Abstract
The native conformation of host-encoded cellular prion protein (PrPC) is metastable. As a result of a post-translational event, PrPC can convert to the scrapie form (PrPSc), which emerges as the essential constituent of infectious prions. Despite thorough research, the mechanism underlying this conformational transition remains unknown. However, several studies have highlighted the importance of the N-terminal region spanning residues 90–154 in PrP folding. In order to understand why PrP folds into two different conformational states exhibiting distinct secondary and tertiary structure, and to gain insight into the involvement of this particular region in PrP transconformation, we studied the pressure-induced unfolding/ refolding of recombinant Syrian hamster PrP expanding from residues 90–231, and compared it with heat unfolding. By using two intrinsic fluorescent variants of this protein (Y150W and F141W), conformational changes confined to the 132–160 segment were monitored. Multiple conformational states of the Trp variants, characterized by their spectroscopic properties (fluorescence and UV absorbance in the fourth derivative mode), were achieved by tuning the experimental conditions of pressure and temperature. Further insight into unexplored conformational states of the prion protein, likely to mimic the in vivo structural change, was obtained from pressure-assisted cold unfolding. Furthermore, salt-induced conformational changes suggested a structural stabilizing role of Tyr150 and Phe141 residues, slowing down the conversion to a β-sheet form.
Keywords: prion protein, high pressure, protein folding, thermodynamic stability, fluorescence variants
A characteristic feature of transmissible spongiform en-cephalopathies (TSE) or prion diseases is a conformational change of PrP protein: The primarily α-helical structure of benign cellular prion protein (PrPC) is refolded into the predominantly β-sheet form of the pathogenic scrapie form (PrPSc) (Prusiner et al. 1998). This structural change causes a modification of the physicochemical properties of PrP protein and promotes its self-association and tissue deposition in the form of amyloid fibrils (Caughey et al. 1991; Pan et al. 1993). The failure of PrP to adopt its correct structure is a major threat to cell function and viability. Nevertheless, we still lack a mechanistic understanding of how the abnormal protein (monomer, oligomer, or amyloid) induces cell death and tissue degeneration. This is particularly challenging, since such processes play also an essential role in a number of other neurodegenerative disorders including Al-zheimer’s, Parkinson’s, and Huntington’s diseases (Taylor et al. 2002). The discovery of proteins with prion-like characteristics in yeast and fungi (Wickner 1994; Coustou et al. 1997) suggests that the ability to form amyloid fibrils is a common property of proteins under appropriate destabilizing conditions. Indeed, the formation of insoluble amyloid structures has been recently described also for other proteins which are not associated with known diseases (Dobson 2002). Therefore the coupled processes of protein misfold-ing and aggregation are of great interest. A better knowledge of the thermodynamic stability of PrPC and the mechanism of conformational changes involved in the conversion from PrPC to PrPSc would help in the understanding of these processes.
Several studies have shown that in vitro transitions of purified recombinant PrPC into aggregated, β-rich, yet not infectious isoforms occur under mildly denaturing conditions, such as acidic pH and the presence of chaotropic agents (Swietnicki et al. 2000; Morillas et al. 2001; Baskakov et al. 2002). These abnormal PrP isoforms are partially folded thermodynamic or kinetic intermediates. Nevertheless, experimental limitations (i.e., low solubility and the unavailability of crystals) have precluded their comprehensive structural and thermodynamic characterization. Thus, the molecular mechanism and energetic basis underlying the structural conversion of PrPC into the infectious protein conformation remain undetermined.
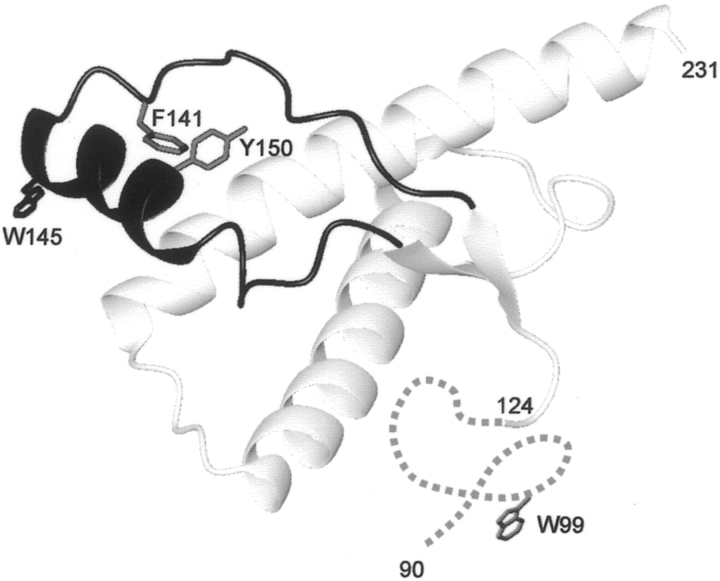
The present study focused on specific structural changes of the recombinant Syrian hamster PrP expanding from residues 90–231 (SHaPrP90–231). Several lines of evidence suggest that either helix 1 (residues 144–154) or the 90–145 part of the PrP protein plays a critical role in the conversion process to a β-sheet structure (Peretz et al. 1997; Zhang et al. 1997; Hornemann and Glockshuber 1998; Morrissey and Shakhnovich 1999; Swietnicki et al. 2000; Viles et al. 2001; Speare et al. 2003). However, as a consequence of the lack of buried tryptophan residues (James et al. 1997; Liu et al. 1999), the spectroscopic probes available for monitoring structural transitions of wild-type PrP protein are limited. Therefore we decided to place additional tryptophan residues on the 132–160 region of the molecule, which is integrated in a long hairpin subdomain comprising residues 124–167. This hairpin subdomain encompasses helix 1 and the β-strands, and is located in the outer shell of the well-structured domain (Fig. 1 ▶). Structural studies with synthetic peptides indicate that this region possesses a high β-sheet propensity (Jamin et al. 2002). Together with the highly flexible 90–123 tail, it is believed to participate in the structural reorganization to PrPSc.
Figure 1.
Ribbon diagram of the 3D structure of SHaPrP90–231 showing the location of the two native Trp residues (W99, W145) and the residues (F141, Y150) that were substituted for a Trp. The 132–160 region is highlighted in dark gray. The figure was produced using MOLMOL (Koradi et al. 1996). The N-terminal segment of residues 90–123 is disordered and displayed schematically as a dotted line.
In addition to conventional denaturation methods (i.e., high temperature or acidic pH) to induce conformational transitions, we employed high hydrostatic pressure. With respect to other denaturing agents, pressure is likely to induce a polypeptide chain to undergo a different conformational change, since it promotes the generation of unfolded protein states retaining significant residual secondary structure, which sometimes exhibit the characteristics of molten-globule structures (Jonas 2002; Silva et al. 2002). Therefore, this less used thermodynamic parameter offers an attractive approach to investigate new aspects of protein folding and misfolding (Silva et al. 2001; Kuwata et al. 2002, 2004; Randolph et al. 2002; Ferrao-Gonzales et al. 2003; Foguel et al. 2003; Marchal et al. 2003; Martins et al. 2003; Torrent et al. 2003, 2004; Jansen et al. 2004).
In a previous study we confirmed that SHaPrP90–231 adopts alternative structural changes under high pressure (Torrent et al. 2003). Recent experiments in our group demonstrated that the high-pressure approach can be used to stabilize prion protein aggregates, and depending on the experimental conditions of time, pressure, and temperature, amyloid fibrils or precursor structures can be obtained (Torrent et al. 2004).
In the present study we examined SHaPrP90–231 and two Trp variants (Y150W and F141W) by UV absorbance spectroscopy in the fourth derivative mode, and fluorescence spectroscopy as a function of both pressure and temperature. The analysis of conformational changes sensed by Tyr and Trp residues provided site-specific details about the conformational changes involved in PrP folding and misfolding.
Results
High pressure or high temperature does not affect significantly the fluorescence spectrum of wild-type SHaPrP90–231. Indeed, the fluorescence emission maximum at 354 nm reflects that the two Trp residues are water-exposed already in the native folded state (Fig. 2A ▶), in good agreement with the NMR structure (James et al. 1997; Liu et al. 1999). Trp99 lies under the disordered N-terminal region (residues 90–123), and Trp145 is situated on helix 1, where the side chain projects into the solvent. To make possible the study by fluorescence spectroscopy of the conformational changes involved in folding and unfolding of SHaPrP90–231, Phe141 and Tyr150 were substituted for a Trp. These residues are both buried, and are located inside the hairpin subdomain spanning residues 124–167.
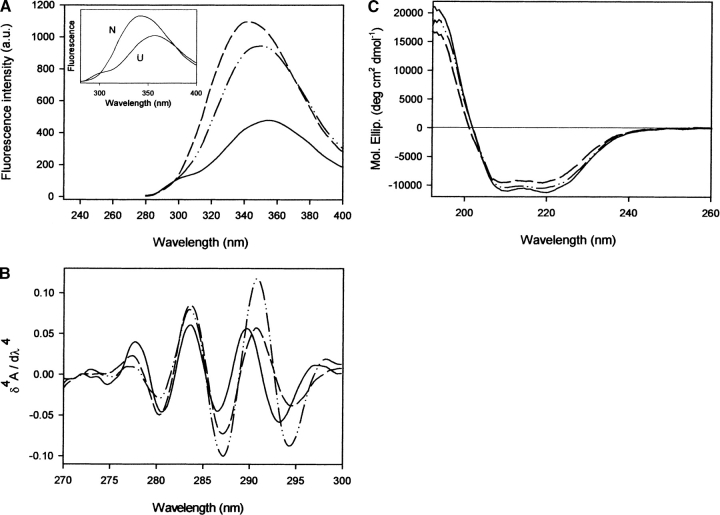
Figure 2.
Spectral properties of SHaPrP90–231 and its variant forms. (A) Fluorescence emission spectra, (B) fourth derivative UV absorbance spectra, and (C) circular dichroism spectra in the aromatic region of Y150W (dashed line) and F141W (dashed and dotted line) variants, in comparison with that of the wild-type protein (solid line). The protein concentration was 0.3 mg mL−1 in Tris-HCl buffer, 20 mM (pH 7.0), at 20°C. In A, fluorescence emission spectra of the folded and unfolded forms are shown as an inset for the variant Y150W. Unfolded state in 5.3 M GdnHCl.
The fluorescence characteristics of a Trp residue are highly dependent on its environment (Eftink 2000). This makes it an ideal residue to report conformational changes and interactions with other molecules. Thus, Trp141 and Trp150 may serve as reporters for the structural reorganization of the hairpin subdomain, specifically of helix 1 and the segment between helix 1 and strand 1, upon unfolding-folding or conformational transition to a scrapie-like form.
Spectral properties of SHaPrP90–231 and its variants
The presence of Trp141 or Trp150 residues in the PrP variants results in different fluorescent properties (Fig. 2A ▶). The emission maxima of Y150W and F141W variants display increased intensity, and are blue-shifted relative to wild-type SHaPrP90–231 by 13 nm and 4 nm, respectively, confirming that the newly introduced Trp residues are in a less polar environment than the two other Trp (Trp99 and Trp149). The emission of both Trp and Tyr residues is evident in the fluorescence emission spectra of the wild-type protein upon excitation at 265 nm. However, the Tyr contribution (centered at 303 nm) is not detectable in the fluorescence spectra of the variants in their native states, suggesting a strong fluorescence quenching, possibly by Tyr→Trp energy transfer. This quenching is lost in both variants upon protein unfolding (inset, Fig. 2A ▶).
Further studies were performed on the surroundings of the aromatic residues by fourth-derivative UV spectroscopy. The fourth derivative spectral features of each protein may be used as an intrinsic probe to sense the average polarity in the vicinity of Tyr and Trp residues (Lange and Balny 2002). The UV absorbance spectra in the fourth derivative mode of wild-type SHaPrP90–231 and the two Trp variants in the native state are shown in Figure 2B ▶. For the wild-type protein, the 289.7-nm band can be assigned to Trp residues in a strongly polar environment. The two variants exhibited an ~1-nm shift of the Trp derivative band to longer wavelengths, consistent with the lower polarity in the average Trp environment observed by fluorescence. The 270–280-nm region reflects mainly Tyr and Phe, without any contribution from Trp. The performed amino acid replacements in both Y150W and F141W variants led to spectral changes in this region. Only an ~0.3-nm blue shift in the position of the band centered in the wild-type protein at 277.7 nm was observed. However, a clear decrease of the amplitude of this band was apparent, in agreement with the amino acid replacements performed (loss of a Tyr or Phe residue).
To gain additional structural insight into the effect of the amino acid substitutions, circular dichroism (CD) spectroscopy was used. The far-UV CD spectra of the variants are very similar to that of the wild-type SHaPrP90–231 (Fig. 2C ▶), suggesting that the variants adopted a native-like fold. All proteins display spectra typical of α-helical peptides with minima at 208 and 222 nm. The minor differences in the spectrum of the Y150W variant could indicate a slight change in the level of secondary structure, probably a destabilization of helix 1.
In the present study, the unfolding of wild-type SHaPrP90–231 and its variants was investigated using high temperature (up to 80°C at 200 MPa) and high pressure (up to 6000 MPa at 25°C, and/or 40°C). In order to compare the pressure- and temperature-induced changes we used fourth derivative UV spectroscopy and fluorescence spectroscopy.
Unfolding and refolding monitored by fourth derivative UV spectroscopy
The fourth derivative spectra of the wild-type protein and the two engineered variants in their native and unfolded states (after heat or pressure treatment) are shown in Figure 3 ▶.
Figure 3.
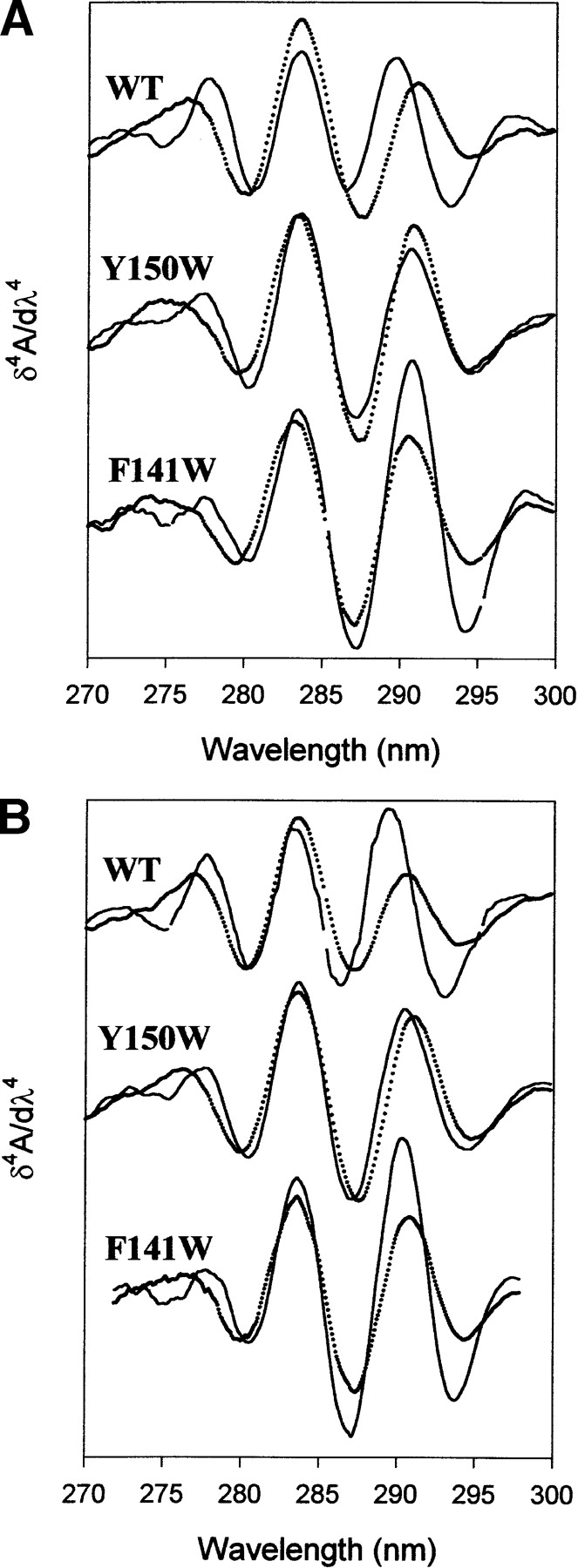
Fourth derivative spectra of the wild-type protein, Y150W and F141W variants under the native (solid line) and unfolded conditions (dotted line). (A) Temperature treatment. Unfolded state was at 200 MPa and 80°C. (B) Pressure treatment. Unfolded state was at 480 MPa and 25°C. Protein concentration was 0.8 mg mL−1 in Tris-HCl buffer, 20 mM (pH 7.0), (pressure experiments) or sodium phosphate buffer, 20 mM (pH 7.0), (temperature experiments).
Upon unfolding, the proteins showed an increased solvent exposure of Tyr side chains, as indicated by a blue shift of the derivative band located within the 270–280-nm region. For wild-type protein, the Trp-specific derivative band was red-shifted (~1.2 nm), indicating that its Trp environment became less polar at high temperature or high pressure, which may be explained by a closer interaction with nonpolar residues. As for the variants, the Trp band, originally located in a less polar environment in the native state, was not significantly affected. However, caution should be used regarding the interpretation of this finding, since the analysis of derivative UV absorbance bands is sometimes complex due to overlapping Trp and Tyr contributions (Lange et al. 1996a; Torrent et al. 2003).
The pressure-induced unfolded states of the three proteins exhibit fourth derivative absorbance spectra with similar λmax values, suggesting that the Tyr and Trp residues are exposed to solvent to the same degree (Table 1). In contrast, from thermal experiments, it is obvious that the heat-unfolded states of these proteins differ in their λmax values of the derivative band corresponding to Tyr. For the variants, these values are significantly blue-shifted compared to that of the wild-type SHaPrP90–231, indicating an increased polarity of the environment of Tyr residues. On the basis of these results, it is also apparent that the thermal treatment leads to an unfolded state with the Tyr residues located in a more polar environment than the one obtained after increasing pressure.
Table 1.
Wavelength of the maximum 4th derivative band
| Temperature-induced unfolding | ||||
| λmax (nm) 270–280 nm region | λmax (nm) 285–295 nm region | |||
| Protein | Native (25°C, 200 MPa) | Unfolded (80°C, 200 MPa) | Native (25°C, 200 MPa) | Unfolded (80°C, 200 MPa) |
| WT | 277.8 | 276.3 | 289.8 | 291.1 |
| Y150W | 277.2 | 275.3 | 290.7 | 290.8 |
| Y141W | 277.4 | 274.0 | 290.8 | 290.5 |
| Pressure-induced unfolding | ||||
| λmax (nm) 270–280 nm region | λmax (nm) 285–295 nm region | |||
| Protein | Native (0.1 MPa, 25°C) | Unfolded (480 MPa, 25°C) | Native (0.1 MPa, 25°C) | Unfolded (480 MPa, 25°C) |
| WT | 277.7 | 276.9 | 289.3 | 290.5 |
| Y150W | 277.6 | 276.2 | 290.4 | 291.0 |
| Y141W | 277.7 | 276.9 | 290.3 | 290.7 |
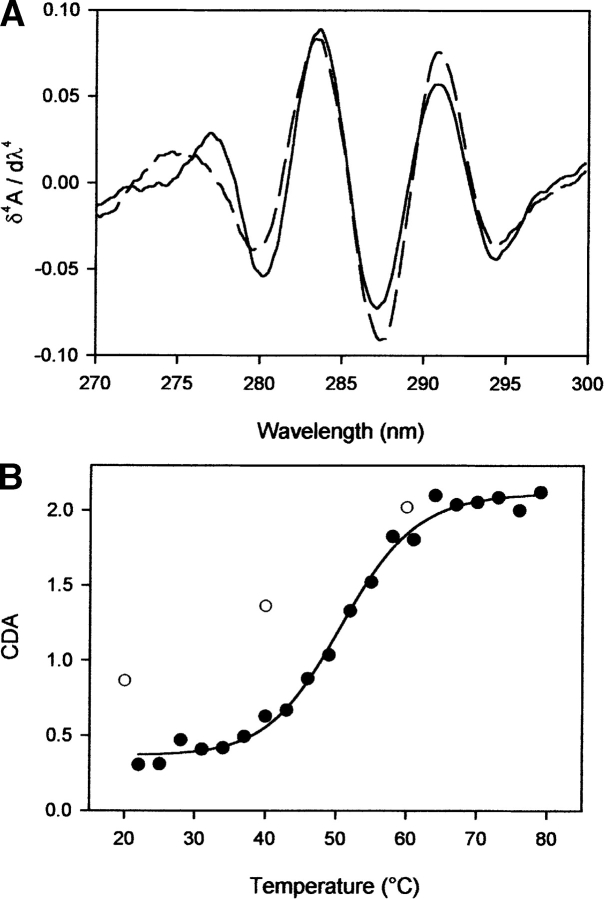
Upon stepwise release of pressure and temperature, the original fourth derivative UV spectra were almost fully recovered. For the two variants, the spectral changes observed upon pressure-induced unfolding were too small to discern a clear unfolding transition. In contrast, temperature-induced unfolding led to clear transition curves, allowing the determination of thermodynamic parameters (Table 2). However, the spectral changes were rather small; they could be resolved only by plotting the cumulative difference amplitudes (CDA) within the 275–290-nm range (Tyr effects) according to Torrent et al. (1999) (Fig. 4 ▶). The two variants exhibited a thermal stability lower than that of wild-type protein, as judged by the ~4°–5°C shifts in the transition midpoint temperature (T1/2). Both variants are slightly less stable than the wild-type protein (see ΔGU values in Table 2).
Table 2.
Thermodynamic parameters of SHaPrP90–231and its variants forms calculated from UV absorbance spectroscopy
| Temperature-induced unfoldinga | |||||
| Protein | ΔST1/2 (J mol−1 K−1) | ΔHT1/2 (KJ mol−1) | T1/2 (°C) | ΔGU200MPa, 25°Cb (KJ mol−1) | ΔGU200MPa, 40°Cb (KJ mol−1) |
| WT | 585 (61) | 193 (20) | 56 | 9.67 | 7.23 |
| Y150W | 508 (56) | 165 (18) | 51 | 7.19 | 4.49 |
| Y141W | 452 (70) | 148 (22) | 54 | 5.66 | 4.52 |
Numbers in parentheses are the standard errors of the data.
a The temperature denaturation profile was recorded at 200 MPa.
b Free energy of unfolding at 200 MPa and 25°C or 40°C.
Figure 4.
(A) Fourth derivative UV spectra of Y150W at 21°C (solid line) and 80°C (dashed line). (B) Temperature unfolding curve for this PrP variant. The CDA was plotted upon increasing (•) and decreasing (○) temperature. Solid line is the nonlinear least-squares fit of the data based on a two-state model. Experimental conditions: Y150W at 0.8 mg mL−1 in sodium phosphate buffer, 20 mM (pH 7.0), P = 200 MPa.
Unfolding and refolding monitored by intrinsic fluorescence changes
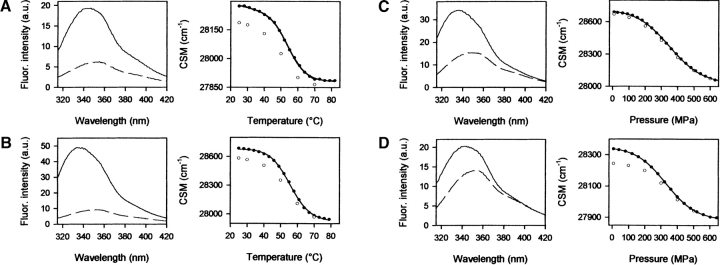
Unfolding of the wild-type protein could not be analyzed by fluorescence spectroscopy, since the spectral changes were too small. However, heat and pressure-induced unfolding and refolding of the two Trp variants were readily monitored by this technique. Figure 5 ▶ shows the fluorescence emission spectra of the native and unfolded states of Y150W and F141W. A substantial spectral change upon unfolding was observed for both variants. The emission maximum was shifted to a higher wavelength because of an increased exposure of the introduced Trp residue, from a partially solvent-shielded environment to the aqueous solvent. The λmax for Y150W was shifted from 336 to 352 nm after high-temperature treatment and to 349 nm after high-pressure treatment. The λmax for F141W was shifted from 344 to 349 nm upon increase of temperature, and to 351 nm at high pressure. Upon unfolding, the Trp fluorescence emission intensity was greatly quenched. The unfolding transition was monitored using changes in center of spectral mass (Fig. 5 ▶). For both variants, the equilibrium unfolding processes were found to be almost fully reversible. The thermodynamic parameters deduced from changes of the center of spectral mass were determined within the frame of a two-state model (Table 3). Since some transitions were not completely reversible, the deduced thermodynamic parameters should be considered as indicative. However they are in good agreement with those derived from absorbance. Both variants show similar stabilities. The transition curves of these proteins are very similar, with comparable transition midpoints and free energies. However, in all cases the values of ΔGU of temperature-induced unfolding transitions are higher (15%–23%) than the corresponding values obtained from pressure experiments. Although—as stated above—the thermodynamic parameters are only indicative (due to a small irreversible component of the transitions), this points to a different mechanism of pressure- and temperature-induced unfolding.
Figure 5.
Left panels: Fluorescence emission spectra of the native and unfolded state for the temperature- and pressure-induced unfolding of Y150W (A and C, respectively) and F141W (B and D, respectively). Right panels: Transition curves for the thermally and pressure-induced unfolding/refolding of Y150W (A and C, respectively) and F141W (B and D, respectively). Experimental conditions: Pressure experiments were performed in Tris-HCl buffer, 20 mM (pH 7.0), at 40°C, and temperature experiments in sodium phosphate buffer, 20 mM (pH 7.0), at 200 MPa. Protein concentration was 0.3 mg mL−1.
Table 3.
Thermodynamic parameters of SHaPrP90–231variants forms calculated from fluorescence spectroscopy
| Pressure-induced unfoldinga | ||||
| Protein | Δ GU0b (KJ mol−1) | Δ VU (ml mol− 1) | P1/2 (MPa) | Δ GU200MPa, 25°C or 40°Cc (KJ mol−1) |
| Y150W 25°C | 9.71 (0.85) | −24.3 (2.2) | 399 | 4.85 |
| 40°C | 8.80 (0.18) | −25.2 (0.5) | 349 | 3.76 |
| F141W 25°C | 9.61 (0.27) | −25.5 (0.8) | 377 | 4.51 |
| 40°C | 9.10 (0.06) | −27.6 (0.19) | 330 | 3.58 |
| Temperature-induced unfoldinga | |||||
| Protein | ΔST1/2 (J mol−1 K−1) | ΔHT1/2 (KJ mol−1) | T1/2 (°C) | ΔGU200MPa, 25°Cc (KJ mol−1) | ΔGU200MPa, 40°Cc (KJ mol−1) |
| Numbers in parentheses are the standard errors of the data. | |||||
| a Unfolding transitions obtained upon increasing pressure were carried out at 25°C or 40°C. The temperature denaturation profile was recorded at 200 MPa. | |||||
| b Free energy of unfolding at atmospheric pressure and 25°C. | |||||
| c Free energy of unfolding at 200 MPa and 25°C or 40°C. | |||||
| Y150W | 458 (14) | 151 (4) | 56 | 5.71 | 5.07 |
| F141W | 459 (24) | 150 (8) | 53 | 5.88 | 4.58 |
The effects of sub-zero temperatures
In order to discern protein structural changes on decreasing temperature, experiments were carried out at a constant pressure of 200 MPa. Under this condition, water stays in the liquid state even at −20°C in the absence of anti-freeze cosolvents (Bridgman 1935). Significant conformational changes and entire cold-induced unfolding transitions were observed at pH 4.0. The major transition took place between 5° and -20°C. Pressure-assisted cold unfolding of both variants was accompanied by a red shift of the maximum of the fluorescence emission spectra, indicating that the Trp residues became more water-exposed. Furthermore, a blue shift of the Tyr band of the fourth derivative UV absorbance spectra was observed, indicating that at low temperatures the Tyr residues are also in a more polar environment (data not shown). From the sigmoidal profile of the unfolding transitions monitored by the center of spectral mass (Fig. 6 ▶), with a midpoint transition temperature of -5.5°C (Y150W) and -8°C (F141W), the thermodynamic parameters were deduced. For Y150W: ΔST1/2 = -386 ± 18 J mol−1 K−1, ΔHT1/2 = -103 ± 5 KJ mol−1. For F141W: 3ST1/2 = -376 ± 33 J mol−1 K−1, ΔHT1/2 = -100 ± 9 KJ mol−1.
Figure 6.
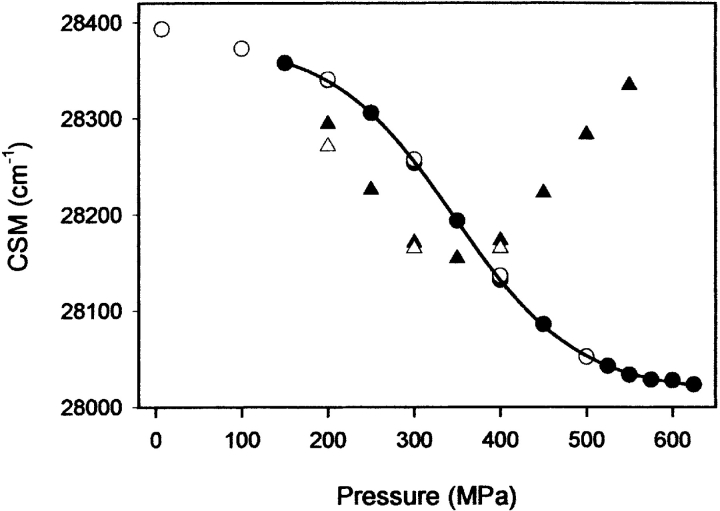
Pressure-assisted cold unfolding (filled circles, temperature scale), and pressure unfolding at -5°C (open circles, pressure scale) and refolding (open triangles, pressure scale) curves for Y150W (A) and F141W (B) variants. The protein conformational changes were recorded by the changes of fluorescence emission spectra, reported as center of spectral mass (CSM). Protein concentration was 0.3 mg mL−1 in sodium acetate buffer, 20 mM (pH 4.0).
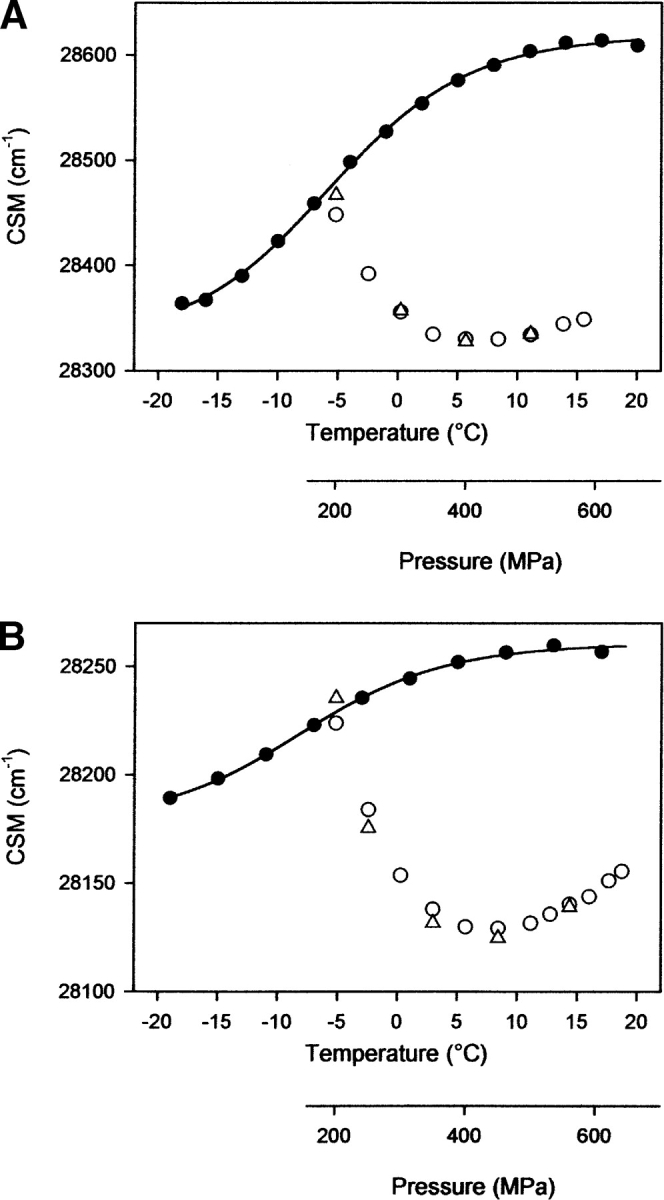
Upon return of the sample to -5°C and further compression up to 450 MPa, additional conformational changes were induced for the F141W variant, leading to an even higher solvent exposure of the Trp residues, as revealed by the decrease in center of mass of fluorescence emission spectra (Fig. 6B ▶). In contrast, for the F150W variant, both cold- and pressure-induced unfolded states had comparable center of spectral mass values (Fig. 6A ▶). In both proteins, the induced transition was completely reversible after decompression. From these experiments, we can conclude that pressure-assisted cold unfolding did not lead to a completely unfolded state of F141W variant. Indeed, cooling the sample to -20°C at 200 MPa induced the transition to a stable conformation, which is only partially unfolded. For both variants, we note that above 450 MPa, the fluorescence emission maximum was slightly blue-shifted, as stated by a small increase in the center of spectral mass. For the Y141W variant, this phenomenon was enhanced at a higher protein concentration (2 mg mL−1) and at pH 5.5 and -10°C (Fig. 7 ▶). Remarkably, the center of spectral mass value of the misfolded state obtained at -10°C and 550 MPa was similar to that monitored for the native state, indicating that the microenvironment of the Trp residues became more solvent-exposed within the initial stages of unfolding, and then became less polar in a second unfolding step. A different trend was seen with F150W: A reversible aggregation process, with almost no change in the center of spectral mass values, was observed above 450 MPa and at identical experimental conditions (data not shown). The above-mentioned decrease in average polarity in the vicinity of the Trp residues at high pressures for F141W is compatible with Trp shielding. Interestingly, no aggregation process was detectable under such conditions, as monitored by the change in light scattering intensity (data not shown). Together with the fact that the spectral changes were fully reversible, this indicates strongly that the specific spectral changes observed upon compression above 450 MPa are attributable only to conformational changes of the protein. Thus, the conformational state of the F141W variant monitored at -10°C and 550 MPa could be an important precursor of later stages of the assembly process. In fact, at physiological pH, the partial shielding of the Trp residues was concomitantly associated with aggregation processes, yielding a sharp increase in light scattering intensity (data not shown). Furthermore, a completely different state of F141W under pressure (550 MPa) with fully solvated Trp residues (λmax = 353 nm) was obtained at 25°C and at pH 5.5 (Fig. 7 ▶).
Figure 7.
Pressure unfolding (filled circles) and refolding (open circles) curves at 25°C. Pressure unfolding (filled triangles) and refolding (open triangles) curves at -10°C. Protein was F141W variant, and concentration was 2 mg mL−1 in sodium acetate buffer, 20 mM (pH 5.5).
Salt-induced conformational conversion
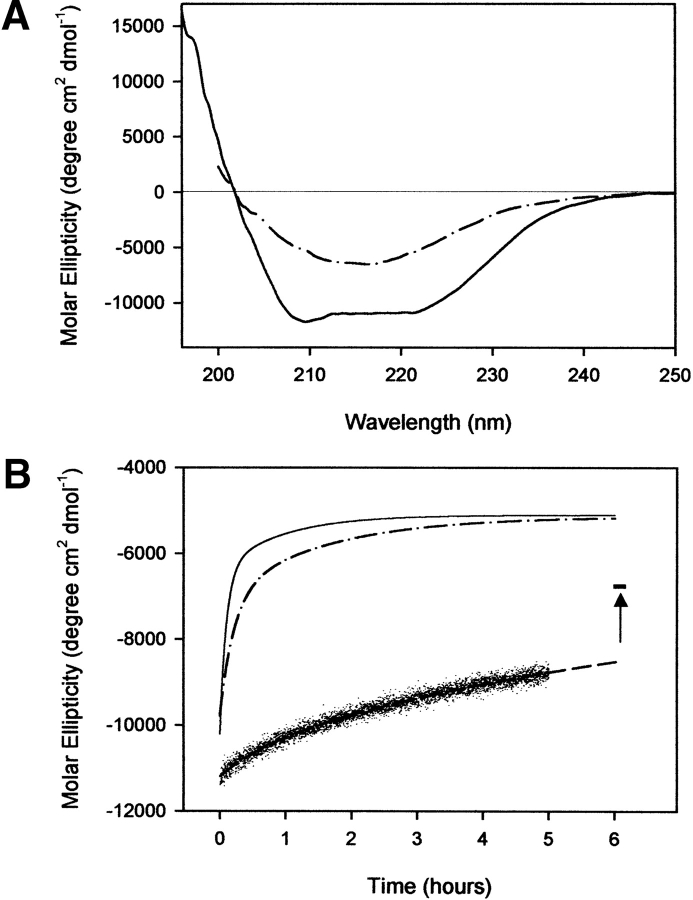
It has been shown that under mildly denaturing conditions such as acidic pH and the presence of chaotropic agents, the recombinant prion protein undergoes a transition to a β-sheet-rich structure having features in common with the infectious isoform (Swietnicki et al. 2000; Morillas et al. 2001; Baskakov et al. 2002). We have observed that wild-type SHaPrP90–231 at pH 4.0, in the absence of denaturant, if incubated at 37°C in the presence of low concentrations of NaCl (150 mM) converts to a β-sheet-rich form. No aggregation processes were observed under these conditions. As monitored by CD (Fig. 8 ▶), the data fit well to a model assuming two exponential relaxation processes. The corresponding relaxation times for wild-type protein are τ1 = 42 min and τ2 = 464 min. The amplitude of the rapid phase is only ~9% of the final molar ellipticity value. The conversion process was altered in both variants, which exhibited significantly faster decays. The resultant relation times for Y150W are τ1 = 6 min and τ2 = 56 min, and τ1 = 13 min and τ2 = 83 min for F141W. For both variants, the amplitude of the faster phase dominates over the slower phase, being about 75% (Y150W) and 54% (F141W) of the final molar ellipticity value. In addition, the final induced conformational states in both variant forms have more β-sheet and less α-helix structure than the wild-type protein, suggesting that the amino acid replacements performed allow for a more profound reorganization of the PrP structure.
Figure 8.
(A) Far-UV circular dichroism spectra of Y150W variant. Spectra were recorded at a 0.2 mg mL−1 protein concentration in sodium acetate buffer, 20 mM (pH 4.0), at 37°C (solid line), and under the same conditions 6 h after adding NaCl 150 mM (dashed/dotted line). (B) Time-dependent change in ellipticity at 222 nm for wild-type protein (dashed line), F141W variant (dashed/dotted line), and Y150W variant (solid line). Lines represent best fits of data to a double exponential decay function. The end point of the conformational transition for wild-type protein was monitored at 28 h. Its molar ellipticity value is shown as a trait marker. The protein concentration was 0.2 mg mL−1 in sodium acetate buffer, 20 mM, NaCl 150 mM (pH 4.0), at 37°C.
Discussion
Analysis of the pressure- and heat-induced unfolding processes
The fourth derivative UV spectra monitor average changes of the microenvironment of Tyr and Trp residues. SHaPrP90–231 contains 10 Tyr and two Trp residues. The three-dimensional structure of the protein shows that while the Tyr residues are homogeneously distributed along the polypeptide chain, with the sole exception of α-helix 2 (which does not contain any Tyr residue), the Trp residues are largely solvent-exposed and located outside the stable protein core (James et al. 1997; Liu et al. 1999). Specifically, Trp99 is positioned at the flexible unstructured N-terminal part of the molecule, and Trp145 at the beginning of the relatively isolated first α-helix. As a result, the information obtained from Tyr depicts overall protein structural events. Since the fluorescence intensity and emission maximum of intrinsic Trp residues of SHaPrP90–231 which are already solvent-exposed in the native state do not change during unfolding, the use of protein variants possessing additional Trp residues (Y150W and Phe141W) was required to study site-specific unfolding/refolding events using fluorescence spectroscopy.
The fluorescence from Tyr residues in wild-type SHaPrP90–231 is evident upon excitation at 265 nm, but is strongly quenched in the native state of the Trp variants. The observed Tyr quenching results from proton transfer in the excited state, most likely because of specific interactions with neighboring residues and with the engineered Trp. Tyr quenching is abolished in the completely unfolded state. This observation provides evidence that the conformational change induced upon unfolding breaks the close contact between the introduced Trp residue and the Tyr residues. The heat- and pressure-induced equilibrium unfolding transitions obtained by monitoring the Trp fluorescence and the UV absorbance changes in the Tyr spectral region lead to comparable free energies, indicating that region-localized and global unfolding exhibit concerted behavior. The small differences observed between parameters determined by absorbance and fluorescence experiments suggests that, under the experimental conditions used, the cooperativity of unfolding might not be complete (especially for variant Y150W). However, given the indicative nature of the thermodynamic parameters, this possibility remains hypothetical.
Unfolding intermediates arise under pressure-assisted cold unfolding
Heremans and Smeller (1998) pointed out that high pressure- and low temperature-induced unfolding are both driven by negative volume changes. Furthermore, several experimental observations suggest a structural similarity between pressure- and cold-induced unfolded states, which retain significant amounts of secondary structure (Jonas 2002; Meersman et al. 2002). In our experiments, PrP variants (0.3 mg mL−1 [pH 4.0]) were first subjected to cold unfolding (200 MPa, -20°C). After return to -5°C, a pressure-induced unfolding (up to 550 MPa) was performed. The main observation here was that cold-induced unfolding of F141W variant led to a structure which was only partially unfolded. A further unfolding step was then achieved by compressing to 550 MPa. Moreover, our results indicated that the heat-induced unfolding led to an even more complete loss of the structure than the one obtained after increasing pressure. This became clearly evident from the stronger polarity of the environment of Tyr residues, and the larger ΔGU values of the heat-induced unfolding transitions.
Additional information on the pressure-induced conformational changes at low temperatures (-10°C) and acidic pH (pH 5.5) came from the use of the F141W variant at a higher protein concentration (2 mg mL−1). Above 450 MPa, after an initial partial unfolding event, the protein experiences a further structural reorganization (monitored by fluorescence spectroscopy) leading to a significant shielding of its Trp residues, without any apparent aggregation process. Since the engineered Trp residue is located within the 132–160 region, we propose that this part of the PrP molecule is concerned in the conformational change observed. This conformational rearrangement does not seem to encompass the whole molecule, as no equivalent process was observed by monitoring average changes of the microenvironment of Tyr residues using UV absorbance spectroscopy. Interestingly, at neutral or basic pH, the above-mentioned structural rearrangement leads to aggregation. This is consistent with the hypothesis that the N-terminal subdomain 90–167, containing the two β-strands and the rather isolated first α-helix, is involved in structural rearrangements and association processes leading to PrPSc formation. Therefore, the aberrant conformational states obtained in this study could represent important precursors or intermediate states of later stages of the assembly process to the pathogenic form. Partially structured intermediates have been previously observed for a number of recombinant PrP proteins carrying amino acid substitutions associated with familial forms of prion disease (Apetri and Surewicz 2002; Vanik and Surewicz 2002; Apetri et al. 2004) or subjected to partially destabilizing conditions (Baskakov et al. 2002; Martins et al. 2003). In addition, conditions expected to increase the population of intermediates are known to stimulate the seeded conversion of PrPC to a PrPSc-like form (Kocisko et al. 1994; Horiuchi and Caughey 1999; Wong et al. 2001). The observed dependence of protein aggregation on pH points to the participation of electrostatic interactions. The assembly of PrP molecules observed at pH 7.0 or higher is likely to be impeded at acidic pH by electrostatic repulsion between positively charged residues in the protein. At pH 5.5, PrP has a net charge of +8, whereas at pH 7.0 its net charge is +4. At a pH near the isoelectric point, it seems likely that hydrophobic patches exposed to solvent at high pressure (Torrent et al. 2004) will favor aggregation by increasing the tendency of the molecules to stick together. Because of the specific structural effects of pressure (studies of model systems show that hydrogen bonds are stabilized by high pressure), it is conceivable as well that new intermolecular hydrogen bonds may contribute in the assembly of the species formed at neutral pH, and at -10°C and 550 MPa.
Formation of a β-sheet-rich isoform is kinetically enhanced by mutation in the N-terminal region
Conditions of acidic pH, 37°C, and the presence of NaCl trigger the conformational transition of SHaPrP90–231 to a form rich in β-sheet structure, without requiring the presence of chemical denaturing agents. Y150W and F141W variants are characterized by a substantially increased propensity to form a β-sheet-rich form under identical conditions. As has been hypothesized (Liu et al. 1999; Calzolai and Zahn 2003), this conformational transition may be explained structurally in terms of a potential secondary structural reorganization of the long hairpin subdomain expanding residues 124–167. Although the amino acid substitution performed does not significantly affect native structure and overall protein stability, it is conceivable that Tyr150 and Phe141 contribute to maintain specific structural interactions to preserve local architecture in the hairpin region. However, under the above-mentioned destabilizing conditions, this region, which does not belong to the well-structured hydrophobic core, may promote the formation of new intra- or intermolecular interactions. It may thus act as a nucleus in the formation of β-sheet structure. The enhanced propensity of both variants to convert into a β-sheet isoform fits well with the large amount of data suggesting heterogeneous conformational propensities of this region (Derreumaux 2001). Interestingly, low hydrogen-deuterium exchange protection factors have been identified for this region (Liu et al. 1999), a fact which further underlines its conformational instability.
Concluding remarks
The fact that PrP possess an inherent tendency to undergo a profound structural reorganization argues strongly in favor of a causal role of protein misfolding in the pathogenic process. However, the mechanisms of conformational modification and aggregation of PrP remains to be proven. In conjunction with earlier observations (Alvarez-Martinez et al. 2003; Torrent et al. 2003, 2004), our results indicate that high pressure arises as a new strategy for approaching their study. The high-pressure approach applied to single-point variants of the 132–160 region revealed an ensemble of conformations between native and unfolded states of PrP. This argues in favor of a central, eventually initial role of this region in the PrPC to PrPSc conversion. Depending on the experimental conditions used, the pressure-induced intermediate states underwent aggregation. Therefore, the elucidation of pressure effects on amyloidogenic proteins could help to understand the conformation and dynamics of the species involved in the initial stages of aggregation. It would be interesting now to carry out nuclear magnetic resonance measurements on such folding intermediates as a function of pressure, and to design drugs susceptible to bind specifically to the 132–160 part of the molecule, in order to confirm its role in PrP misfolding, and further on to inhibit the misfolding reaction.
Materials and methods
Prion protein
Recombinant SHaPrP90–231 was prepared from Escherichia coli BL21(DE3) as described (Alvarez-Martinez et al. 2003). The sequence of this protein corresponds to the sequence of the PK-resistant core of PrPSc, denoted PrP 27–30, which transmits the disease. Site-directed mutagenesis using a QuikChange kit (Strata-gene) was performed on the gene, giving rise to two PrP variants in which phenylalanine 141 or tyrosine 150 was replaced by tryp-tophan. Both mutations were verified with DNA sequencing. The production and purification of the variant proteins were carried out as described for wild-type SHaPrP90–231. Protein concentration was routinely determined spectrophotometrically using, for wild-type protein, a molar extinction coefficient at 278 nm of 25327 M−1 cm−1. The extinction coefficients for Y150W and F141W variants were estimated according to the method of Gill and von Hippel (1989), and at 278 nm were found to be 29,527 M−1 cm−1 and 30,927 M−1 cm−1, respectively.
UV absorbance spectroscopy
For high-pressure experiments, the proteins were dissolved in 20 mM Tris-HCl buffer at pH 7.0. For experiments as a function of temperature, the proteins were dissolved in 20 mM sodium phosphate buffer, pH 7.0. These buffers were selected for their relatively small pressure and thermal pH dependencies, respectively (Kitamura and Itoh 1987). Heat-induced unfolding studies were performed at a pressure of 200 MPa to avoid PrP aggregation (Torrent et al. 2003). The final protein concentration was 0.8 mg mL−1. Absorbance spectra between 260 and 305 nm were recorded in steps of 0.1 nm (1-nm bandpath) as a function of temperature and pressure, using a modified Cary3 (Varian Inc.) absorption spectrometer (Lange et al. 1996b). Following each pressure or temperature change, typically in steps of 20 MPa or 3°C, the sample was allowed to equilibrate for 6 min before the next measurement. Each spectrum was corrected for pressure and temperature dependence of the sample volume, and the fourth derivative spectra were determined as described (Lange et al. 1996a,b). Transitions between two spectral forms were quantified within the 275–290-nm range, typical of Tyr effects, by cumulative difference amplitude (CDA) as reported (Torrent et al. 1999).
Fluorescence spectroscopy
Fluorescence emission spectra were recorded on an Aminco-Bow-mann Series 2 luminescence spectrometer (SLM Aminco), modified to accommodate a thermostated pressure cell. Protein concentrations of 0.3 mg mL−1 in the same buffers as for absorption were used for pressure and temperature measurements, unless stated otherwise. Solutions were placed in a cylindrical 5-mm-diameter quartz cuvette. Cold-induced unfolding studies were carried out at a pressure of 200 MPa, which allows the possibility of lowering the temperature down to -20°C without freezing (Bridgman 1935). Trp fluorescence was measured by exciting at 295 nm (4-nm slit) and emission (8-nm slit) being recorded between 315 and 420 nm (average of three scans). Spectra were analyzed by calculating the center of spectral mass (Silva et al. 1986; Ruan and Weber 1989) between 315 and 400 nm. For monitoring Tyr and Trp fluorescence, excitation was at 265 nm (4-nm slit), and emission spectra (8-nm slit) were collected (accumulation of three scans) between 280 and 400 nm. Fluorescence was observed to equilibrate well within a 6-min pause before each measurement. Specially, during cooling the samples were allowed to equilibrate for 10 min at each condition, and the windows of the pressure cell were flushed with pure dry nitrogen to avoid condensation.
Protein aggregation was followed by monitoring the changes in light scattering intensity at 340 nm (4-nm slit widths).
Thermodynamics
The thermodynamic parameters of the pressure- and temperature-induced spectral transitions were determined by using a simple two-state model,
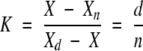 |
(1) |
where K is the equilibrium constant between the denatured (d) and the native (n) protein conformational states, which are characterized by the spectral CDA values Xd and Xn, respectively. X is the measured CDA value. Introducing thermodynamic relationships, equation 1 transforms to
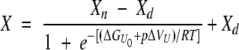 |
(2) |
and
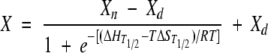 |
(3) |
for the pressure and temperature denaturation, respectively. The fit of X = f(p) and X = f(1/T) in equations 2 and 3 permitted then the determination of the thermodynamic parameters. ΔGU for temperature-induced unfolding has been calculated using equations 3–5, and the fitted values of ΔHT1/2 and ΔST1/2. A fixed ΔCp value (change in heat capacity for the denaturation reaction) of 5.63 kJ K−1 mol−1 was used (Torrent et al. 2003).
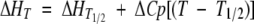 |
(4) |
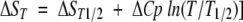 |
(5) |
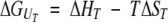 |
(6) |
Circular dichroism spectroscopy
CD spectra of native PrP proteins were recorded at 20°C using a J810 spectropolarimeter (Jasco). A 0.1-cm optical path-length quartz cell was used to record spectra of proteins in the far UV region (190–260 nm). The protein concentration was 0.3 mg mL−1, and the buffer was 20 mM Tris-HCl buffer at pH 7.0. Baseline corrected CD spectra were acquired at a scan speed of 20 nm min−1, a 1-nm bandwidth, and a response time of 1 sec. The sample compartment was purged with pure dry nitrogen. Spectra were signal-averaged over four scans. The kinetics of conformational transition to β-sheet structure were followed by changes in ellipticity at 222 nm. The proteins (0.2 mg mL−1) were incubated in 20 mM sodium acetate buffer (pH 4.0), containing 150 mM NaCl, at 37°C.
Acknowledgments
This work was supported by GIS-prions (Ministry of Research), ATC-prions (INSERM), and Human Science Frontier Program (HSFP). J.T. acknowledges an INSERM postdoctoral fellowship. We are grateful to F. Heitz for providing CD equipment.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04989405.
References
- Alvarez-Martinez, M.T., Torrent, J., Lange, R., Verdier, J.M., Balny, C., and Liautard, J.P. 2003. Optimized overproduction, purification, characterization and high-pressure sensitivity of the prion protein in the native (PrP(C)-like) or amyloid (PrP(Sc)-like) conformation. Biochim. Biophys. Acta 1645 228–240. [DOI] [PubMed] [Google Scholar]
- Apetri, A.C. and Surewicz, W.K. 2002. Kinetic intermediate in the folding of human prion protein. J. Biol. Chem. 277 44589–44592. [DOI] [PubMed] [Google Scholar]
- Apetri, A.C., Surewicz, K., and Surewicz, W.K. 2004. The effect of disease-associated mutations on the folding pathway of human prion protein. J. Biol. Chem. 279 18008–18014. [DOI] [PubMed] [Google Scholar]
- Baskakov, I.V., Legname, G., Baldwin, M.A., Prusiner, S.B., and Cohen, F.E. 2002. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 277 21140–21148. [DOI] [PubMed] [Google Scholar]
- Bridgman, P.W. 1935. The pressure-volume-temperature relations of the liquid, and the phase diagram of heavy water. J. Chem. Phys. 3 597–605. [Google Scholar]
- Calzolai, L. and Zahn, R. 2003. Influence of pH on NMR structure and stability of the human prion protein globular domain. J. Biol. Chem. 278 35592–35596. [DOI] [PubMed] [Google Scholar]
- Caughey, B.W., Dong, A., Bhat, K.S., Ernst, D., Hayes, S.F., and Caughey, W.S. 1991. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry 30 7672–7680. [DOI] [PubMed] [Google Scholar]
- Coustou, V., Deleu, C., Saupe, S., and Begueret, J. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anse-rina behaves as a prion analog. Proc. Natl. Acad. Sci. 94 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derreumaux, P. 2001. Evidence that the 127–164 region of prion proteins has two equi-energetic conformations with β or α features. Biophys. J. 81 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, C.M. 2002. Getting out of shape. Nature 418 729–730. [DOI] [PubMed] [Google Scholar]
- Eftink, M.R. 2000. Use of fluorescence spectroscopy as thermodynamics tool. Methods Enzymol. 323 459–473. [DOI] [PubMed] [Google Scholar]
- Ferrao-Gonzales, A.D., Palmieri, L., Valory, M., Silva, J.L., Lashuel, H., Kelly, J.W., and Foguel, D. 2003. Hydration and packing are crucial to amyloido-genesis as revealed by pressure studies on transthyretin variants that either protect or worsen amyloid disease. J. Mol. Biol. 328 963–974. [DOI] [PubMed] [Google Scholar]
- Foguel, D., Suarez, M.C., Ferrao-Gonzales, A.D., Porto, T.C., Palmieri, L., Einsiedler, C.M., Andrade, L.R., Lashuel, H.A., Lansbury, P.T., Kelly, J.W., et al. 2003. Dissociation of amyloid fibrils of α-synuclein and trans-thyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc. Natl. Acad. Sci. 100 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Heremans, K. and Smeller, L. 1998. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta 1386 353–370. [DOI] [PubMed] [Google Scholar]
- Horiuchi, M. and Caughey, B. 1999. Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the pro-tease-resistant state. EMBO J. 18 3193–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann, S. and Glockshuber, R. 1998. A scrapie-like unfolding intermediate of the prion protein domain PrP(121–231) induced by acidic pH. Proc. Natl. Acad. Sci. 95 6010–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, T.L., Liu, H., Ulyanov, N.B., Farr-Jones, S., Zhang, H., Donne, D.G., Kaneko, K., Groth, D., Mehlhorn, I., Prusiner, S.B., et al. 1997. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. 94 10086–10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin, N., Coic, Y.M., Landon, C., Ovtracht, L., Baleux, F., Neumann, J.M., and Sanson, A. 2002. Most of the structural elements of the globular domain of murine prion protein form fibrils with predominant β-sheet structure. FEBS Lett. 529 256–260. [DOI] [PubMed] [Google Scholar]
- Jansen, R., Grudzielanek, S., Dzwolak, W., and Winter, R. 2004. High pressure promotes circularly shaped insulin amyloid. J. Mol. Biol. 338 203–206. [DOI] [PubMed] [Google Scholar]
- Jonas, J. 2002. High-resolution nuclear magnetic resonance studies of proteins. Biochim. Biophys. Acta 1595 145–159. [DOI] [PubMed] [Google Scholar]
- Kitamura, Y. and Itoh, T. 1987. Reaction volume of protonic ionization for buffering agents: Prediction of pressure-dependence of pH and pOH. J. Sol. Chem. 16 715–725. [Google Scholar]
- Kocisko, D.A., Come, J.H., Priola, S.A., Chesebro, B., Raymond, G.J., Lans-bury, P.T., and Caughey, B. 1994. Cell-free formation of protease-resistant prion protein. Nature 370 471–474. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Kuwata, K., Li, H., Yamada, H., Legname, G., Prusiner, S.B., Akasaka, K., and James, T.L. 2002. Locally disordered conformer of the hamster prion protein: A crucial intermediate to PrPSc? Biochemistry 41 12277–12283. [DOI] [PubMed] [Google Scholar]
- Kuwata, K., Kamatari, Y.O., Akasaka, K., and James, T.L. 2004. Slow conformational dynamics in the hamster prion protein. Biochemistry 43 4439–4446. [DOI] [PubMed] [Google Scholar]
- Lange, R. and Balny, C. 2002. UV-visible derivative spectroscopy under high pressure. Biochim. Biophys. Acta 1595 80–93. [DOI] [PubMed] [Google Scholar]
- Lange, R., Bec, N., Mozhaev, V.V., and Frank, J. 1996a. Fourth derivative UV-spectroscopy of proteins under high pressure. II. Application to reversible structural changes. Eur. Biophys. J. 24 284–292. [Google Scholar]
- Lange, R., Frank, J., Saldana, J.L., and Balny, C. 1996b. Fourth derivative UV-spectroscopy of proteins under high pressure. I. Factors affecting the fourth derivative spectrum of the aromatic amino acids. Eur. Biophys. J. 24 277–283. [Google Scholar]
- Liu, H., Farr-Jones, S., Ulyanov, N.B., Llinas, M., Marqusee, S., Groth, D., Cohen, F.E., Prusiner, S.B., and James, T.L. 1999. Solution structure of Syrian hamster prion protein rPrP(90–231). Biochemistry 38 5362–5377. [DOI] [PubMed] [Google Scholar]
- Marchal, S., Shehi, E., Harricane, M.C., Fusi, P., Heitz, F., Tortora, P., and Lange, R. 2003. Structural instability and fibrillar aggregation of non-expanded human ataxin-3 revealed under high pressure and temperature. J. Biol. Chem. 278 31554–31563. [DOI] [PubMed] [Google Scholar]
- Martins, S.M., Chapeaurouge, A., and Ferreira, S.T. 2003. Folding intermediates of the prion protein stabilized by hydrostatic pressure and low temperature. J. Biol. Chem. 278 50449–50455. [DOI] [PubMed] [Google Scholar]
- Meersman, F., Smeller, L., and Heremans, K. 2002. Comparative Fourier transform infrared spectroscopy study of cold-, pressure-, and heat-induced unfolding and aggregation of myoglobin. Biophys. J. 82 2635–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas, M., Vanik, D.L., and Surewicz, W.K. 2001. On the mechanism of α-helix to β-sheet transition in the recombinant prion protein. Biochemistry 40 6982–6987. [DOI] [PubMed] [Google Scholar]
- Morrissey, M.P. and Shakhnovich, E.I. 1999. Evidence for the role of PrP(C) helix 1 in the hydrophilic seeding of prion aggregates. Proc. Natl. Acad. Sci. 96 11293–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, K.M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehl-horn, I., Huang, Z., Fletterick, R.J., Cohen, F.E., et al. 1993. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. 90 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz, D., Williamson, R.A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R.B., Rozenshteyn, R., James, T.L., Houghten, R.A., Cohen, F.E., et al. 1997. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J. Mol. Biol. 273 614–622. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B., Scott, M.R., DeArmond, S.J., and Cohen, F.E. 1998. Prion protein biology. Cell 93 337–348. [DOI] [PubMed] [Google Scholar]
- Randolph, T.W., Seefeldt, M., and Carpenter, J.F. 2002. High hydrostatic pressure as a tool to study protein aggregation and amyloidosis. Biochim. Biophys. Acta 1595 224–234. [DOI] [PubMed] [Google Scholar]
- Ruan, K. and Weber, G. 1989. Hysteresis and conformational drift of pressure-dissociated glyceraldehydephosphate dehydrogenase. Biochemistry 28 2144–2153. [DOI] [PubMed] [Google Scholar]
- Silva, J.L., Miles, E.W., and Weber, G. 1986. Pressure dissociation and conformational drift of the β dimer of tryptophan synthase. Biochemistry 25 5780–5786. [DOI] [PubMed] [Google Scholar]
- Silva, J.L., Foguel, D., and Royer, C.A. 2001. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem. Sci. 26 612–618. [DOI] [PubMed] [Google Scholar]
- Silva, J.L., Oliveira, A.C., Gomes, A.M., Lima, L.M., Mohana-Borges, R., Pacheco, A.B., and Foguel, D. 2002. Pressure induces folding intermediates that are crucial for protein-DNA recognition and virus assembly. Biochim. Biophys. Acta 1595 250–265. [DOI] [PubMed] [Google Scholar]
- Speare, J.O., Rush 3rd, T.S., Bloom, M.E., and Caughey, B. 2003. The role of helix 1 aspartates and salt bridges in the stability and conversion of prion protein. J. Biol. Chem. 278 12522–12529. [DOI] [PubMed] [Google Scholar]
- Swietnicki, W., Morillas, M., Chen, S.G., Gambetti, P., and Surewicz, W.K. 2000. Aggregation and fibrillization of the recombinant human prion protein huPrP90–231. Biochemistry 39 424–431. [DOI] [PubMed] [Google Scholar]
- Taylor, J.P., Hardy, J., and Fischbeck, K.H. 2002. Toxic proteins in neurode-generative disease. Science 296 1991–1995. [DOI] [PubMed] [Google Scholar]
- Torrent, J., Connelly, J.P., Coll, M.G., Ribo, M., Lange, R., and Vilanova, M. 1999. Pressure versus heat-induced unfolding of ribonuclease A: The case of hydrophobic interactions within a chain-folding initiation site. Biochemistry 38 15952–15961. [DOI] [PubMed] [Google Scholar]
- Torrent, J., Alvarez-Martinez, M.T., Heitz, F., Liautard, J.P., Balny, C., and Lange, R. 2003. Alternative prion structural changes revealed by high pressure. Biochemistry 42 1318–1325. [DOI] [PubMed] [Google Scholar]
- Torrent, J., Alvarez-Martinez, M.T., Harricane, M.C., Heitz, F., Liautard, J.P., Balny, C., and Lange, R. 2004. High pressure induces scrapie-like prion protein misfolding and amyloid fibril formation. Biochemistry 43 7162–7170. [DOI] [PubMed] [Google Scholar]
- Vanik, D.L. and Surewicz, W.K. 2002. Disease-associated F198S mutation increases the propensity of the recombinant prion protein for conformational conversion to scrapie-like form. J. Biol. Chem. 277 49065–49070. [DOI] [PubMed] [Google Scholar]
- Viles, J.H., Donne, D., Kroon, G., Prusiner, S.B., Cohen, F.E., Dyson, H.J., and Wright, P.E. 2001. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry 40 2743–2753. [DOI] [PubMed] [Google Scholar]
- Wickner, R.B. 1994. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264 566–569. [DOI] [PubMed] [Google Scholar]
- Wong, C., Xiong, L.W., Horiuchi, M., Raymond, L., Wehrly, K., Chesebro, B., and Caughey, B. 2001. Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J. 20 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Stockel, J., Mehlhorn, I., Groth, D., Baldwin, M.A., Prusiner, S.B., James, T.L., and Cohen, F.E. 1997. Physical studies of conformational plasticity in a recombinant prion protein. Biochemistry 36 3543–3553. [DOI] [PubMed] [Google Scholar]