Abstract
The solution form of IIANtr from Escherichia coli and its interaction with its partner protein, NPr, were characterized by nuclear magnetic resonance (NMR) spectroscopy. The diffusion coefficient of the protein (1.13 × 10−6 cm/sec) falls between that of HPr (`9 kDa) and the N-terminal domain of E. coli enzyme I (~30 kDa), indicating that the functional form of IIANtr is a monomer (~18 kDa) in solution. Thus, the dimeric structure of the protein found in the crystal is an artifact of crystal packing. The residual dipolar coupling data of IIANtr (covering residues 11–155) measured in the absence and presence of a 4% polyethyleneglycol-hexanol liquid crystal alignment medium fit well to the coordinates of both molecule A and molecule B of the dimeric crystal structure, indicating that the 3D structures in solution and in the crystal are indeed similar for that protein region. However, only molecule A possesses an N-terminal helix identical to that derived from chemical shifts of IIANtr in solution. Further, the 15N heteronuclear nuclear Overhauser effect (NOE) data also support molecule A as the representative structure in solution, with the terminal residues 1–8 and 158–163 more mobile. Chemical shift mapping identified the surface on IIANtr for NPr binding. Residues Gly61, Asp115, Ser125, Thr156, and nearby regions of IIANtr are more perturbed and participate in interaction with NPr. The active-site His73 of IIANtr for phosphoryl transfer was found in the Nδ1-H tautomeric state. This work lays the foundation for future structure and function studies of the signal transducing proteins from this nitrogen pathway.
Keywords: NMR, translational diffusion coefficient, heteronuclear NOE, residual dipolar coupling, IIANtr, NPr, Escherichia coli, structure-based functional discovery
The bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS), which controls the use of carbohydrates by signal transduction via phosphorylation (Postma et al. 1993), has been well-characterized from the functional, regulatory (catabolite repression and inducer exclusion), and structural standpoints. In these pathways, the phosphate group is transferred from the high-energy molecule phosphoenolpyruvate (PEP), to two common proteins enzyme I (EI) and HPr followed by delivery of the phosphate group from HPr to a variety of sugar-specific enzyme IIA proteins such as IIAGlc, and finally to membrane-bound transporters. Interestingly, a nitrogen regulatory pathway, paralogous to the PTS, was discovered (Powell et al. 1995) and suggested to function in the linkage of nitrogen to carbon metabolism. These nitrogen-related proteins such as NPr and IIANtr were found to show sequence homology with the classic PTS carbon-related proteins HPr and IIA (for fructose and mannitol), respectively. In this paralogous PTS nitrogen pathway, the phosphoryl group, also originated from PEP, flows sequentially from nitrogen-related enzyme I (EINtr) to NPr, then to IIANtr (Rabus et al. 1999), although the downstream partner of IIANtr has not been found. IIANtr homologs have been identified in several other Gram-negative bacteria, further supporting the importance of the corresponding ptsN gene in nitrogen metabolism. NPr and IIANtr are cotran-scribed with σ54 (Powell et al. 1995) and negatively regulate this sigma factor (Merrick and Coppard 1989). Therefore, these proteins play a role in the regulation of nitrogen assimilation. A further understanding of such a bacterial signal network would require structural data for both the free and bound forms of these proteins at atomic resolution. While the 3D structures of the three protein–protein complexes from the glucose pathway of the PTS have already been elucidated (Garrett et al. 1999; Wang et al. 2000a; Cai et al. 2003), only the structure of free IIANtr in this nitrogen pathway has been determined by X-ray crystallography (Bordo et al. 1998). The crystal structure offers an excellent starting point for us to understand the function of this protein. On the basis of crystal structure, we utilize solution NMR spectroscopy to explore additional intriguing questions. For example, does the dimeric form found in the crystal also occur in solution? If it does, would dimerization suggest a novel regulatory mechanism by restricting its interaction with other partner proteins? This possibility is based on the finding that the active site of IIANtr for phosphoryl transfer is located at the dimer interface (Bordo et al. 1998). If IIANtr is not a dimer in solution, will any molecule in the crystal correspond to the functional form in solution? What residues of the protein are required for interaction with its partner protein NPr? What is the tautomeric state of histidines? To answer these questions, we have cloned and purified both IIANtr and NPr. Here we report the results obtained from multidimensional NMR studies. This study provides a basis for elucidation of the downstream protein partner of IIANtr.
Results and Discussion
IIANtr is a dimer in the crystal but a monomer in solution
The dimeric crystal structure of IIANtr (Bordo et al. 1998) is illustrated in Figure 1A ▶. In molecule A, the coordinates for the first seven and the last six residues are not available, whereas only the C-terminal residues 158–163 are absent in the coordinates of molecule B. The N-terminal helix of molecule B shows extensive contacts with molecule A. If this dimer also exists in solution, one would expect that IIANtr behaves like a 36-kDa protein. To evaluate this prediction, we measured its translational diffusion coefficient by 1D NMR spectroscopy. The translational diffusion coefficient of a molecule in solution is inversely proportional to its molecular weight, i.e., the smaller the molecule, the faster it diffuses. For comparison, we also measured the diffusion coefficients for a variety of proteins with varying sizes. Figure 1B ▶ shows a linear correlation between the inverse cubic root of the molecular weights and diffusion coefficients of these proteins. The translational diffusion coefficient of IIANtr was found to be slightly smaller than that of lysozyme (14.4 kDa), but greater than that of the 30-kDa N-terminal domain of EI from Escherichia coli. In addition, NPr (with a C-terminal five-residue extension, 10.2 kDa) and HPr (9.1 kDa) from E. coli have similar sizes and diffusion coefficients, which are greater than that of lysozyme. Because IIANtr diffuses as an 18 kDa protein, we conclude that it exists as a monomer in solution (Fig. 1B ▶). The proposed regulation model alluded to above for IIANtr does not occur at the concentration (~1.0 mM) used for our NMR studies.
Figure 1.
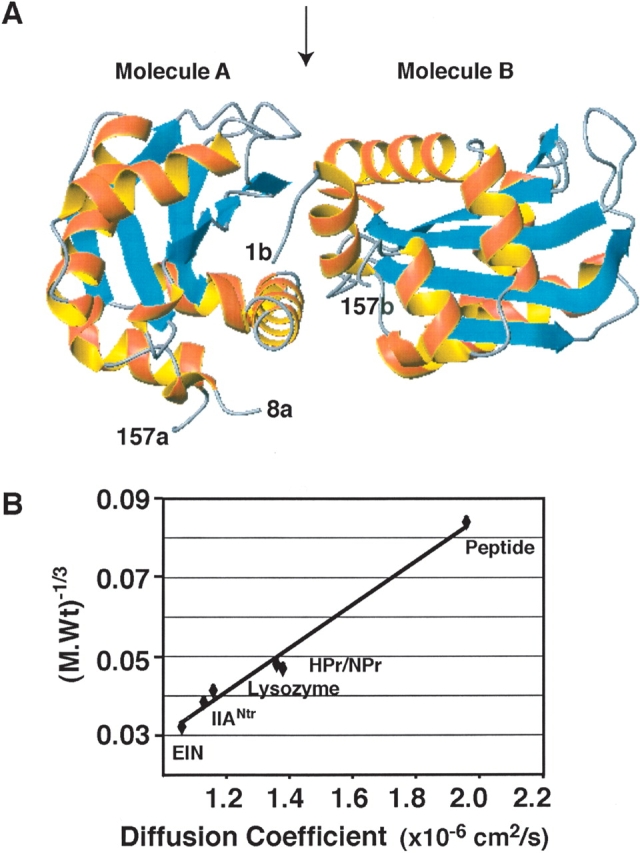
IIANtr is a dimer in the crystal but a monomer in solution. (A) In the crystal structure (PDB code: 1A6J), the coordinates for molecule A cover residues 8–157 (labeled 8a and 157a), while the coordinates for molecule B include residues 1–157 (labeled 1b and 157b). In both molecules, α-helices are in red and β-strands are in cyan. The interface between molecules A and B is indicated by an arrow at the top of the ribbon view of the structure (Bordo et al. 1998). This figure was made using MOLMOL (Koradi et al. 1996). (B) The molecular weight and diffusion coefficient of the proteins used in this study are a 15-residue peptide (1.7 kDa, 1.96 × 10−6 cm/sec) corresponding to the N-terminal amphitropic domain of the E. coli enzyme IIAGlc (Wang et al. 2003); HPr (~9 kDa, 1.36 × 10−6 cm/sec) (Garrett et al. 1999); NPr (with a C-terminal five-residue extension, ~10.2 kDa, 1.38 × 10−6 cm/sec); lysozyme (14.4 kDa, 1.16 × 10−6 cm/sec); IIANtr (~18 kDa, 1.13 × 10−6 cm/sec); and the N-terminal domain of enzyme I (EIN) (~30 kDa, 1.06 × 10−6 cm/sec) from E. coli (Garrett et al. 1999). A linear correlation was found between the cubic root of molecular weight and diffusion coefficient of these proteins with a correlation coefficient of 0.98.
It is interesting to note that IIANtr possesses three free cysteines. Because the protein was not prepared in an absolute oxygen-free atmosphere, there is the possibility that this protein forms dimers via intermolecular disulfide bonds. This can be tested using chemical shifts, because the Cβchemical shifts of cysteines in the reduced (~28.9 ppm) and oxidized (~43.7 ppm) forms differ significantly (Cornilescu et al. 1999). The Cβchemical shifts of the three cysteines in IIANtr are 29.83 ppm for Cys18, 29.54 ppm for Cys25, and 26.52 ppm for Cys137, respectively (Li et al. 2003). Since these chemical shifts are comparable to those in the reduced form, it is concluded that all the cysteines in IIANtr are in the reduced form under the conditions we measured, i.e., in the absence of reducing agents such as dithiothreitol (DTT). The functional role of these reduced Cys residues remains to be elucidated. It is possible that they help maintain a reduced environment for nitrogen metabolism within E. coli.
Secondary structures, residual dipolar coupling, and heteronuclear NOE data of IIANtr in solution
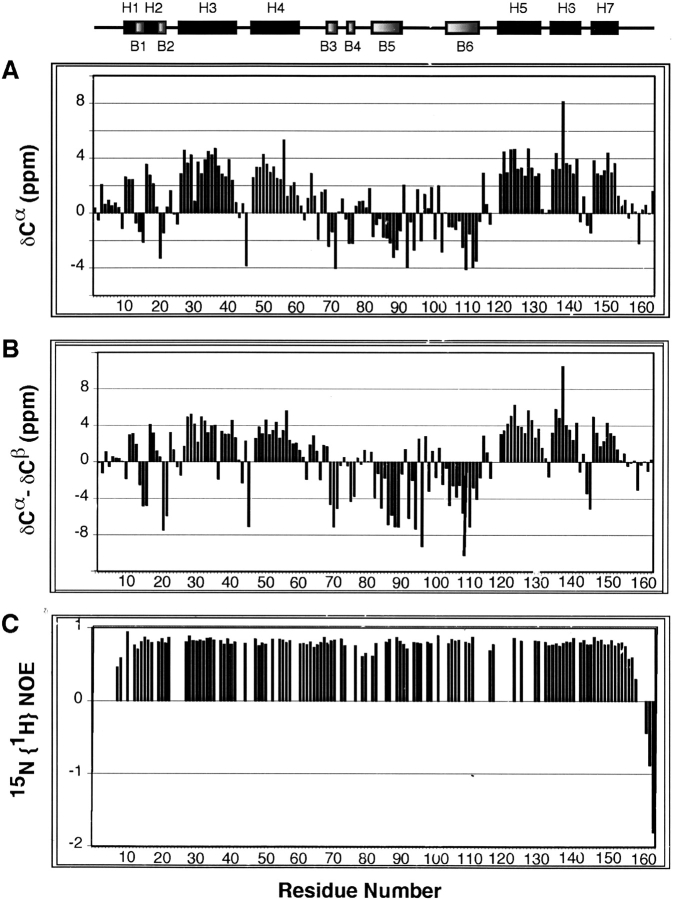
Since the NMR diffusion data indicate a monomer in solution, the IIANtr dimer observed in the crystal can be attributed to crystal packing. Then, which molecule in the dimeric crystal form has a structure resembling that in solution? We employed NMR to answer this question. The backbone signal assignments for IIANtr in solution were achieved by triple-resonance NMR techniques using stable isotope-enriched protein samples (Li et al. 2003). On the basis of these assignments, the secondary chemical shifts of both Cα (Fig. 2A ▶) and Cβ resonances (not shown) for each residue in the protein were calculated. Secondary shifts are the chemical shift differences between measured and random-coil shifts (Wishart and Sykes 1994). A group of positive bars in Figure 2A ▶ for Cαsuggests helical structures and a train of negative bars suggests β-strands (Spera and Bax 1991; Wishart and Sykes 1994). We also calculated the differences between Cα and Cβ secondary shifts (Fig. 2B ▶), which provide a clearer definition of structured regions (Metzler et al. 1993). Thus, the chemical-shift-derived helical regions in IIANtr are located at residues 10–12 (H1), 16–18 (H2), 26–41 (H3), 47–60 (H4), 119–131 (H5), 134–141 (H6), and 146–153 (H7) while β-strands are found for residues 13–15 (B1), 20–21 (B2), 69–71 (B3), 75–76 (B4), 82–94 (B5), and 104–113 (B6) (Fig. 2 ▶, top). Structural analysis of both molecules in the crystal by MOLMOL (Koradi et al. 1996) revealed that they have essentially the same secondary structures as found in solution. The existence of a β-strand between residues Gly64 and Gly66 in the crystal is supported by Hα secondary shifts for these residues in solution. Also of note is that Gly64 amide protons showed strong NOEs to the aromatic ring of Phe96, indicating that the aromatic ring-backbone amide proton interaction (De Marco et al. 1982; Toth et al. 2001) in the crystal structure (Bordo et al. 1998) also occurs in solution. Such an interaction may be critical to stabilize the structure of this glycine-rich segment (residues 61–68) in the vicinity of the active residue of IIANtr. Some differences, however, were observed. In solution, the first helix covers residues 10–12. In the crystal, the first helical region spans residues 5–12 in molecule B, but residues 10–12 in molecule A. It is evident that only molecule A in the crystal shares the identical structure at the N terminus as that found in solution (Fig. 2A ▶). Because the longer helix at the N terminus of molecule B shows multiple contacts with the active site of molecule A (Fig. 1A ▶), the additional structure in molecule B may be ascribed to crystal packing.
Figure 2.
Secondary shifts and heteronuclear NOEs of IIANtr in solution at pH 7.3 and 35°C. (A) A plot of the secondary shifts of Cα (δCα) for IIANtr as a function of residue number. (B) The plot of the shift differences between δCαand δCβ vs. the residue number of IIANtr. The NMR-derived secondary structures of IIANtr (see text) are illustrated at the top of the panels, using black boxes for α-helices (H1 to H7) and shaded boxes for β-strands (B1 to B6), respectively. (C) 15N{1H} NOE values of IIANtr measured using an 15N-labeled protein without a His-Tag.
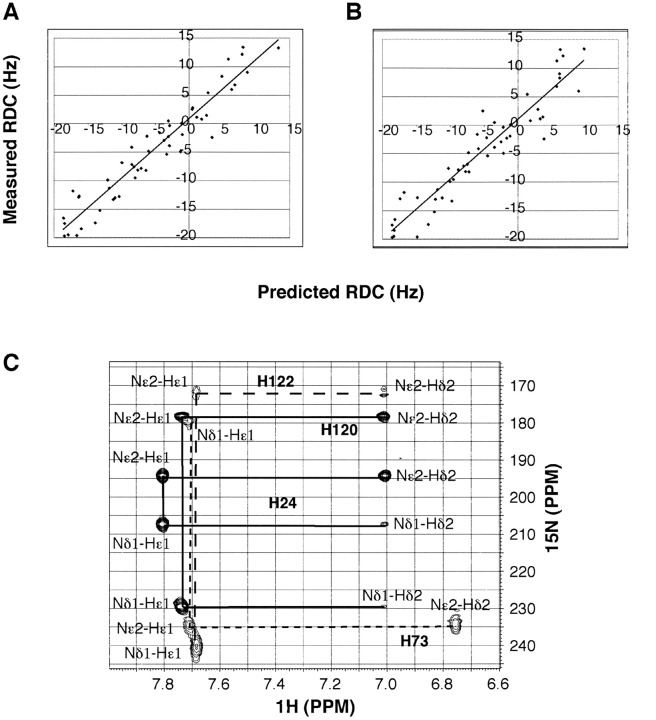
To determine whether the 3D structure of this protein in crystal form is the same as that in solution, we have also measured one-bond residual dipolar couplings (RDC) for the backbone amide HN vectors of IIANtr using the liquid crystal system developed by Ruckert and Otting (2000). RDC results from small alignments of the protein in the liquid crystal phase. For covalently linked vectors such as HN, the RDC for each vector of the protein provides long-range angle information relative to the molecular alignment tensor (Tolman et al. 1995; Tjandra and Bax 1997). Such structural information is very different from the traditional short-range NOE restraints, thereby enabling the determination of the solution structure of biomolecules to a high accuracy. When the RDC data for 51 well-resolved cross-peaks were used to fit the molecular coordinates in the crystal using the single value decomposition (SVD) method (Zweckstetter and Bax 2000), almost identical fits were obtained for both molecules (RSVD = 0.96 for molecule A and RSVD = 0.95 for molecule B) (Fig. 3A,B ▶). Since the RDC data measured in solution fit well to both structures in the dimeric crystal form, we conclude that the 3D structures in the crystal and in solution are indeed similar (Wang et al. 2000a). Taken together with chemical shift data above, molecule A in the crystal is identified as more closely representative of the structure of IIANtr in solution (see molecule A in Fig. 1A ▶).
Figure 3.
Residual dipolar coupling data and histidine tautomeric states for free IIANtr in solution at pH ~7 and 35°C. (A) Shown here is the fit between the measured RDC data and those calculated based on molecule A (A) and molecule B (B) in the crystal (Bordo et al. 1998). The correlation coefficients of the fits between measured (51 used covering residues 11 to 155) and calculated RDC data are 0.92 and 0.90 for molecules A and B in the crystal, respectively. Protons were added to the crystal coordinates by using Xplor-NIH (Schwieters et al. 2003). (C) Both NMR (Pelton et al. 1993) and site-directed mutagenesis (see Materials and Methods) were used to achieve the tautomeric state assignments of histidines in IIANtr. The spectrum was obtained from a protein without His-Tag. The cross-peaks are labeled with solid lines for His24 and His120 and dotted lines for His73 and His122.
To shed further light on the structural differences of II-ANtr in the crystal and in solution, we also measured heteronuclear nuclear Overhauser effects (NOEs) for the protein. The values of 15N{1H} NOEs for residues 9–157 in solution are > 0.5, indicative of a rigid folded domain with backbone motions at picosecond-to-nanosecond time scales (Kay et al. 1989; Stuart et al. 2003). Residues 161–163 at the C terminus show negative NOEs (Fig. 2C ▶), indicating that these residues are highly mobile (Wang et al. 1999). Residues 8 and 157 have values < 0.5, indicating more conformational states. NOE values for residues 1–7 were not obtained due to line broadening. This line broadening for residues 1–7 as well as a heteronuclear NOE value < 0.5 for residue 8 may be caused by an exchange between different conformational states of the N-terminal tail of IIANtr. The structural differences in the crystal state for the N-terminal region of the protein may reflect two possible conformational states: helical (in molecule B) and random coils (in molecule A). Multiple conformational states at both backbone and side chain levels are one of the fundamental aspects for a signal transduction protein to be able to recognize several other partner proteins as observed previously in the glucose pathway of the PTS (Wang et al. 2000a; Cai et al. 2003). Thus, these observations suggest that the N-terminal region of IIANtr is potentially important for interactions with other proteins yet to be identified.
Interactions between IIANtr and NPr
An important function of signal transducing proteins such as IIANtr and NPr is to allow phosphoryltransfer between them. Such a process necessarily involves a binary protein–protein association and disassociation process similar to the carbon PTS proteins (Wang et al. 2000c). To map the surface on IIANtr for NPr binding, we recorded the HSQC spectra for both the free and bound IIANtr (Fig. 4A ▶). While the majority of the cross-peaks superimposed nicely, a select set of peaks such as Arg57, Gly61, and Gly66 shifted, indicating a selective interaction between the two proteins. In addition, cross-peaks for residues Ile65, His73, Leu126, and Ala132 of the protein disappeared in complex with NPr (Fig. 4A ▶), presumably due to line broadening from the exchange between the free and bound states. To identify the shifted residues, we calculated the combined chemical shift differences of both 1HN and 15N (Wang et al. 2000a) for IIANtr in the absence and presence of NPr and the results are plotted in Figure 4B ▶. The chemical shifts of residues Arg57, Glu58, Gly61, Thr63, Gly64, Gly66, Lys75, Asp79, Ile99, Val112, Ala114, Asp115, Ser125, Val127, Ala128, Lys129, Arg130, Ile136, Ile153, Asp155, and Thr156 are more perturbed (up to 120 Hz) by NPr than those of other residues (< 40 Hz). Because the overall chemical shift alterations of IIANtr from the free to the bound states are small, no significant structural change to the protein occurred as a result of NPr binding (Wang et al. 2000a). When all the perturbed residues are mapped to the 3D structure of molecule A of the crystal structure, it becomes apparent that the shifted residues (purple), including nondetected residues in the complex His73, Ile65, Leu126, and Ala132 (gold), are clustered on the same protein surface (Fig. 4B ▶). Consequently, these residues, including the active site His73, constitute the NPr binding surface on IIANtr. Interestingly, IIANtr uses the same surface to pack as a dimer in the crystal (Bordo et al. 1998). As a result, the local structures for residues 8–11, 77–82, 120–125, and 154–157 are more variable than other regions in the crystal. It appears that molecule B mimics NPr to some extent by interacting with molecule A of IIANtr in the crystal (Fig. 1A ▶).
Figure 4.
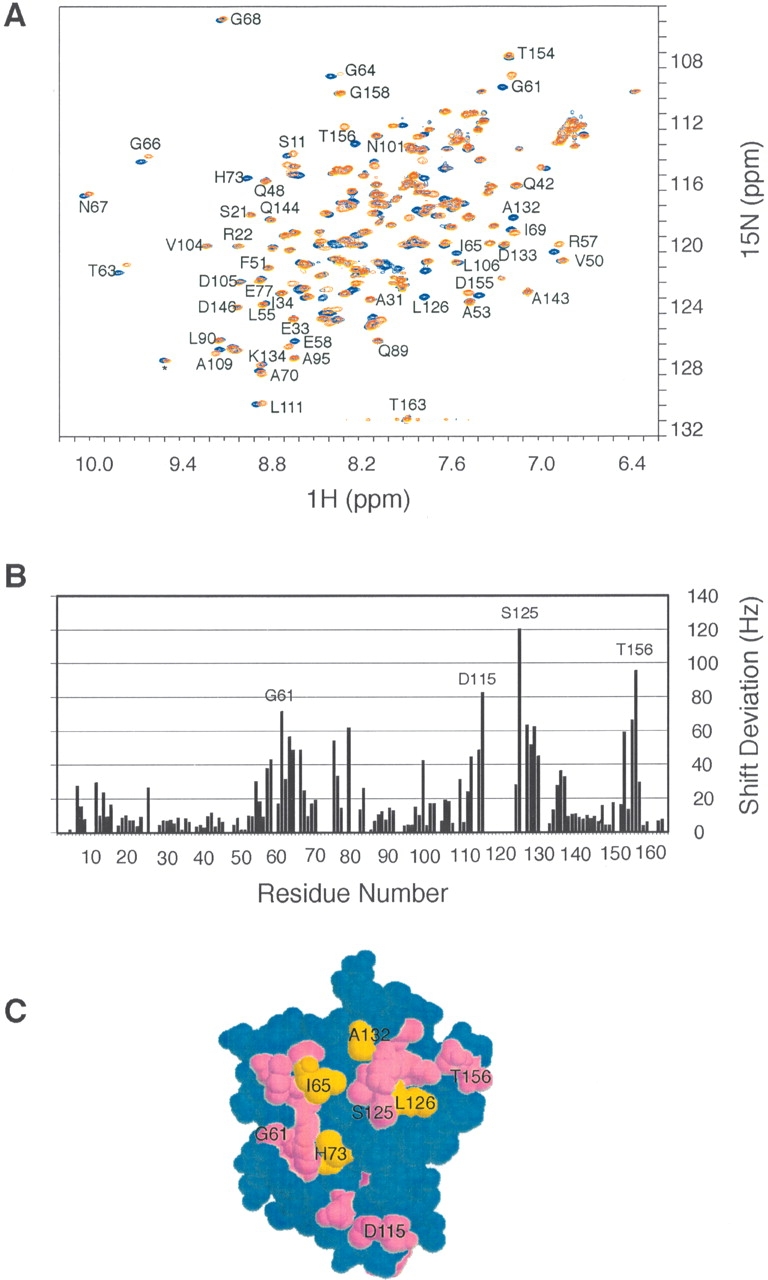
Chemical shift mapping of the surface on IIANtr for interaction with NPr. (A) Superimposed view of the HSQC spectra of IIANtr in the presence (red) and absence (blue) of NPr in 10 mM phosphate buffer at pH 7.3, 35°C. Data were collected using ~1.0 mM 15N-labeled IIANtr without His-Tag purified in the same manner as described elsewhere for IIAGlc (Wang et al. 2000a). In the presence of NPr, the molar ratio between the two proteins is 1:1. For clarity, only well-resolved cross-peaks are labeled. (B) Chemical shift deviations of IIANtr caused by NPr binding. For a more complete list, the chemical shift data were extracted from 3D HNCO experiments for both the free and bound IIANtr. The chemical shift deviation (in Hz) was calculated as the square root of the sum of 1HN and 15N chemical shift deviations squared. Only those residues with the largest deviations are labeled. (C) Residues with chemical shift deviations > 35 Hz (in purple) and those not detected due to line broadening (in gold) are mapped to the space-filling model of the 3D structure of molecule A in the crystal. These residues are clustered and form the binding surface on this protein for NPr. According to the crystal structure, this surface is concave.
To confirm whether the N-terminal tail of IIANtr is required for interaction with NPr, we also made an N-terminal eight-residue truncated form of IIANtr (IIANtr[9–163]). Because the majority of the cross-peaks in the HSQC spectrum of this mutant overlay nicely with the wild type, deletion of the disordered N-terminal region from IIANtr had little impact on the protein conformation, further substantiating the notion that molecule A represents the solution structure (molecule A, Fig. 1A ▶). Moreover, when mixed with unlabeled NPr at a 1:1 molar ratio, the HSQC spectrum of IIANtr(9–163) bound to NPr superimposed nicely upon the spectrum of the wild-type IIANtr bound to the same protein, with only Ser10, Gly61, and D115 displaying slightly additional shifts. Therefore, the N-terminal tail of IIANtr is not required for interaction with NPr. More importantly, after IIANtr(9–163) was isolated from E. coli, mass spectrometry found that 70% of the mutant protein isolated from E. coli was phosphorylated, indicating that this mutant form is capable of phosphoacceptance in vivo. Such a phosphorylated form of the protein is not stable in solution and the phosphate group is lost with time. This dephosphorylation process is even faster (in hours) in the presence of its partner protein, NPr. Therefore, the NMR data reported here for both the free and bound IIANtr correspond to the dephosphorylated forms.
The phosphotransfer between these nitrogen-related signal-transducing proteins is mediated via histidines (Powell et al. 1995; Rabus et al. 1999). Interestingly, histidine kinases have also been found in eukaryotes (Wolanin et al. 2002). To provide additional insight into the chemical form of the active site residue, we also utilized NMR to characterize the tautomeric states of the histidines in IIANtr. This was done by following an established procedure (Pelton et al. 1993). In addition, we also made the H120A/H122A mutant of IIANtr to verify the assignments. The double histidine mutant gave essentially the same HSQC spectrum as the wild type, indicating that there is no global conformational change, although a few peaks in the vicinity of the mutations did show some small deviations. In addition, after isolation from the expression system of E. coli, this mutant was partially phosphorylated, indicating that it is capable of accepting a phosphate group in vivo. The long-range 1H-15N HSQC spectrum for IIANtr (Fig. 3C ▶) provides correlations between the imidazole nitrogen and the carbon-attached protons of histidines. As observed previously for other PTS proteins (Pelton et al. 1993; Garrett et al. 1998), the Nɛ2–Hɛ1, Nɛ2–Hδ2, and Nδ1–Hɛ1 cross-peaks are much stronger than Nδ1–Hδ2 (Fig. 3C ▶). The protonation or deprotonation of the histidine ring influences its chemical shifts. A neutral form has a nitrogen chemical shift at ~168 ppm, while the unprotonated form appears at ~250 ppm. In contrast, both nitrogen nuclei in a positively charged histidine have similar chemical shifts at ~175 ppm with a ~1 ppm separation between them (Pelton et al. 1993). While the chemical shift separations between the two imidazole nitrogens of His73, His120, and His122 are > 50 ppm, the difference for residue His24 is only slightly larger than ~1 ppm, indicating that only His24 is protonated and other histidines are neutral in IIANtr. Note that the same conclusion was also arrived at by using the Cα and Cβ chemical shifts of these histidines (Cornilescu et al. 1999). For histidine, the more stable tautomeric state is the Nɛ2-form (Garrett et al. 1998). Indeed, three out of four histidines in IIANtr are in the Nɛ2-tautomeric state. Only the active-residue His73 is in the Nδ1-tautomeric state (Fig. 3C ▶). Considering the concave surface on IIANtr (Bordo et al. 1998), such a tautomeric state is necessary, because the Nα 2 atom is more exposed on the protein surface, allowing the access of NPr and transfer of its phosphoryl group to its partner protein. The tautomeric state of His73 in nitrogen-related IIANtr is consistent with that of the active histidine (His90) in glucose-related enzyme IIAGlc (Pelton et al. 1993), which also possesses a concave surface (Liao et al. 1991; Feese et al. 1997) for interacting with the convex surface of HPr (Wang et al. 2000a).
Conclusions
We have identified the functional form of IIANtr in solution by multidimensional NMR spectroscopy. In the crystal, this protein is in a dimeric form as a result of crystal packing. In solution, however, it is a monomer. The RDC data covering residues 11–155 fit well to both molecules in the crystal. Only molecule A, however, possesses the same N-terminal helix (residues 10–12) as found in solution. A monomeric form of IIANtr would allow a direct interaction with its partner proteins for phosphoryl transfer or other functions. The binding surface on IIANtr for NPr was identified by chemical shift mapping. The inclusion of the active site residue into the mapped binding interface is fully consistent with the biological experiments that identified His73 as the active site residue, which is phosphorylated by NPr (Powell et al. 1995). NMR experiments also determined the tautomeric states of all histidines in the protein. Such information is not always clear from the crystal structure because of the difficulty in distinguishing between carbon and nitrogen at 2.35 Å (Bordo et al. 1998). Our work indicates that the N-terminal tail of this protein is not required for interaction with NPr. Therefore, this study provides additional valuable insight into the functional form of IIANtr in solution. It also lays the basis for us to determine the structure of the protein–protein complex between NPr and IIANtr. The small shifts of IIANtr in complex with NPr indicate that little structural change is required for complex formation. Thus, the coordinates of molecule A in the crystal can be directly employed for the structure determination of the protein complex between NPr and IIANtr using the established approach (Wang et al. 2000a) in the next step. Because the downstream partner of IIANtr has not been identified, this work also offers an approach to search for another potential partner protein by using the IIANtr with and without the N-terminal tail as baits. Will it directly interact with the sigma factor σ54? Additional structure determinations for this nitrogen pathway will also offer a clue to functional discovery as demonstrated previously by the finding of the membrane anchor role of the N-terminal tail of IIAGlc (Wang et al. 2000b).
Materials and methods
Cloning and purification of IIANtr
E. coli chromosomal DNA was used as a template to amplify, by PCR, the coding sequence for IIANtr. The cloning and purification of the wild-type IIANtr has been described previously (Li et al. 2003). In addition, two mutant forms of IIANtr were made to facilitate this study.
H120A/H122A mutant of IIANtr
pREHistag-IIANtr (Li et al. 2003) was used as a template for a two-step PCR mutagenesis experiment (Higuchi 1989) to convert both His120 and His122 to Ala. The PCR product, verified by DNA sequencing, was gel-purified and then digested with NdeI and XbaI and ligated into pREI (Reddy et al. 1989). The ligation mixture was used for transformation of strain GI698Δpts (Nosworthy et al. 1998). The isolated strain (no. 1674), after induction with tryptophan, showed good expression of the mutated form of IIANtr.
Strain 1674 was grown at 30°C in GI Rich medium (LaVallie et al. 1993), with glycerol as carbon source, to A600 = 0.5, then transferred, after washing, to minimal medium (Sondej et al. 2002) containing 15N-NH4Cl and induced with tryptophan for expression of IIANtr. The harvested cells were ruptured by two passages through a French press at 10,000 psi. The extract was ultracentrifuged, and fractionated on a DE-52 column (Reddy et al. 1991). Fractions enriched in IIANtr were pooled, concentrated and further purified on a gel filtration column (AcA44) (Reddy et al. 1991). Mass analysis of the purified protein indicated that most of the protein corresponded to residues 2–163 and was 95% substituted with 15N. About 10% of the purified protein was in the phospho-form.
The IIANtr(9–163) mutant
pREHistag-IIANtr was used as the template for a PCR reaction designed to create a new NdeI site at the position corresponding to residue 9 of IIANtr. The 620 bp product, verified by DNA sequencing, was gel purified and digested with NdeI and XbaI. This fragment was cloned into pET-Duet-1 (Novagen) restricted with NdeI and AvrII. The ligation mixture was used for transformation of strain ER2566 (New England Biolabs). The isolated strain (no. 1682), after induction with IPTG, showed good expression of the truncated form of IIANtr.
Strain 1682 was grown at 37°C in a minimal medium (Sondej et al. 2002) supplemented with glycerol and 15N-NH4Cl to an A600 of ~0.5, then induced with IPTG for protein expression overnight. The cells were collected by centrifugation and treated as described above for the H120A/H122A mutant for French press treatment, ultracentrifugation, and DE-52 column fractionation. The pooled fractions were essentially pure based on SDS-PAGE. Mass analysis of the purified protein indicated that it corresponded to residues 9–163 and was 95% substituted with 15N. In addition, roughly 70% of the purified protein was in the phosphorylated form.
Cloning and purification of NPr
Overexpression of unmodified NPr was problematic since it seemed to be degraded. Consequently, the NPr was cloned to produce a fusion protein, which was found to be stable. An expression vector that encodes E. coli NPr with a His-Tag at the N terminus was used as the template for PCR amplification of the complete gene with an engineered XhoI site immediately after the last codon of NPr to facilitate the cloning to construct an intein fusion. The sequence of the PCR product (383 bp) was verified by DNA sequencing. Both the PCR product and the expression vector pTYB1 (New England Biolabs) were digested with NdeI and XhoI. The purified fragments were ligated using T4 DNA ligase (New England Biolabs) and used for transformation by electroporation of strain ER2566 (New England Biolabs). The resultant clone (pTYB1-NPr) encodes a fusion protein of NPr linked via an intein sequence to a chitin-binding domain.
To facilitate protein location from the column and quantification by UV spectroscopy, site-directed mutagenesis was carried out to replace a serine residue by a tyrosine in the intein sequence between NPr and the chitin-binding domain. The 35-mer oligonucleotide, 5′-GAAGATCTCGAGGGCTATTCGTGCTTTGCC AAGGG-3′ and its complement were synthesized (Qiagen operon) and used to effect the designated change in pTYB1-NPr using the Quik-Change Mutagenesis system (Stratagene). The resultant vector pTYB1-NPr(LEGYS) was moved into ER2566 for protein expression studies. This construct contains an additional tail (LEGYS) at the C terminus of NPr.
Strain ER2566, harboring the expression vector pTYB1-NPr(LEGYS), was grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/mL). To produce 13C and/or 15N-labeled NPr, the cells were cultured in a minimal medium containing 2.5 g/L of 13C-glucose and/or 1.0 g/L 15N-NH4Cl. IPTG (0.3 mM) was added at A600 of 0.5 to induce NPr expression. The culture was shifted to a 15°C incubator for continued shaking overnight.
The harvested cells, resuspended in 20 mL of 25 mM Tris (pH 8.0)/200 mM NaCl/1 mM EDTA/1 mM PMSF, were lysed by French press twice at 10,000 psi. The broken cell suspension was ultracentrifuged and the supernatant solution was adjusted to 1% streptomycin and incubated at 4°C for 1 h. After centrifugation, the supernatant solution was applied to a chitin bead (New England Biolabs) column (20-mL bed volume). The column was washed with 200 mL of Tris (pH 8.0)/1 M NaCl followed by 60 mL of the same buffer supplemented with 100 mM mercaptoethanol.
The column flow was then stopped to effect overnight cleavage of the fusion protein. After cleavage, the column was washed with six 10-mL aliquots of Tris (pH 8.0)/200 mM NaCl. Fractions containing NPr(LEGYS) were pooled, concentrated, and further purified on a FPLC MonoQ 10/10 column. The NPr(LEGYS) mutant was then sequentially dialyzed against 25 mM Tris buffer (pH 8.0), then 25 mM phosphate buffer (pH 7.0), then 2 mM phosphate buffer (pH 7.0). The purified protein was essentially homogeneous as judged by SDS-PAGE and NMR spectra. Mass analysis confirmed the expected mass (10,228.7) corresponding to the protein with the N-terminal Met deleted. NMR analysis revealed that the C-terminal extension LEGYS on NPr left from the molecular cloning strategy is disordered while the rest of the protein is folded.
NMR spectroscopy
All data were collected at 35°C on a four-channel Varian INOVA 600 MHz NMR instrument with waveform generators and triple-axis pulsed-field gradient accessories. The diffusion coefficients (Df) of a series of proteins and a peptide were measured using a longitudinal eddy-current delay (LED) pulse sequence (Gibbs and Johnson 1991; Altieri et al. 1995) modified to improve water suppression by using the WET (Smallcombe et al. 1995) technique (WET-LED) (Keifer et al. 2004). Df was obtained by plotting the intensity ratio of the NMR signals (I/Io) versus the strength of pulsed field gradients using the following equation:
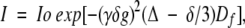 |
where γ is the gyromagnetic ratio of the nucleus (1H), δ is the gradient duration (5 msec), and Δ is the diffusion time (100 msec) between the gradient pulses. The gradient strengths (g) were arrayed by 20 levels from 2 to 45 Gauss/cm using Z-axis gradients. Gradient data were processed using VNMR software (Varian Inc.). For each data set, both integral and peak heights were obtained to fit the equation and the results are similar. The NMR sample conditions for the peptide and proteins used for diffusion studies are as follows: the synthetic peptide with a sequence corresponding to the first 15 residues of enzyme IIAGlc (> 95% pure from Genemed Synthesis, Inc.), 1 mM in water at pH 5.4; HPr, 1 mM at pH 7.1; NPr, 1 mM at pH ~7; IIANtr, 1 mM at pH 7.3; the N-terminal domain of EI, 1 mM at pH 7. All proteins in 10 mM phosphate buffer were expressed and purified as above or detailed elsewhere (Garrett et al. 1999). No DTT was added to any NMR samples. The lysozyme (1 mM) is a sealed standard NMR sample solubilized in water.
Backbone chemical shifts of IIANtr were assigned using a set of triple-resonance experiments, including HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO, C(CO)NH, H(CCO)NH, HNHA, HNHB, and HCCH-TOCSY (Bax and Grzesiek 1993) as described elsewhere (Li et al. 2003). A 3D HBHA(CO)NH experiment was also performed on the 13C,15N-labeled protein to further corroborate the assignments achieved previously (Li et al. 2003) after the installation of the cryoprobe and NMR software update. The chemical shift assignments for the NPr-bound state of IIANtr were confirmed by triple resonance experiments. Residue-specific heteronuclear 15N{1H} NOE values for IIANtr were obtained from 2D (1H, 15N) correlated spectroscopy with and without proton saturation.
RDC between the backbone amide proton and nitrogen were measured using IPAP-HSQC (Ottiger et al. 1998) in a liquid crystalline medium of 4% polyethyleneglycol-hexanol mixture (Ruckert and Otting 2000). The RDC data were fit to both molecules in the crystal. For molecule A, the magnitutes of axial and rhombic components of the alignment tensor are Da = −9.4 Hz and R = 0.489, respectively. The orientation of the alignment tensor is depicted by four sets of Euler angles, one of which is α = 122.58°, β = 156.24°, and γ = 28.61°. For molecule B, the magnitude and orientation for the alignment tensor are depicted by Da = −9.4 Hz, R = 0.532, and three Euler angles (one of the sets) α = 151.52°, β = 122.83°, and γ = 44.82°.
In all 2D NMR experiments such as HSQC, the sweep width for the 15N dimension was typically 2200 Hz with 100–200 increments, whereas 1024 complex points were collected in the 1H-detected dimension with a spectral width of 8510.6 Hz. 1H chemical shifts were referenced indirectly to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) and 13C and 15N chemical shifts were referenced as recommended (Markley et al. 1998). Data were processed using NMRPipe (Delaglio et at. 1995) and analyzed by PIPP (Garrett et al. 1991).
Acknowledgments
This research was supported by the startup fund from the Eppley Institute of the University of Nebraska Medical Center to G.W., including the accessibility to the NMR Core Facility. We are grateful to Lewis Kay’s laboratory (U. Toronto) for pulse sequences, and Frank Delaglio and Dan Garrett (NIDDK, NIH) for NMR software. A special thank also goes to Dan Garrett for providing his program for generating Figure 4A ▶. We thank Rodney Levine (NHLBI, NIH) for performing the mass spectrometric analyses of the proteins. We declare that there is no competing interest with this article. This structural study does not require an approval from an ethics committee.
Abbreviations
PTS, phosphoenolpyruvate:sugar phosphotransferase system
PEP, phosphoenolpyruvate
IIAGlc, glucose-specific enzyme IIA
HPr, histidine-containing phosphocarrier protein
EI, enzyme I
EINtr, nitrogen-related enzyme I
NPr, nitrogen-related HPr
IIANtr, nitrogen-related enzyme IIA
IIANtr(9–163), IIANtr with N-terminal residues 1–8 deleted
NMR, nuclear magnetic resonance
IPAP, in-phase antiphase
HSQC, heteronuclear single quantum coherence
NOE, nuclear Overhauser effect
RDC, residual dipolar coupling
SVD, single value decomposition
PCR, polymerase chain reaction
IPTG, isopropyl-β-D-thiogalactopyranoside
PMSF, phenylmethylsulfonyl fluoride
SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041232805.
References
- Altieri, A.S., Hinton, D.P., and Byrd, R.A. 1995. Association of biomolecular systems via pulsed field gradient NMR self-diffusion measurements. J. Am. Chem. Soc. 117 7566–7567. [Google Scholar]
- Bax, A. and Grzesiek, S. 1993. Methodological advances in protein NMR. Acc. Chem. Res. 26 131–138. [Google Scholar]
- Bordo, D., van Monfort, R.L.M., Pijning, T., Kalk, K.H., Reizer, J., Saier Jr., M.H., and Dijkstra, B.W. 1998. The three-dimensional structure of the nitrogen regulatory protein IIANtr from Escherichia coli. J. Mol. Biol. 279 245–255. [DOI] [PubMed] [Google Scholar]
- Cai, M., Williams, D.G., Wang, G., Lee, B.R., Peterkofsky, A., and Clore, G.M. 2003. Solution structure of the phosphoryl transfer complex between the signal-transducing protein IIAGlucose and the cytoplasmic domain of the glucose transporter IICBGlucose of the Escherichia coli glucose phosphotransferase system. J. Biol. Chem. 278 25191–25206. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- De Marco, A., Menegatti, E., and Guarneri, M. 1982. 1H-NMR studies of the structure and stability of the bovine pancreatic secretory trypsin inhibitor. J. Biol. Chem. 257 8337–8342. [PubMed] [Google Scholar]
- Feese, M.D., Comolli, L., Meadow, N.D., Roseman, S., and Remington, S.J. 1997. Structural studies of the Escherichia coli signal transducing protein IIAGlc: Implications for target recognition. Biochemistry 36 16087–16096. [DOI] [PubMed] [Google Scholar]
- Garrett, D.S., Powers, R., Gronenborn, A.M., and Clore, G.M. 1991. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95 214–220. [DOI] [PubMed] [Google Scholar]
- Garrett, D.S., Seok, Y.-J., Peterkofsky, A., Clore, G.M., and Gronenborn, A.M. 1998. Tautomeric state and pKa of the phosphorylated active site histidine in the N-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. Protein Sci. 7 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, D.S., Seok, Y.-J., Peterkofsky, A., Gronenborn, A.M., and Clore, G.M. 1999. Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat. Struct. Biol. 6 166–173. [DOI] [PubMed] [Google Scholar]
- Gibbs, S.J. and Johnson, C.S. 1991. A PFG NMR experiment for accurate diffusion and flow studies in the presence of eddy currents. J. Magn. Reson. 93 395–402. [Google Scholar]
- Higuchi, R. 1989. Using PCR to engineer DNA. In PCR technology: Principles and applications for DNA (ed. H.A. Ehrlich), pp. 61–70.
- Kay, L.E., Torchia, D.A., and Bax, A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28 8972–8979. [DOI] [PubMed] [Google Scholar]
- Keifer, P.A., Peterkofsky, A., and Wang, G. 2004. Effects of detergent alkyl chain length and chemical structure on the properties of a micelle-bound bacterial membrane targeting peptide. Anal. Biochem. 331 33–39. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- LaVallie, E.R., DiBlasio, E.A., Kovacic, S., Grant, K.L., Schendel, P.F., and McCoy, J.M. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology 11 187–193. [DOI] [PubMed] [Google Scholar]
- Li, X., Peterkofsky, A., and Wang, G. 2003. 1H, 15N, and 13C chemical shift assignments of the Escherichia coli nitrogen regulatory phosphocarrier II-ANtr. J. Biomol. NMR 27 401–402. [DOI] [PubMed] [Google Scholar]
- Liao, D.L., Kapadia, G., Reddy, P., Saier Jr., M.J., Reizer, J., and Herzberg, O. 1991. Structure of the IIA domain of the glucose permease of Bacillus subtilis at 2.2 Å resolution. Biochemistry 30 9583–9594. [DOI] [PubMed] [Google Scholar]
- Markley, J.L., Bax, A., Arata, Y., Hilbers, C.W., Kaptein, R., Sykes, B.D., Wright, P.E., and Wüthrich, K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J. Biomol. NMR 12 1–23. [DOI] [PubMed] [Google Scholar]
- Merrick, M.J. and Coppard, J.R. 1989. Mutations in genes downstream of the rpoN gene (encoding σ54) of Klebsiella pneumoniae affect expression from σ54-dependent promoters. Mol. Microbiol. 3 1765–1775. [DOI] [PubMed] [Google Scholar]
- Metzler, W.J., Constantine, K.L., Friedrichs, M.S., Bell, A.J., Ernst, E.G., Lavoie, T.B., and Mueller, L. 1993. Characterization of the three-dimensional solution structure of human profilin: 1H, 13C, and 15N NMR assignments and global folding pattern. Biochemistry 32 13818–13829. [DOI] [PubMed] [Google Scholar]
- Nosworthy, N.J., Peterkofsky, A., Konig, S., Seok, Y.-J., Szczepanowski, R.H., and Ginsburg, A. 1998. Phosphorylation destabilizes the amino-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. Biochemistry 37 6718–6726. [DOI] [PubMed] [Google Scholar]
- Ottiger, M., Delaglio, F., and Bax, A. 1998. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Mag. Reson. 131 373–378. [DOI] [PubMed] [Google Scholar]
- Pelton, J.G., Torchia, D.A., Meadow, N.D. and Roseman, S. 1993. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 2 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, P.W., Lengeler, J.W., and Jacobson, G.R. 1993. Phosphoenolpyruvate-:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, B.S., Court, D.L., Inada, T., Nakamura, Y., Michotey, V., Cui, X., Reizer, A., Saier Jr., M.H., and Reizer, J. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270 4822–4839. [DOI] [PubMed] [Google Scholar]
- Rabus, R., Reizer, J., Paulsen, I., and Saier Jr., M.H. 1999. Enzyme INtr from Escherichia coli. A novel enzyme of the phosphoenolpyruvate-dependent phosphotransferase system exhibiting strict specificity for its phosphoryl acceptor, NPr. J. Biol. Chem. 274 26185–26191. [DOI] [PubMed] [Google Scholar]
- Reddy, P., Peterkofsky, A., and McKenney, K. 1989. Hyperexpression and purification of Escherichia coli adenylate cyclase using a vector designed for expression of lethal gene products. Nucleic Acids Res. 17 10473–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, P., Fredd-Kuldell, N., Liberman, E., and Peterkofsky, A. 1991. Overproduction and rapid purification of the phosphoenolpyruvate:sugar phosphotransferase system proteins enzyme I, HPr and Protein IIIGlc of Escherichia coli. Protein Expr. Purif. 2 179–187. [DOI] [PubMed] [Google Scholar]
- Ruckert, M. and Otting, G. 2000. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122 7793–7797. [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160 65–73. [DOI] [PubMed] [Google Scholar]
- Smallcombe, S.H., Patt, S.L., and Keifer, P.A. 1995. A WET solvent suppression and its applications to LC NMR and high-resolution NMR spectroscopy. J. Magn. Reson. 117A 295–303. [Google Scholar]
- Sondej, M., Weinglass, A.B., Peterkofsky, A., and Kaback, H.R. 2002. Binding of enzyme IIAGlc, a component of the phosphoenolpyruvate:sugar phosphotransferase system, to the Escherichia coli lactose permease. Biochemistry 41 5556–5565. [DOI] [PubMed] [Google Scholar]
- Spera, S. and Bax, A. 1991. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 113 5490–5492. [Google Scholar]
- Stuart, A.C., Gottesman, M.E., and Palmer III, A.G. 2003. The N-terminus is unstructured, but not dynamically disordered, in the complex between HK022 Nun protein and γ-phage BoxB RNA hairpin. FEBS Lett. 553 95–98. [DOI] [PubMed] [Google Scholar]
- Tjandra, N. and Bax, A. 1997. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278 1111–1114. [DOI] [PubMed] [Google Scholar]
- Tolman, J.R., Flanagan, J.M., Kennedy, M.A., and Prestegard, J.H. 1995. Nuclear magnetic dipole interactions in field-oriented proteins: Information for structure determination in solution. Proc. Natl. Acad. Sci. 92 9279–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, G., Watts, C.R., Murphy, R.F., and Lovas, S. 2001. Significance of aromatic-backbone amide interactions in protein structure. Proteins 43 373–381. [DOI] [PubMed] [Google Scholar]
- Wang, G., Wylie, G.P., Twigg, P.D., Caspar, D.L.D., Murphy, J.R., and Logan, T.M. 1999. Solution structure and peptide binding studies of the C-terminal Src homology 3-like domain of the diphtheria toxin repressor protein. Proc. Natl. Acad. Sci. 96 6119–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Louis, J.M., Sondej, M., Seok, Y.-J., Peterkofsky, A., and Clore, G.M. 2000a. Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIAglucose of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J. 19 5635–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Peterkofsky, A., and Clore, G.M. 2000b. A novel membrane anchor function for the N-terminal amphipathic sequence of the signal-transducing protein IIAGlucose of the Escherichia coli phosphotransferase system. J. Biol. Chem. 275 39811–39814. [DOI] [PubMed] [Google Scholar]
- Wang, G., Sondej, M., Garrett, D.S., Peterkofsky, A., and Clore, G.M. 2000c. A common interface on histidine-containing phosphocarrier protein for interaction with its partner proteins. J. Biol. Chem. 275 16401–16403. [DOI] [PubMed] [Google Scholar]
- Wang, G., Keifer, P.A., and Peterkofsky, A. 2003. Solution structure of the N-terminal amphitropic domain of Escherichia coli glucose-specific enzyme IIA in membrane-mimetic micelles. Protein Sci. 12 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D.S. and Sykes, B.D. 1994. The 13C chemical shift index: A simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 4 171–180. [DOI] [PubMed] [Google Scholar]
- Wolanin, P.M., Thomason, P.A., and Stock, J.B. 2002. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 3 REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweckstetter, M. and Bax, A. 2000. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J. Am. Chem. Soc. 122 3791–3792. [Google Scholar]