Abstract
Protein subunits of several RNA viruses are known to undergo post-assembly, autocatalytic cleavage that is required for infectivity. Nudaurelia capensis ω virus (Nω V) is one of the simplest viruses to undergo an autocatalytic cleavage, making it an excellent model to understand both assembly and the mechanism of autoproteolysis. Heterologous expression of the coat protein gene of Nω V in a baculovirus system results in the spontaneous assembly of virus-like particles (VLPs) that remain uncleaved when purified at neutral pH. After acidification to pH 5.0, the VLPs autocatalytically cleave at residue 570, providing an in vitro control of the cleavage. The crystal structure of Nω V displays three residues near the scissile bond that were candidates for participation in the reaction. These were changed by site-directed mutagenesis to conservative and nonconservative residues and the products analyzed. Even conservative changes at the three residues dramatically reduced cleavage when the subunits assembled properly. Unexpectedly, we discovered that these residues are not only critical to the kinetics of Nω V autoproteolysis, but are also necessary for proper folding of subunits and, ultimately, assembly of Nω V VLPs.
Keywords: autocatalysis, autoproteolysis, virus maturation, Nudaurelia capensis ω virus, tetravirus, nodavirus
Nudaurelia capensis ω virus (Nω V) is the prototype of the ω-tetraviruses; nonenveloped, single-stranded, positive-sense RNA viruses with T = 4 icosahedral symmetry and bipartite genomes. The two RNA molecules of Nω V are copackaged in each viral capsid; RNA1 is 5.3 kb and codes for the RNA-dependent RNA polymerase, while RNA2 is 2.4 kb and encodes the 70-kDa coat protein precursor termed α. Tetraviruses exclusively infect insects of the Lepidopteran order and, with the exception of Providence Virus (a β-tetravirus) (Pringle et al. 2003), a cell culture system has not been established to support propagation of authentic tetravirus particles (Hanzlik and Gordon 1997). However, when Nω V RNA2 is expressed in a recombinant baculovirus expression system, 240 chemically identical copies of protein α assemble into a porous but stable procapsid intermediate that, after acidification (pH 5.0), undergoes a large-scale conformational change into the mature capsid (Canady et al. 2000). Assembly and maturation in the expression system occurs in the absence of RNA1 with mostly cellular RNA packaged in the virus-like particle (VLP) (Agrawal and Johnson 1995). The conformational change is followed by an autoproteolytic cleavage that requires pH 5.0. The cleavage prevents the capsid from reverting to the procapsid form even if the pH is raised back to 7.0 or higher. The conformational change is rapid (<100 msec) while the cleavage of the α coat protein to 62 kDa β and 8 kDa γ products is slow (t1/2 ~1 h) (Agrawal and Johnson 1992; Canady et al. 2001). We previously demonstrated that the asparagine at the scissile bond (Asn-570–Phe-571) is required for cleavage, and that a point mutation to threonine (N570T) allows the particles to undergo the conformational change from procapsid to capsid reversibly as a function of pH (Taylor et al. 2002).
The crystal structure of the mature capsid of authentic NωV virion revealed that the coat protein subunit is comprised of three different regions: the outer Ig-like domain, the canonical virus β-barrel observed in the majority of viral capsid proteins, and an interior helical domain (Munshi et al. 1996; Fig. 1A ▶). The scissile bond of the autocatalytic cleavage site lies in the helical region and is juxtaposed to side chains of residues from intrasubunit β-strands (see Fig. 1B ▶). Like many virus capsid proteins, the termini of the NωV coat proteins are disordered in the crystal structure. Located at the interior portion of the protein shell, both the N- and C-terminal regions are arginine-rich, suggesting an involvement in electrostatic interactions with packaged RNA.
Figure 1.
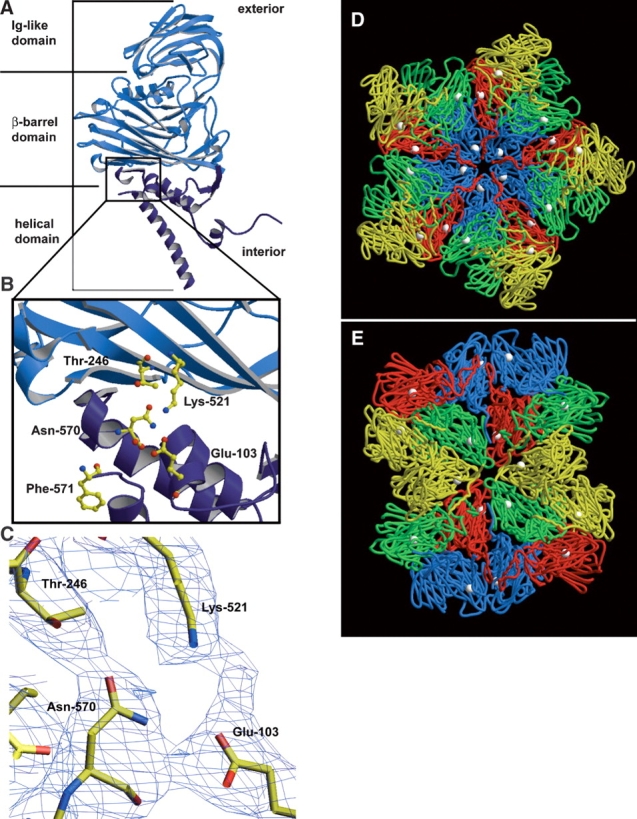
The crystal structure of NωV reveals that the cleavage site is located on the interior of the capsid in the helical region, near the β-barrel domain of the coat protein subunit. The helical region is colored dark blue in A and B. (A) A ribbon diagram of the X-ray structure of the NωV A subunit. The NωV capsid is composed of 60 copies each of A, B, C, and D subunits, which are from the same gene and related by quasi-equivalence. (B) Magnified view of the cleavage site of the NωV coat protein with ball-and-stick representations of residues believed to be involved in the autocatalytic cleavage that occurs between Asn-570 and Phe-571. (C) Electron density of intrasubunit residues near the cleavage site of NωV contoured at 0.8 σ. (D) The crystal structure of NωV capsid at the icosahedral fivefold axis of symmetry and (E) the icosahedral twofold axis of symmetry from the inside of the capsid looking out. A subunits are displayed in blue, B subunits in red, C subunits in green, D subunits in yellow, and asparagine-570 of each coat protein subunit as white spheres in D and E. The ordered N termini of neighboring subunits are approximately 10–14 Å from carbonyl carbons of Asn-570. A, B, D, and E were made with Molscript (Kraulis 1991) and Raster3D (Merritt and Murphy 1994). C was made with the program O (Jones et al. 1991).
The similarity in atomic structures in the region of the scissile bond of different RNA viruses that undergo auto-catalytic cleavage at an asparagine residue is striking, suggesting similar mechanisms of autoproteolysis. In addition to the asparagine, there are nearby residues that are comparable to NωV in the side chain and location relative to the scissile bond in nodaviruses (Fisher and Johnson 1993) and reoviruses (Liemann et al. 2002). The electron density among these residues in NωV suggests some important intrasubunit interactions during assembly and autoproteolysis (Fig. 1C ▶). The crystal structure of NωV also shows that the cleavage site is exposed on the interior of the virus capsid and within 10–14 Å of ordered residues in neighboring coat protein subunits (Fig. 1D,E ▶). We targeted three residues in NωV that are proximal to the cleavage site in the crystal structure—Glu-103, Thr-246, and Lys-521—by site-directed mutagenesis in an attempt to understand their role in the cleavage mechanism. Cleavage was clearly affected in those mutants that properly assembled; however, we discovered that point mutations to these three residues also altered, and in some instances inhibited, assembly of NωV VLPs.
Materials and methods
Construction of NωV VLP mutants
A 5′–BamHI and 3′–XbaI restriction site and all point mutations to the NωV coat protein were generated by site-directed mutagenesis and the PCR primer extension method as previously described (Taylor et al. 2002). The mutated coat protein gene was inserted into the multiple cloning site of the baculovirus transfer vector, pBacPAK-9 (GibcoBRL). pBacPAK-9 was transfected with pBacPAK-6 and the two were allowed to recombine in vivo as described by the protocol for the BAK-to-BAK recombinant baculo-virus expression system (GibcoBRL). Positive constructs were screened by immunoblotting as previously described (Taylor et al. 2002) and positive constructs were plaque purified, amplified, and titered.
Infection of cells
Spodoptera frugiperda 21 (Sf21) cells were infected as a previously described (Taylor et al. 2002). Briefly, a monolayer of cells at a density of 8 × 106 cells/dish was infected in 100-mm dishes at a multiplicity of infection of 5. A total of 64 × 106 cells were infected for each construct. After 1 h of gentle rocking, 5 mL of warm 10% fetal bovine serum supplemented TC100 with 1× penicillin and 1× streptomycin was added to each dish. Cells were incubated for 6 days at 26°C.
Purification of VLPs
Each construct was purified at pH 7.6 in Tris-HCl (condition for wild-type procapsid) and at pH 5.0 (condition for wild-type capsid) in sodium acetate. At 6 d postinfection, cells were washed off dishes with gentle pipetting and lysed with the addition of 0.5% Nonidet P-40 and gentle rocking for 15 min on ice. Cellular debris was removed by centrifugation for 10 min at 4°C and 10,000 rpm in a Beckman JA-14 rotor. Supernatant was loaded in Beckman 50.2 Ti tubes with 2.5 mL of a 30% sucrose cushion prepared in 50 mM of either Tris-HCl (pH 7.6) or sodium acetate (pH 5.0) and 250 mM NaCl. Samples were centrifuged at 30,000 rpm in a Beckman 50.2 Ti rotor for 3 h at 11°C. Supernatant was discarded and 400 μ1 of either 50 mM Tris-HCl (pH 7.6) or 50 mM sodium acetate (pH 5.0) and 250 mM NaCl was added to the pellets which were allowed to resuspend overnight at 4°C. The next day, pellets were completely resuspended with pipetting and insoluble material was removed by low-speed centrifugation.
Each sample was sedimented on a 10% to 40% sucrose gradient prepared with 250 mM NaCl and 50 mM Tris-HCl (pH 7.6) or 50 mM sodium acetate (pH 5.0). Sucrose gradients were made with a Biocomp Gradient Master and centrifuged in a Beckman SW41 rotor at 40,000 rpm for 1.25 h at 11°C. After centrifugation, sucrose gradients were fractionated on an ISCO fractionator with constant OD254 reading, a pump setting of 0.75 mL/min, and collections at 30-sec intervals.
Sample analysis by electron microscopy
The OD254 indicated the fraction containing the highest concentration of VLPs, which was used for negative stained electron microscopy. Parlodium covered copper grids (400 mesh) were first treated with 2 mM β-octyl glucoside and samples were stained in 1% uranyl acetate (Ted Pella) as previously described (Dong et al. 1998). Five microliters of VLPs were allowed to adsorb to the grids for 2 min before staining. Samples were observed with a Phillips CM100 transmission electron microscope operating at a voltage of 80 kV.
Gel electrophoresis
Sucrose-purified fractions containing the highest concentration of VLPs were mixed with Laemmli buffer (Laemmli 1970) and heated to 95°C for 10 min. Samples were then loaded on a precast, NuPage 4%–12% Bis-Tris gel (Invitrogen) and run at 200 V for approximately 35 min in MES buffer following the protocol from Invitrogen.
Results
Wild-type phenotype
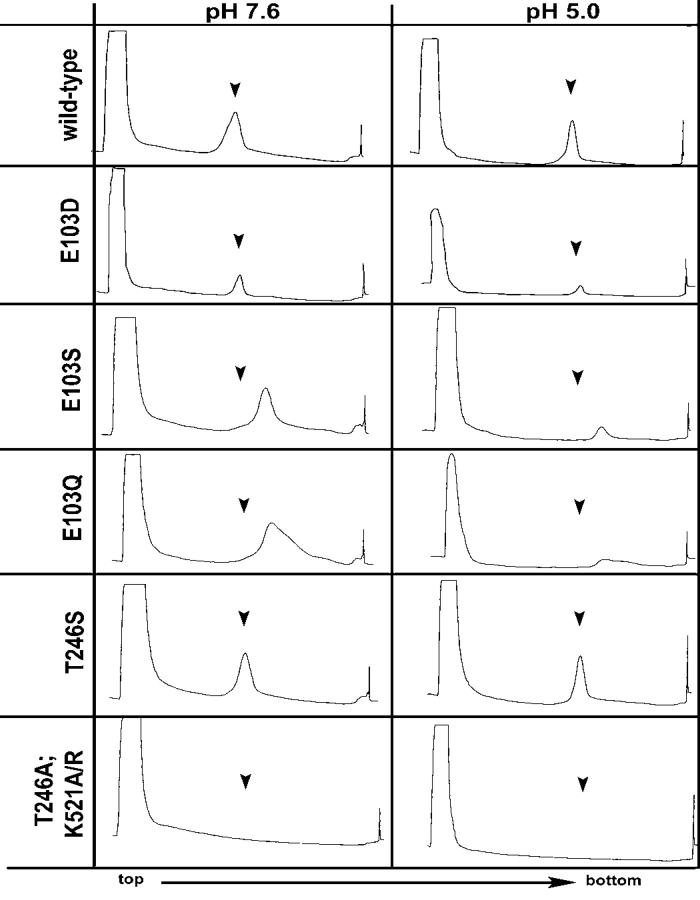
Wild-type particles were produced for positive controls as procapsid at pH 7.6, and as capsid at pH 5.0. Radioisotope differential labeling indicated that the procapsid and capsid populations of NωV sediment at different rates on a velocity sucrose gradient (Taylor et al. 2002). Following purification as described above, the procapsid particles sediment at fractions 17 and 18 (Fig. 2A ▶) on a 10%–40% sucrose gradient. The capsid reproducibly sediments at fraction 19 on an equivalent gradient prepared at pH 5.0 (Fig. 2B ▶). Negative-stained electron micrographs revealed that the procapsid population of wild-type particles was porous and penetrable to the stain. Procapsids were also labile, with several particles breaking under the staining conditions (Fig. 3A ▶). Wild-type capsid particles were more robust than procapsid and were generally impenetrable to the stain (Fig. 3B ▶).
Figure 2.
Constant UV absorbance (OD254) of 10%–40% sucrose gradients used to purify wild-type and mutant NωV VLPs at pH 7.6 and pH 5.0. Arrows in each window indicate where wild-type VLPs sediment under identical conditions.
Figure 3.
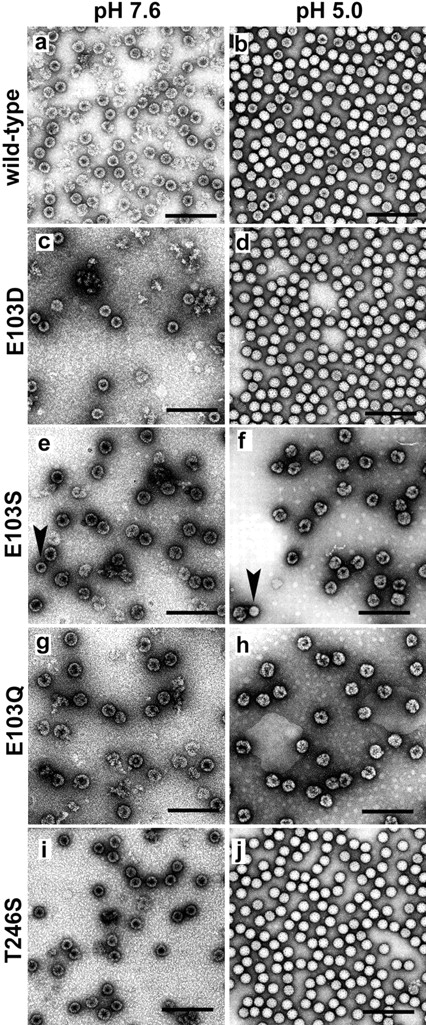
Negative stained electron micrographs of purified wild-type and mutant NωV VLPs at pH 7.6 and pH 5.0. E103Q (g,h) and E103S (e, f ) formed aberrant assembly products. E103D (c,d) and T246S (i, j ) formed particles that resembled wild type (a,b). No particles were detected for T246A, K521A, and K521R. Arrows in (e) and ( f ) indicate E103S mutant particles that resemble wild-type particles in size and appearance. Scale bar is 200 nm.
Different point mutations to Glu103 result in different phenotypes
Table 1 summarizes the point mutations made to the NωV coat protein and the consequent phenotypes observed in particle morphology and coat protein cleavage. When Glu-103 was mutated to an aspartic acid (E103D), particles displayed the wild-type phenotype for procapsid and capsid on a 10% to 40% sucrose gradient (Fig. 2C,D ▶) and by negative stain electron microscopy (Fig. 3C,D ▶). The yield of particles, both procapsid and capsid, was reduced ~10-fold when compared to wild type.
Table 1.
Results of point mutations made near asparagine 570 in the NωV coat protein
| Mutation | Coat protein phenotype | Cleavage after maturation |
| K521A | insoluble | — |
| K521R | insoluble | — |
| T246A | insoluble | — |
| T246S | wild type | yesa |
| E103D | wild type | yesa |
| E103Q | aberrant assembly | no |
| E103S | aberrant assembly | no |
Phenotypes were characterized by velocity gradients and negative stained electron microscopy at pH 7.6 and pH 5.0. Cleavage at pH 5.0 was detected using gel electrophoresis.
aCleavage occurs, but at a significantly reduced rate compared to wild type.
Glu-103 mutations to a serine (E103S) resulted in particles that sediment faster than wild-type procapsid (Fig. 2E ▶) and capsid (Fig. 2F ▶) at pH 7.6 and pH 5.0, respectively. Negative-stained electron micrographs revealed that the E103S mutation prevented proper assembly with larger than normal particles formed at pH 7.6 (Fig. 3E ▶) and pH 5.0 (Fig. 3F ▶). In general, the diameter of E103S particles were approximately 150% larger than wild type at pH 7.6 and pH 5.0. A few E103S particles resembled expected wild-type particles in size and morphology at pH 7.6 and 5.0, but less than one in a hundred (see arrows in Fig. 3E,F ▶) were identified. Mutations of Glu-103 to glutamine (E103Q) resulted in the E103S EM phenotype. Sucrose sedimentation, however, revealed that E103Q particles sediment in a broader band on the gradient suggesting more heterogeneity than wild-type or E103S particles (Fig. 2G,H ▶). Except for the significant increase in diameter, both the E103S and E103Q particles resembled wild-type procapsids at pH 7.6 by forming large, round particles that were penetrated by the stain (Fig. 3E,G ▶). In contrast to wild type, E103S and E103Q particles form misshapen, heterogeneous aggregates of coat proteins after reducing the pH to 5.0 (Fig. 3F,H ▶). The acid maturation does cause the E103S and E103Q particles to sediment faster on the sucrose gradient, suggesting a pH-dependent rearrangement of coat protein subunits is preserved in the mutants, analogous to wild type.
T246S resembles wild-type phenotype, but T246A, K521A, and K521R result in insoluble coat protein
Thr-246 and Lys-521are two residues in the NωV coat protein that are (1) close to the cleavage site in the X-ray model; (2) conserved in all the tetraviruses; and (3) similar or identical to residues in the X-ray model of other virus structural proteins with asparagine at the scissile bond, i.e., nodaviruses (Fisher and Johnson 1993; Wery et al. 1994), reovirus μ1 (Liemann et al. 2002), picornaviruses (Page et al. 1988; Kim et al. 1989); and (4) believed to have a role in the autocatalytic cleavage event (Taylor et al. 2002). Replacing Thr246 with serine (T246S) resulted in particles that sediment on a sucrose gradient at the same rate as wild-type particles at both pH 7.6 and pH 5.0 (Fig. 2I,J ▶). Negative-stained micrographs revealed particles that displayed wild-type phenotypes at pH 7.6 for procapsids and pH 5.0 for capsids (Fig. 3I,J ▶).
Mutations of Thr-246 to Ala (T246A) resulted in no detectable particle assembly by sucrose sedimentation and UV absorbance of the gradient (Fig. 2K,L ▶). Immunoblotting detected very low levels of NωV coat protein in the high-speed sucrose pellet and high levels of the coat protein in the low-speed spin used to remove insoluble cellular debris after lysis (data not shown). The same results were observed when Lys-521 was mutated to either an alanine (K521A) or to an arginine (K521R). The phenotypes of mutations at Thr-246 and Lys-521 are summarized in Table 1.
Autocatalysis monitored by gel electrophoresis
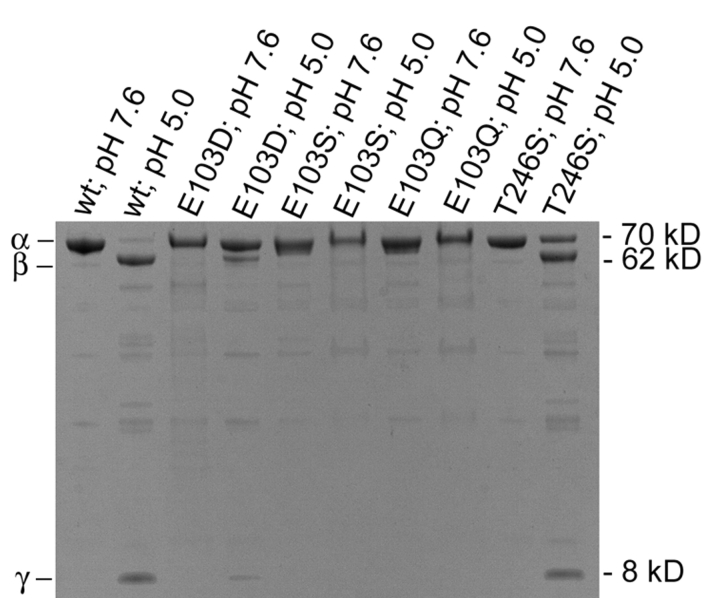
A 4%–12% Bis-Tris denaturing gel was employed to test for cleavage of the coat protein α into the 62 kDa β and 8 kDa γ polypeptides (Fig. 4 ▶). As expected, none of the mutants cleaved at pH 7.6. Cleavage of wild-type VLPs occurred only after the pH was reduced to 5.0. Analogous to wild type, both E103D and T246S mutants cleaved at pH 5.0, but to a lesser degree. Incubation at room temperature for 3 h revealed that the coat protein of wild-type capsid was almost entirely cleaved. Conversely, only about half of the T246S mutant subunits and much less of the E103D mutant subunits cleaved after 3 h incubations at room temperature. Both the E103S and E103Q mutations displayed no cleavage as detected by Coomassie staining, even after incubating the samples at room temperature overnight at pH 5.0.
Figure 4.
4% to 12% Bis-Tris gel used to detect cleavage of the NωV coat protein, α(70 kDa), into β(62 kDa) and γ(8 kDa) polypeptides. Cleavage of wild-type coat protein occurs only after acidification to pH 5.0. E103D and T246S mutants cleaved at pH 5.0, but significantly less than wild type under identical conditions. No cleavage was detected in the aberrantly assembled E103Q and E103S mutants, even at pH 5.0.
Discussion
Based on the refined crystal structure of mature NωV capsids (Munshi et al. 1996; Helgstrand et al. 2004), we examined residues that were candidates for catalyzing the maturation cleavage. The residues were chosen due to their proximity to the scissile bond between Asn-570 and Phe-571 and include Lys-521, Glu-103, and Thr-246. We discovered that this region not only affects cleavage in the mature NωV virions, but that the residues adjacent to this site are also important for proper assembly of particles. Mutations to the conserved Lys-521 of NωV to alanine or arginine, or Thr-246 to Ala, resulted in the expression of coat protein, but assembly of VLPs was not detected. Instead, the expressed viral protein aggregated and was insoluble, indicating misfolding of the polypeptide. Other point mutations that alter protein–protein interactions such as sickle cell anemia, the result of a glutamate to valine mutation in hemoglobin, are well documented (Ingram 1956). The hydrophobic point mutation in deoxygenated hemoglobin S tetramers causes abnormal clumping, and ultimately polymerization into long, aggregated protein fibers (Briehl 1980). While the point mutations, K521A, K521R, and T246A, to the NωV coat protein are more conservative than the glutamate to valine in hemoglobin S, they still lead to protein aggregation. It is possible that the T246A, K521A, and K521R point mutations lead to aggregation of the NωV coat protein by altering local folding enough to change surface hydrophobicity of the subunits near the cleavage site.
We previously reported that mutations of the scissile asparagine to threonine (N570T) yielded production of particles with about three times the efficiency of wild type (Taylor et al. 2002). The sensitivity of the active site of NωV cleavage is further demonstrated by mutations to Glu-103. Expression of E103S and E103Q constructs resulted in aberrant assembly products. The E103D mutation resulted in particles with wild-type phenotypes, but yields of VLPs were about an order of magnitude less than that of wild type. It is apparent that the structure of NωV has evolved to optimize the residues proximal to the scissile bond. This is not surprising, considering that the fine-tuned function of this area generates the relatively rapid cleavage of the coat protein at Asn-570. One cause for cataracts is a well-documented deamidation or cleavage at asparagine residues in the protein crystalline (Voorter et al. 1988) and the mechanism proposed for this is analogous to that described for NωV (Patel and Borchardt 1990; Perler et al. 1997). Fortunately, this process takes decades in crystalline while tetra-viruses like NωV have adapted the local environment of the coat protein to catalyze the cleavage with a half-life of about 1 h (Canady et al. 2001).
Given the relatively large size of the coat protein (644 residues) it is interesting that a single-point mutation would have such a dramatic impact on folding or the assembly of VLPs. The crystal structure of the mature capsid shows that the cleavage site is highly exposed on the interior, helical portion of the subunits, which also contain the disordered termini of the coat proteins (Fig. 1 ▶). Umashankar et al. (2003) have reported single-point mutations to residues involved with intersubunit interactions of Physalis mottle tymovirus coat proteins that lead to partial misfolding. The misfolded proteins remained soluble, but were incapable of assembling particles. The crystal structure of mature NωV shows that the ordered regions of the nearest neighboring subunit is more than 10 Å from the α carbon of Asn-570 (Fig. 1D,E ▶). This distance suggests that Lys-521, Glu-103, and Thr-246 are not directly involved in intersubunit contacts. However, it is difficult to directly interpret the crystal structure of the mature capsid for two important reasons: (1) The point mutations here affect assembly of the much larger procapsid (16% larger diameter than the mature form viewed in the crystal structure), and (2) most changes during maturation occur in the helical region where the point mutations were made (Canady et al. 2000). The helical region contains the molecular switch (residues ~600 to ~644) which defines either a bent (~138°) or flat (~180°) subunit contact in the T = 4 capsid. We propose that local folding is slightly disrupted near the point mutations, which consequently alters those dihedral angles. Assembly requires 240 copies of the subunit in the T = 4 capsid, thus dramatically amplifying the effect of any single change in the polypeptide (Johnson and Reddy 1998). The point mutations near the cleavage site could slightly alter this geometry of subunit–subunit contacts, resulting in abnormally sized VLPs. The fact that the E103S and E103Q mutants assemble as significantly larger particles supports this conclusion.
Asparagine at the cleavage site of NωV, and the conserved residues proximal to it, resemble the autocatalytic cleavage site in several, diverse RNA viruses including the coat protein of nodaviruses (Fisher and Johnson 1993) and the μ13σ13 heterohexamer of reovirus (Liemann et al. 2002). The coat proteins of picornaviruses such as human rhinovirus (Kim et al. 1989) and poliovirus (Page et al. 1988) also have an asparagine at the site of cleavage, but a different mechanism has been proposed that depends on a critical histidine residue proximal to the scissile bond (Hindiyeh et al. 1999). The role of cleavage in NωV is believed to play a role in cell entry and/or infection. Without a cell culture system available this hypothesis cannot be tested, but analogies with related RNA viruses support this assumption. The μ1 protein of reovirus forms the μ13σ13 heterohexameric complex that is responsible for the penetration of the host cell membrane in infections (Borsa et al. 1979). The catalysis of the μ1 protein depends on the presence of asparagine and proline residues at positions 42 and 43, respectively, at the cleavage site (Tillotson and Shatkin 1992). In addition to the asparagine residue at the scissile bond and analogous to NωV, the reovirus heterohexamer has conserved lysine and glutamic acid residues proximal to the cleavage site (Liemann et al. 2002); however, the mechanism of μ1 cleavage remains unclear. In the case of the nodavirus, Flockhouse virus (FHV), the asparagine is required for cleavage and essential for virion infectivity (Schneemann et al. 1992). It has been hypothesized that cleavage of the C terminus in FHV coat protein is required to release γ peptides, which are structurally similar to the γ peptides of NωV, and that these alter cellular membranes for translocation of the RNA into the infected host (Cheng et al. 1994).
We proposed a mechanism for the cleavage of tetraviruses based on the crystal structure at the cleavage site (Taylor et al. 2002; Fig. 5 ▶). In NωV, the carbonyl group of Glu-103 is believed to hydrogen bond with the amide proton on the side chain of Asn-570. This interaction polarizes the nitrogen of the asparagine side chain, enabling it to perform a nucleophilic attack on its own main chain carbonyl carbon resulting in the formation of a succinimide intermediate (Fig. 5 ▶). The succinimide intermediate is quickly opened by hydrolysis of a water molecule into either asparagine or iso-asparagine. Because of proximity to the scissile bond in NωV, Thr-246 is believed to facilitate the cleavage by also hydrogen bonding with the side chain of Asn-570. Lys-521 is conserved throughout the known coat protein sequences of tetraviruses and could have a similar role to Thr-246 by forming favorable interactions with the side chain of asparagine to facilitate cleavage.
Figure 5.
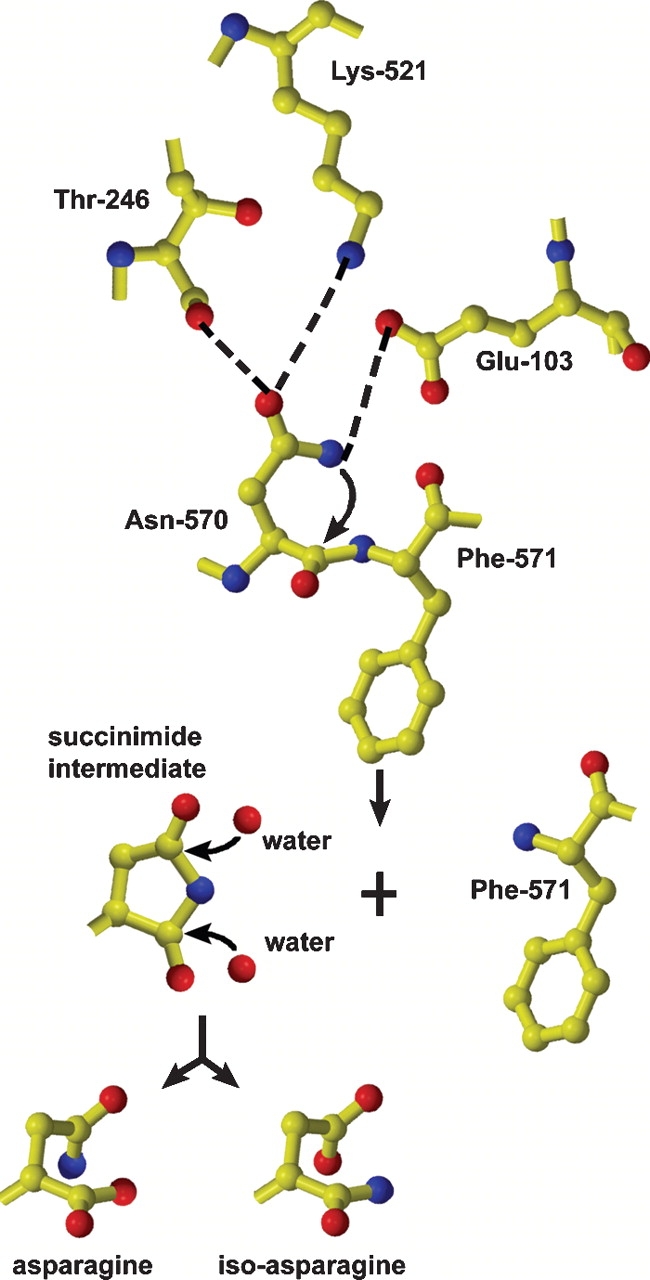
Proposed autocatalytic cleavage mechanism of NωV coat protein based on the deamidation mechanism of the protein crystalline. See text for details.
The difficulty in testing this theory arises in that the mutant NωV VLPs must first assemble properly before cleavage can be determined. Only two mutations, E103D and T246S, met this criterion and were further analyzed to test the proposed cleavage mechanism. Mutations of Asp-75 in the FHV coat protein (analogous to Glu-103 of NωV) to valine, glutamic acid, asparagine, or threonine resulted in cleavage-defective FHV particles (Zlotnick et al. 1994). Mutations to the acidic residue (Glu-103) of NωV are more sensitive than Asp-75 of FHV because the only point mutation at Glu-103 in NωV to properly assemble particles was E103D. The side chain of aspartate is slightly shorter than that of glutamate, but the chemical properties of the two side chains are conserved. Aspartate at position 103 of NωV could still hydrogen bond to the amide group of the asparagine but with less efficiency since its shorter side chain would increase the distance for the hydrogen bond to form. This anticipation is confirmed by gel electrophoresis, which indicates that after 3 h incubation at room temperature (pH 5.0) the E103D mutant cleaves significantly less than the wild-type capsid (Fig. 4 ▶).
The T246S mutation was also less efficient for autocatalysis than the wild type, supporting a role for Thr-246 in the cleavage mechanism. We hypothesize that threonine at position 246 hydrogen bonds more efficiently to the asparagine further constraining the asparagine side chain. This interaction is likely not essential for autoproteolysis, but increases the rate by minimizing movement of the asparagine side chain and holding it in a position that is optimal for nucleophilic attack of the carbonyl carbon. After 3 h incubation at pH 5.0 and room temperature, cleavage of the T246S coat protein is about half complete, whereas wild-type capsids cleave almost entirely (Fig. 4 ▶). We speculate that cleavage of the T246S mutant is more efficient than that of E103D because the residue at position 103 is directly involved with polarizing the side chain amide of Asn-570, while Thr-246 simply aids the process by orienting the Asn-570 side chain. This approach to determining the amount of cleavage in the mutants is strictly a qualitative observation, and a more thorough experiment will have to be performed to provide kinetic analyses of the cleavage mechanism.
The two mutations to Glu-103, E103S, and E103Q, that resulted in aberrant assembly products, did not cleave as determined by gel electrophoresis. It is unclear whether the inhibition of cleavage resulted from improper assembly of the VLPS or is a direct result from the point mutation to Glu-103. Although the E103S and E103Q mutations resulted in aberrant particles, sucrose sedimentation revealed that a reduction in pH from 7.6 to 5.0 caused the particles to sediment faster indicating a structural transition (Fig. 2E–H ▶). This observation reveals that even though proper assembly is inhibited, the region responsible for the pH sensitive switch characterized for NωV is preserved.
Acknowledgments
We thank Dr. Anette Schneemann for assistance with cell culture and advice and Dr. Vijay Reddy for assistance in generating figures. This work was supported by NIH grant GM54076 (J.E.J.).
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041054605.
References
- Agrawal, D. and Johnson, J. 1992. Sequence and analysis of the capsid protein of Nudaurelia capensis ω Virus, an insect virus with T = 4 icosahedral symmetry. Virology 190 806–814. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Assembly of the T = 4 Nudaurelia capensis ω virus capsid protein, post translational cleavage, and specific encapsidation of its mRNA in a baculovirus expression system. Virology 207 89–97. [DOI] [PubMed] [Google Scholar]
- Borsa, J., Morash, B.D., Sargent, M.D., Copps, T.P., Lievaart, P.A., and Szekely, J.G. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45 161–170. [DOI] [PubMed] [Google Scholar]
- Briehl, R.W. 1980. Solid-like behaviour of unsheared sickle haemoglobin gels and the effects of shear. Nature 288 622–624. [DOI] [PubMed] [Google Scholar]
- Canady, M.A., Tihova, M., Hanzlik, T.N., Johnson, J.E., and Yeager, M. 2000. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV). J. Mol. Biol. 299 573–584. [DOI] [PubMed] [Google Scholar]
- Canady, M., Tsuruta, H., and Johnson, J. 2001. Analysis of rapid, large-scale protein quaternary structural changes: Time-resolved X-ray solution scattering of Nudaurelia capensis ω virus (NωV) maturation. J. Mol. Biol. 311 803–814. [DOI] [PubMed] [Google Scholar]
- Cheng, R.H., Reddy, V.S., Olson, N.H., Fisher, A.J., Baker, T.S., and Johnson, J.E. 1994. Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X.F., Natarajan, P., Tihova, M., Johnson, J.E., and Schneemann, A. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 72 6024–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, A.J. and Johnson, J.E. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361 176–179. [DOI] [PubMed] [Google Scholar]
- Hanzlik, T.N. and Gordon, K.H. 1997. The tetraviridae. Adv. Virus Res. 48 101–168. [PubMed] [Google Scholar]
- Helgstrand, C., Munshi, S., Johnson, J.E., and Liljas, L. 2004. The refined structure of Nudaurelia capensis ω virus reveals control elements for a T = 4 capsid maturation. Virology 318 192–203. [DOI] [PubMed] [Google Scholar]
- Hindiyeh, M., Li, Q.H., Basavappa, R., Hogle, J.M., and Chow, M. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73 9072–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, V.M. 1956. A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature 178 792–794. [DOI] [PubMed] [Google Scholar]
- Johnson, J. and Reddy, V. 1998. Structural studies of noda and tetraviruses. In The insect viruses (eds. L. Miller and L. Ball), pp. 171–223. Plenum, New York.
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt 2): 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, S.S., Smith, T.J., Chapman, M.S., Rossmann, M.G., Pevear, D.C., Dutko, F.J., Felock, P.J., Diana, G.D., and McKinlay, M.A. 1989. Crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 210 91–111. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Liemann, S., Chandran, K., Baker, T.S., Nibert, M.L., and Harrison, S.C. 2002. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell 108 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, E.A.M. and Murphy, M.E.P. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol Crystallogr. 50 869–873. [DOI] [PubMed] [Google Scholar]
- Munshi, S., Liljas, L., Cavarelli, J., Bomu, W., McKinney, B., Reddy, V., and Johnson, J.E. 1996. The 2.8 Å structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 261 1–10. [DOI] [PubMed] [Google Scholar]
- Page, G.S., Mosser, A.G., Hogle, J.M., Filman, D.J., Rueckert, R.R., and Chow, M. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62 1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, K. and Borchardt, R.T. 1990. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm. Res. 7 703–711. [DOI] [PubMed] [Google Scholar]
- Perler, F.B., Xu, M.Q., and Paulus, H. 1997. Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1 292–299. [DOI] [PubMed] [Google Scholar]
- Pringle, F.M., Johnson, K.N., Goodman, C.L., McIntosh, A.H., and Ball, L.A. 2003. Providence virus: A new member of the Tetraviridae that infects cultured insect cells. Virology 306 359–370. [DOI] [PubMed] [Google Scholar]
- Schneemann, A., Zhong, W., Gallagher, T.M., and Rueckert, R.R. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66 6728–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D.J., Krishna, N.K., Canady, M.A., Schneemann, A., and Johnson, J.E. 2002. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J. Virol. 76 9972–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson, L. and Shatkin, A.J. 1992. Reovirus polypeptide sigma 3 and N-terminal myristoylation of polypeptide mu 1 are required for site-specific cleavage to mu 1C in transfected cells. J. Virol. 66 2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umashankar, M., Murthy, M.R., and Savithri, H.S. 2003. Mutation of interfacial residues disrupts subunit folding and particle assembly of Physalis mottle tymovirus. J. Biol. Chem. 278 6145–6152. [DOI] [PubMed] [Google Scholar]
- Voorter, C.E., de Haard-Hoekman, W.A., van den Oetelaar, P.J., Bloemendal, H., and de Jong, W.W. 1988. Spontaneous peptide bond cleavage in aging α-crystallin through a succinimide intermediate. J. Biol. Chem. 263 19020–19023. [PubMed] [Google Scholar]
- Wery, J.P., Reddy, V.S., Hosur, M.V., and Johnson, J.E. 1994. The refined three-dimensional structure of an insect virus at 2.8 Å resolution. J. Mol. Biol. 235 565–586. [DOI] [PubMed] [Google Scholar]
- Zlotnick, A., Reddy, V., Dasgupta, R., Schneemann, A., Ray, W., Rueckert, R., and Johnson, J. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single asparatic acid residue. J. Biol. Chem. 269 13680–13684. [PubMed] [Google Scholar]