α-helix; avidin (CATH = 2.40.128.30) contains 50% β-sheet and 7% helix; serum albumin (CATH = 1.10.246.10) contains 72% helix and 0% β-sheet; glycogen phosphorylase (two domains: CATH = 3.90.270.10 and 3.40.670.10) contains 49% helix and 15% β-sheet. We have tried similar analyses (although not all permutations with all proteins) (data not shown) for a wide range of other proteins (> 70) whose spectra we have collected for a CD protein fold database (Wien et al. 2005) and find similar trends with respect to spectral magnitude for other proteins in the same classes.
CD spectra
Both CD and synchrotron radiation circular dichroism (SRCD) spectra were collected for the test proteins, for comparison. The SRCD spectra did not differ from the conventional CD (cCD) spectra of these proteins to any measurable extent over the wavelengths regions used in this study (Lees and Wallace 2002), but were used in preference to the CD spectra in the analyses, as they allowed lower wavelength data to be collected, and thus enabled the use of all the available reference databases, even those extending to 178 nm.
The magnitude of a CD spectrum, Δɛ, is defined as θ/3298cL, where θ is the measured ellipticity in milli-degrees, c is the concentration of the protein, and L is the pathlength of the cell. Hence, spectral magnitudes depend on a number of factors including the accurate determination of pathlength and concentration, as well as machine calibration (Miles et al. 2003). In this paper, the sum of all these potential sources of variations is represented by a single overall change in magnitude coefficient, the scale factor. Where the scale factor is 1.0, this corresponds to the “correct” magnitude based on careful calibration of the CD instrument and cell pathlength and determination of the protein concentration by (duplicate) quantitative amino acid (QAA) analyses.
Spectral magnitude effect on the accuracy of the structure determined
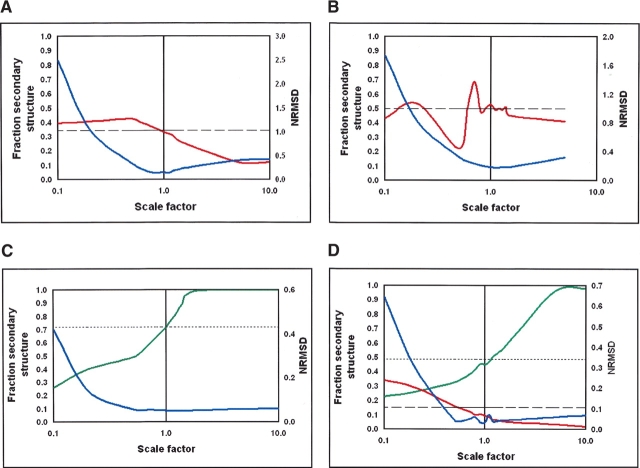
In this paper we have concentrated on the accurate determination of the principal secondary structure component, that is, helix for mainly helical proteins and sheet for mainly sheet proteins. It was found that errors in the magnitude adversely affected the secondary structure analyses for all examples examined. Figure 1 ▶ includes plots of the calculated secondary structures from the CD analyses as a function of scale factor; on these plots the actual secondary structure values determined from the crystal structures are shown as dotted and dashed lines for comparison. The plots show the trends observed for examples of mainly helical, mainly sheet, and mixed proteins. Because β-sheet proteins differ considerably not only in their structure but especially in their spectra, we chose to display the results from two mainly β-proteins that are very different spectrally, ceruloplasmin (Fig. 1A ▶) and avidin (Fig. 1B ▶). Similar trends are seen for these two widely diverse β-structures. For helical and mixed proteins, one example of each is shown in Figure 1, C and D ▶, respectively. The mixed proteins seem to be dominated by the helical components, and thus both the mostly helical (Fig. 1C ▶) and mixed proteins (Fig. 1D ▶) show the same sorts of trends. In all types of proteins, the closest correspondence with the actual secondary structure occurs when the structure factor is equal, or very close, to 1.0. In all cases, when the scale factor is >1.0, helix tends to be over-predicted and sheet underpredicted; when the scale factor is <1.0, the opposite is true. For all examples except avidin (which has an unusual spectrum (Wallace et al. 2004) and is relatively insensitive to differences in scale factors near 1.0), there is a strong, nearly linear, dependence as a function of the log of the scale factor near to 1.0, thus indicating the importance of correct magnitude values for the analyses.
Figure 1.
The effects of spectral magnitude on analyses using the CONTINLL method and reference dataset 1 for (A) ceruloplasmin (mainly β), (B) avidin (mainly β), (C) serum albumin (mainly β), and (D) glycogen phosphorylase (mixed). The calculated fractions of the β-strand are shown in red, the calculated fractions of the helix are shown green, and the NRMSD parameter is in blue. The dashed lines are the fractions of the strand, and the dotted lines are the fractions of the helix determined from the crystal structure.
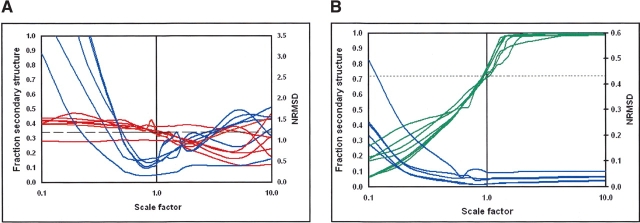
To examine whether the effects were reference database dependent, we examined the magnitude dependence using the seven available reference databases (Sreerama and Woody 2000), which have different protein components and cover different wavelength ranges. Figure 2, A and B ▶, shows that regardless of which reference database is used, the most correct answer is obtained with a scale factor of or very close to 1.0.
Figure 2.
The effects of spectral magnitude on analyses using the CONTINLL method with seven difference reference datasets for (A) ceruloplasmin, and (B) serum albumin. The color key is the same as used in Figure 1 ▶.
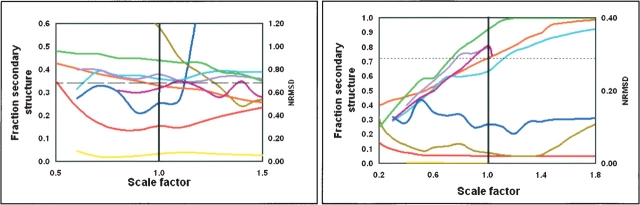
In this study, most of the analyses were done using CONTINLL, but to examine whether the effects were dependent on the method used, we tested five different algorithms, encompassing principal component, variable selection, and neural network methods. As seen in Figure 3, A and B ▶ (in which the central regions around scale factors of 1.0 are expanded to emphasize the differences), generally similar trends were obtained using all methods, although the actual results differed considerably. It should be noted that in a number of cases when the scale factor deviated significantly from 1.0, one or more of the methods failed to produce a solution. In the figures shown, this is indicated when a plot prematurely ends relative to the other plots. The method that appeared to be least sensitive to magnitude in terms of accuracy of secondary structure determined was CDSSTR, but this method was also the most intolerant to large deviations from correct scaling, producing no solution at all in many cases.
Figure 3.
The effects of spectral magnitude on analyses using five different algorithms to analyze (A) ceruloplasmin, and (B) serum albumin. In A, the calculated fractions of sheet are from the following: CONTINLL (orange), SELCON3 (light blue), CDSSTR (purple), VARSLC (pink), and K2d (light green). The NRMSD values are for CONTINLL (red), SELCON3 (dark blue), CDSSTR (yellow), and K2d (dark green). The dashed line is the fraction of strand calculated from the crystal structure. In B, the calculated fractions of helix are from the following: CONTINLL (orange), SELCON3 (light blue), CDSSTR (purple), VARSLC (pink), and K2d (light green). The NRMSD values are for CONTINLL (red), SELCON3 (dark blue), CDSSTR (yellow), and K2d (dark green). The dotted line is the fraction of the helix calculated from the crystal structure.
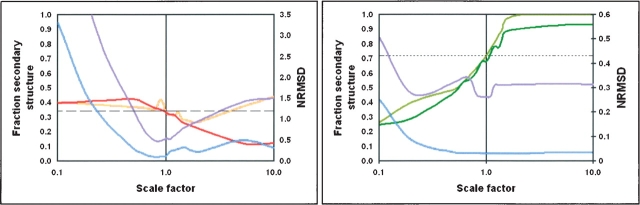
Finally, to examine the sensitivity of the calculations to the wavelength range of the data used in the analysis, comparisons were done using calculations done with the full spectral range (down to 178 nm), and with data truncated at 190 nm at the lower end (more similar to the data routinely obtained on a cCD instrument). While the plots were slightly different (Fig. 4 ▶), the trends for both the sheet and helical proteins were identical for the two wavelength ranges.
Figure 4.
The effects of spectral magnitude on analyses using the CONTINLL method with different wavelength ranges of data for (A) ceruloplasmin and (B) serum albumin. For A, the calculated sheet fractions for data to 178 nm (orange) and for data to 190 nm (red) are shown, as are the corresponding NRMSD values (blue and purple, respectively). For B, the calculated helix fractions for data to 178 nm (light green) and for data to 190 nm (dark green) are shown, as are the corresponding NRMSD values (blue and purple, respectively).
Spectral magnitude effect on goodness-of-fit
The normalized root-mean-square deviation (NRMSD) parameter is a useful means of comparing how well the best calculated structure correlates with the experimental data (Mao et al. 1982). It is defined as: Σ[( θexp − θcal)2/(θexp)2]1/2, summed over all wavelengths, where θexp and θcal are, respectively, the experimental ellipticities and the ellipticities of the back-calculated spectra for the derived structure. The NRMSD value is also plotted in all the figures as a function of scale factor. It is clear that in the case of mainly β-proteins, the NRMSD plots have a minimum at scale factor values at or close to 1.0, and coincide with the correct secondary structure. For mainly helical and mixed proteins, the NRMSD values are very high at low scale factors, but flatten out as the scale factor approaches 1.0; they do not significantly increase at increasing scale factors above 1.0. The principal exception to these trends is with the CDSSTR method (Fig. 3A,B ▶), where the NRMSD values are always low, and the plots are nearly featureless, thus providing little aid in identifying the correct magnitudes.
Discussion
It was previously shown mathematically and experimentally (Mao 1984; Wallace and Teeters 1987) that normalized least-squares–based methods of secondary structural analyses are essentially insensitive to protein concentration, while constrained and unconstrained least-squares analyses are highly sensitive to spectral magnitude (Wallace and Teeters 1987). Recently a procedure that normalized all spectra to a single wavelength prior to deconvolution was shown to similarly minimize magnitude sensitivity (Raussens et al. 2003). Both this and the normalized least-squares methods are effective, as they rely primarily on the relative spectral magnitudes and positions of the various transition peaks rather than absolute magnitudes. To test magnitude effects on analyses using the principle component, neural network, and variable selection algorithms, which are the methods most commonly in use today, we selected a number of proteins whose structures were known from crystallography, but which were not present in the existing databases used for empirical analyses. Their relative magnitudes were varied from 0.1 to 10 times the value that had been determined from the QAA analyses. For these studies, we had previously calibrated the instrument (Miles et al. 2003) and cell pathlength (Miles et al. 2005) accurately.
What was very clear was that the values of the calculated secondary structures tend to be closest to the actual values near the correct (1.0) scale factor, and that there is a strong relationship between the accurately calculated secondary structure and the scale factor. Perhaps more usefully for unknown proteins, the NRMSD values also seem to have minima near the correct scale factor. This would seem to suggest that a scan through potential scale factors to find the lowest NRMSD would be a way of determining the correct scaling for a spectrum collected without knowledge of the correct pathlength or protein concentration. However, while there is a general correlation, we believe it would be unwise to use this as the sole criterion for magnitude determination. While the NRMSD results for the mostly β-sheet are dramatically dependent on the correct magnitude, reaching a minimum at both the correct scale and the correct secondary structure, the cases for the mixed and mostly helical proteins are not as clear cut: In those cases when the scale factors are too small, the magnitude asymptotically reaches a minimum well before the scale factor is correct, while the secondary structure only reaches the correct value when the scale factor is correct. Hence, the lowest NRMSD may be a necessary but not sufficient condition for determining correct scale factor. However, while the NRMSD may not be an absolute determinant, it could form the basis of a useful test for correctness, or more importantly, incorrectness.
It is important to note that in this study we have not examined cases where unusual types of secondary structure are present in the unknown protein. In those cases the spectra are not well fit empirically even when the magnitude is correct.
It was observed that when the scale factor was >1.0, helical content was overpredicted, and β-sheet was under-predicted. This is understandable since the major differences between the sheet and helical spectral signatures are the magnitudes of the negative peaks between 210 and 230 nm, and the positive peaks around 190 nm, with the peaks in a helical spectrum having roughly five times the magnitudes of those in a sheet spectrum. Thus, it is not unexpected that if the magnitude is too high, the sheet content calculated would be sacrificed in favor of too much helix content. However, we expect that if in the future very low wavelength data to ~165 nm can be measured (i.e., using SRCD) and corresponding low wavelength reference data bases become available, the methods may be less sensitive to magnitude since at wavelengths between 165 and 178 nm, sheet and helix spectra differ not only in magnitude but in sign (Wallace 2000).
Factors that can affect magnitude include instrument calibration, cell pathlength (not always as reported by the manufacturer), protein concentration (not always that determined by gravimetric methods, and especially not as determined by colorimetric assays such as the BCA [Smith et al. 1985] and Lowry [Lowry et al. 1951] methods), and protein purity. In this study we have shown that errors in magnitude of spectra (from whatever source) can cause significant errors in empirical secondary structure analyses using principal component, neural network, and variable selection calculation algorithms.
Several algorithms have previously been shown to be useful for samples in which the protein concentration is unknown, including normalized least squares (Wallace and Teeters 1987), g-factor analyses (McPhie 2001), and a quadratic scaling method (Raussens et al. 2003). In the absence of accurate magnitude information, these methods can produce reasonable results, but if such information is available, other methods tend to produce more accurate analyses.
In the limited number of examples in present study, CONTINLL appeared to give the most accurate results when the magnitude was correct, and more importantly, its results were reasonably well correlated with the NRMSD value, which could provide the basis for testing of the magnitude effects. On the other hand, CDSSTR seemed to be the method least sensitive to magnitude variations, although it did not necessarily produce the most accurate results. However, any statistically valid trend discriminating between algorithms would have to be confirmed using a larger sample of proteins.
In summary, the current simple study has examined the effects of magnitude changes on the accuracy of empirical calculations using singular value deconvolution, neural network, and principal component analysis methods. It has demonstrated that correct knowledge of the parameters that contribute to the magnitude calculation, including path-length, protein concentration, and instrument calibration, are essential to produce accurate values for such empirical protein secondary structure analyses.
Materials and methods
Materials
Human serum ceruloplasmin and chicken avidin were purchased from Calbiochem. Human serum albumin and rabbit glycogen phosphorylase b were from Sigma Aldrich Chemical Co. All were of >95% purity. Prior to use, all were extensively dialyzed versus water using slide-A-lyzer mini dialysis units (MW 10 K cutoff) from Pierce. The dialysates were used for the spectral baselines. All samples were degassed under vacuum prior to use, and then centrifuged briefly to remove any undissolved material. Quantitative amino acid analyses were done in duplicate at the Protein and Nucleic Acid Chemistry Facility located at the University of Cambridge (UK).
CD spectra
SRCD spectra were collected at station CD12 located at the SRS Daresbury. Protein samples at ~10 mg/mL protein (the final protein concentrations were determined according to quantitative amino acid analysis) were examined in a circular demountable 0.0015 cm pathlength Suprasil cell (Hellma UK, Ltd.), which had been previously calibrated using both interferometry and chromate dilution methods (Miles et al. 2005). The instrument was calibrated using camphor sulfonic acid (CSA) at two wavelengths, using the recently redetermined (Miles et al. 2004) A285 value for this hygroscopic compound. Three spectra and three baselines were collected at 1 nm intervals over the wavelength range from 280 to 168 nm at 4°C. Measurements were only made down to wavelengths where the HT (high tension) indicated the detector was still in its linear range. CD spectra were collected on an Aviv 62ds instrument under similar conditions using the same cell. In this case, data were collected down to 185 nm.
CD spectral analyses
CD spectra were processed using CDtool software (Lees et al. 2004). The spectra were averaged, baseline subtracted, and smoothed with a Savitsky-Golay filter (Savitsky and Golay 1964), and zeroed between 263 and 270 nm. To calculate Δɛ values each spectrum was calibrated by a CSA file at two points. The mean residue weight value for each protein was calculated from its sequence, as follows: ceruloplasmin, 114.8; avidin, 112.1; serum albumin, 113.6; glycogen phosphorylase, 115.7.
Secondary structure analyses were performed with the DICHROWEB Web server (Lobley et al. 2002; Whitmore and Wallace 2004) using the following algorithms: CONTINLL (Provencher and Glockner 1981; van Stokkum et al. 1990), SELCON3 (Sreerama et al. 1999; Sreerama and Woody 2000), CDSSTR (Manavalan and Johnson 1987; Sreerama and Woody 2000), VARSLC (Compton and Johnson 1986; Manavalan and Johnson 1987), and K2d (Andrade et al. 1993), and seven different reference datasets (Sreerama and Woody 2000). Unless otherwise noted, reference dataset 1 (which uses data down to 178 nm) was used in the analyses (except for with VARSLC and K2d, which did not use external reference datasets). To change the effective spectral magnitude, the “optional scaling factor” function in DICHROWEB was used, enabling the multiplication of the input spectra by factors ranging from 0.5 to 1.5×. For larger variations in scale factor, the spectra were scaled using the CDtool software package (Lees et al. 2004).
A goodness-of-fit parameter (the NRMSD) (Mao et al. 1982) was calculated for all methods that produce back-calculated spectra (CONTINLL, SELCON3, CDSSTR, and K2d). Smaller values of NRMSD indicate closer correspondence between calculated structures and the experimental data. In addition to this parameter, because DICHROWEB plots the differences between the calculated and experimental spectra plus the difference spectra, the spectral features (i.e., magnitude) that are not well reproduced were evident in these plots (data not shown) and thus provide a further visual means of assessing the consequences of the error in magnitude.
Secondary structure calculations from crystal structures
The DSSP algorithm (Kabsch and Sander 1983) was applied to the PDB files for each of the proteins. Although there are a variety of ways of calculating secondary structure (King and Johnson 1999), this method was used because it was the method used to define the secondary structures as they appear in the reference databases (Sreerama et al. 2000). Here we report on the total helix content (corresponding to “helix 1” plus “helix 2” in some reference datasets, or α-plus distorted helices in others) and, likewise, the total β-sheet content, as the sum of all sheet types. For the examples shown, the PDB files used were as follows: ceruloplasmin (1kcw; Zaitseva et al. 1996), avidin (1rav; Nardone et al. 1998), serum albumin (1ao6; Sugio et al. 1999), glycogen phosphorylase (1gpb; Leonidas et al. 1992).
Acknowledgments
This work was supported by grants B14225 and B02959 from the BBSRC (to B.A.W.). The SRCD beamtime was provided by a grant from the CLRC (to B.A.W.). A.J.M. is the recipient of an MRC studentship. We thank Dr. F. Wien (Birkbeck College, London) for help with the SRCD data collection, and Dr. R.W. Janes (Queen Mary College, London) for helpful discussions. We thank Peter Sharratt of PNAC for the quantitative amino acid analyses.
Abbreviations
CD, circular dichroism
cCD, conventional circular dichroism
CSA, camphor sulfonic acid
NRMSD, normalized root-mean-square deviation
QAA, quantitative amino acid analysis
SRCD, synchrotron radiation circular dichroism
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041019905.
References
- Andrade, M.A., Chacón, P., Merelo, J.J., and Morán, F. 1993. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng. 6 383–390. [DOI] [PubMed] [Google Scholar]
- Compton, L.A. and Johnson Jr., W.C. 1986. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155 155–167. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- King, S.M. and Johnson Jr., W.C. 1999. Assigning secondary structure from protein coordinate data. Proteins 35 313–320. [PubMed] [Google Scholar]
- Lees, J. and Wallace, B.A. 2002. Synchrotron radiation circular dichroism and conventional circular dichroism spectroscopy: A comparison. Spectroscopy 16 121–125. [Google Scholar]
- Lees, J.G., Smith, B.R., Wien, F., Miles, A.J., and Wallace, B.A. 2004. CDtool—An integrated software package for circular dichroism spectroscopic data processing, analysis and archiving. Anal. Biochem. 332 285–289. [DOI] [PubMed] [Google Scholar]
- Leonidas, D.D., Oikonomakos, N.G., Papageorgiou, A.C., Acharya, K.R., Barford, D., and Johnson, L.N. 1992. Control of phosphorylase-b conformation by a modified cofactor—Crystallographic studies on R-state glycogen-phosphorylase reconstituted with pyridoxal 5′-diphosphate. Protein Sci. 1 1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobley, A., Whitmore, L., and Wallace, B.A. 2002. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18 211–212. [DOI] [PubMed] [Google Scholar]
- Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193 265–275. [PubMed] [Google Scholar]
- Mao, D. 1984. “An analysis of membrane protein structures using circular dichroism spectroscopy.” Ph.D. thesis, Columbia University, New York.
- Mao, D., Wachter, E., and Wallace, B.A. 1982. Folding of the H+-ATPase proteolipid in phospholipid vesicles. Biochemistry 21 4960–4968. [DOI] [PubMed] [Google Scholar]
- McPhie, P. 2001. Circular dichroism studies on proteins in films and in solution: Estimation of secondary structure by g-factor analysis. Anal. Biochem. 293 109–119. [DOI] [PubMed] [Google Scholar]
- Miles, A.J., Wien, F., Lees, J.G., Rodger, A., Janes, R.W., and Wallace, B.A. 2003. Calibration and standardisation of synchrotron radiation circular dichroism and conventional circular dichroism (cCD) spectrophotometers. Spectroscopy 17 653–661. [Google Scholar]
- Miles, A.J., Wien, F., and Wallace, B.A. 2004. Redetermination of the extinction coefficient of camphor-10-sulfonic acid, a calibration standard for circular dichroism spectroscopy. Anal. Biochem. 335 338–339. [DOI] [PubMed] [Google Scholar]
- Miles, A.J., Wien, F., Lees, J.G., and Wallace, B.A. 2005. Calibration and standardisation of synchrotron radiation and conventional circular dichroism spectrometers. Part 2: Factors affecting magnitude and wavelength. Spectroscopy (in press).
- Nardone, E., Rosano, C., Santambrogio, P., Curnis, F., Corti, A., Magni, F., Siccardi, A.G., Paganelli, G., Losso, R., Apreda, B., et al. 1998. Biochemical characterization and crystal structure of a recombinant hen avidin and its acidic mutant expressed in Escherichia coli. Eur. J. Biochem. 256 453–460. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B., and Thornton, J.M. 1997. CATH—A hierarchic classification of protein domain structures. Structure 5 1093–1108. [DOI] [PubMed] [Google Scholar]
- Provencher, S.W. and Glockner, J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20 33–37. [DOI] [PubMed] [Google Scholar]
- Raussens, V., Ruysschaert, J.-M., and Goormaghtigh, E. 2003. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 319 114–121. [DOI] [PubMed] [Google Scholar]
- Savitsky, A. and Golay, M.J.E. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36 1627–1639. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. [DOI] [PubMed] [Google Scholar]
- Sreerama, N. and Woody, R.W. 2000. Estimation of protein secondary structure from CD spectra: Comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal. Biochem. 282 252–260. [DOI] [PubMed] [Google Scholar]
- Sreerama, N., Venyaminov, S.Y., and Woody, R.W. 1999. Estimation of the number of helical and strand segments in proteins using CD spectroscopy. Protein Sci. 8 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Estimation of protein secondary structure from CD spectra: Inclusion of denatured proteins with native protein in the analysis. Anal. Biochem. 287 243–251. [DOI] [PubMed] [Google Scholar]
- Sugio, S., Kashima, A., Mochizuki, S., Noda, M., and Kobayashi, K. 1999. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 12 439–446. [DOI] [PubMed] [Google Scholar]
- van Stokkum, I.H.M., Spoelder, H.J.W., Bloemendal, M., van Grondelle, R., and Groen, F.C.A. 1990. Estimation of protein secondary structure and error analysis from CD spectra. Anal. Biochem. 191 110–118. [DOI] [PubMed] [Google Scholar]
- Wallace, B.A. 2000. Synchrotron radiation circular dichroism spectroscopy as a tool for investigating protein structures. J. Synch. Rad. 7 289–295. [DOI] [PubMed] [Google Scholar]
- Wallace, B.A. and Teeters, C.L. 1987. Differential absorption flattening optical effects are significant in the circular dichroism spectra of large membrane fragments. Biochemistry 26 65–70. [DOI] [PubMed] [Google Scholar]
- Wallace, B.A., Wien, F., Miles, A.J., Lees, J.G., Hoffman, S.V., Evans, P., Wistow, G.J., and Slingsby, C. 2004. Biomedical applications of synchrotron radiation circular dichroism spectroscopy: Identification of mutant proteins associated with disease and development of a reference database for fold motifs. Faraday Discuss. 17 653–661. [DOI] [PubMed] [Google Scholar]
- Whitmore, L., and Wallace, B.A. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32 W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wien, F., Miles, A.J., Lees, J., Cuff, A.L., Janes, R.W., and Wallace, B.A. 2005. A new circular dichroism reference dataset covering foldspace. Biophys. J. (in press).
- Zaitseva, I., Zaitsev, V., Card, G., Moshkov, K., Bax, B., Ralph, A., and Lindley, P. 1996. The X-ray structure of human serum ceruloplasmin at 3.1 angstrom: Nature of the copper centres. J. Biol. Inorg. Chem. 1 15–23. [Google Scholar]