Abstract
Overproduction of proteins from cloned genes using fusion protein expression vectors in Escherichia coli and eukaryotic cells has increased the quantity of protein produced. This approach has been widely used in producing soluble recombinant proteins for structural and functional analysis. One major disadvantage, however, of applying this approach for clinical or bioindustrial uses is that proteolytic removal of the fusion carrier is tedious, expensive, and often results in products with additional amino acid residues than the native proteins. Here we describe a new method for productions of native proteins with original amino termini in vivo via intracellular self-cleavage of the fusion protein using tobacco etch virus (TEV) protease. Our design allows one to simultaneously clone any gene into multiple fusion protein vectors using two unique cloning sites (i.e., SnaBI and XhoI) without restriction digestion, and then rapidly identifies those constructs producing soluble native proteins. This method will make the fusion protein approach more feasible for protein drug research.
Keywords: tobacco etch virus protease, sticky-end PCR, fusion protein approach
The fusion protein approach has been widely applied in modern biology and protein science research. More than 20 carrier proteins or affinity tags are available now for this approach to produce soluble heterologous proteins in various host organisms (Sambrook and Russell 2000). Although the use of these carrier proteins has resulted in successful overexpression of many heterologous proteins, each must be tested empirically, and certainly may not possess maximal solubility. Moreover, each expression scenario requires a specific vector. Recloning of the passenger protein gene into each of specific vectors is extremely labor-intensive. Recombinational cloning methods (Liu et al. 1998; Hartley et al. 2000) and sticky-end PCR cloning strategy (Shih et al. 2002; Wang and Wang 2004) were applied to minimize efforts required for alternate expression, and also allowed one to develop a high-throughput system to screen for soluble recombinant proteins.
Because of concerns about the impact of carrier proteins or affinity tags on the structure or activity of a passenger target protein, it is ordinarily desirable to remove them. Typically, passenger target protein is separated from the fusion carrier by site-specifically proteolysis after affinity chromatography. It is this step that was considered to be the Achilles’ heel of fusion protein approach, particularly in applications such as structural biology or protein–drug production. It is relatively common to encounter a situation in which fusion carriers cannot be processed effectively because of the steric hindrance at the cleavage site. Tedious optimization of cleavage conditions in conjunction with the high cost of proteases (e.g., Factor Xa and enterokinase) often prevent further use of this approach. In the worst circumstances, the cleaved products aggregate immediately after removal from their fusion carriers. In some situations, affinity tags in fusion proteins fail to interact efficiently with their immobilized ligands. Finally, the resulting cleavage products may contain extraneous amino acid residues due to the introduction of a protease-specific recognition site as well as the restriction enzyme cloning sites in the engineered linker region. Although the last scenario can be overcome by the use of Factor Xa or tobacco etch virus protease (TEVP) (Sambrook and Russell 2000), a rather long PCR forward primer must be used for addition of the protease recognition site at the 5′ end of the passenger protein gene.
Because of its higher stringent sequence specificity, TEVP is more often used than other proteases, including Factor Xa or enterokinase. Recent biochemical and structural studies indicate that TEVP specifically cleaves the amino acid sequence -Glu(P6)-P5-P4-Tyr(P3)-P2-Gln(P1)-↓-P1′- in a fusion protein, where P2, P4, and P5 positions are nonconserved amino acids (Dougherty et al. 1989; Kapust et al. 2002). It had been shown that almost all side chains (except Pro) can be accommodated in the P1′ position with little impact on the efficiency of processing (Phan et al. 2002). An intracellular fusion protein processing system had been developed and exhibited high specificity in processing in Escherichia coli. This system used two compatible expression vectors to separately produce TEVP and a maltose binding protein (MBP) fusion protein containing the TEV recognition site (rsTEV) (Kapust and Waugh 2000). However, this intracellular processing system still will encounter most problems of the in vitro cleavage methods described above.
In an effort to further ameliorate the TEVP intracellular processing system, we found out that an MBP-TEVP-rsTEV-GFP-His6 fusion protein is able to carry out near 100% site-specific autonomous cleavage in vivo, and generates MBP-TEVP and GFP-His6 with a large quantity and high solubility. Sticky-end PCR cloning strategy (Shih et al. 1998; Wang and Wang 2004) was applied to further modify this fusion protein construct so that it could successfully yield one of 20 otherwise identical GFP-His6 proteins with different amino acids in the P1′ position. Therefore, this method allows one to produce recombinant proteins with the native amino termini from MBP fusion proteins. The same design was also utilized here to modify other affinity tag vectors, including MBP, NusA, thioredoxin (Trx), glutathione S-transferase (GST), calmodulin binding protein (CBP), and hexahistidine tag (His6). Taken together, this method makes it possible to quickly clone and screen multiple affinity tag or carrier protein vectors that yield native proteins in vivo.
Results
Intracellular processing of MBP-TEVP-rsTEV-GFP-His6 fusion protein
In this study, we devised a new method in which fusion proteins could carry out site-specific and autonomous processing in intact cells. MBP-TEVP fusion protein (Fig. 1A ▶) was first chosen because of the following considerations. MBP is a far more effective solubilizing agent than most other fusion carriers or affinity tags (Shih et al. 1998; Kapust and Waugh 1999). TEVP exhibits high sequence stringency (Dougherty et al. 1989; Phan et al. 2002), and can be overexpressed in E. coli or eukaryotic cells without interfering with cell viability (Kapust and Waugh 2000; Gruber et al. 2003).
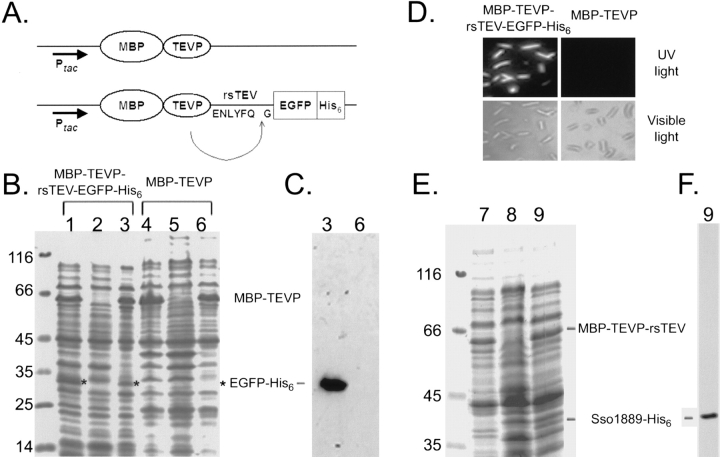
Figure 1.
In vivo cleavage of MBP-TEVP-rsTEV-EGFP-His6 and MBP-TEVP-rsTEV-Sso1889-His6 fusion proteins. (A) Schematic map of the MBP-TEVP and MBP-TEVP-rsTEV-EGFP-His6 fusion protein expression vector. The amino acid sequence of the TEVP recognition site is indicated. Ptac: The tac promoter is used for IPTG induction. Solubility tests were carried out as previously described (Shih et al. 2002). Samples of the total protein and soluble protein fractions were separated by 10% SDS-PAGE under reducing conditions and stained with Coomassie blue (B,E): lanes 1, 4, and 7, whole-cell lysates of E. coli cells induced with IPTG; lanes 2, 5, and 8, whole-cell lysates of uninduced cells; lanes 3, 6, and 9, soluble proteins from IPTG-induced cells. The molecular weight standards are shown on the left. MBP-TEVP is indicated by a bar on the right. EGFP-His6 protein bands are marked by an asterisk (*) on the right. Soluble proteins from IPTG-induced cells (lanes 3,6,9) were analyzed by Western blot using the anti-His6 antibody to verify EGFP-His6 (C) and Sso1889-His6 (E). Note that both EGFP-His6 and Sso1889-His6 are completely cleaved from the MBP-TEVP-rsTEV-EGFP-His6 and MBP-TEVP-rsTEV-Sso1889-His6 fusion proteins, since the latter could not be detected by Western blot. (D) Visualization of EGFP-His6 in the IPTG-induced E. coli cells. Images of living cells were taken by a fluorescence microscopy using either UV light or visible light.
The MBP-TEVP fusion vector was further modified to express the MBP-TEVP-rsTEV-EGFP-His6 fusion protein (Fig. 1A ▶). The E. coli strain JM109(DE3) was used for protein expression. After 24 h of IPTG induction at 18°–20°C, cells were harvested and lysed for the protein solubility test (Shih et al. 2002; Wang and Wang 2004). To increase the accuracy of solubility testing, an ultracentrifugal force (90,000g) was applied to eliminate both partially folded protein aggregates and insoluble materials from total lysates. SDS-PAGE was used to separate the proteins in total cell lysates from cells either induced with IPTG (Fig. 1B ▶, lanes 1,4) or not (Fig. 1B ▶, lanes 2,5), and in the soluble protein fraction from IPTG-induced cells (Fig. 1B ▶, lanes 3,6). We found that the MBP-TEV fusion protein (apparent molecular weight ∼70,000) was well induced and soluble (Fig. 1B ▶, lane 6). The MBP-TEVP-rsTEV-EGFP-His6 fusion protein not only was well induced but also correctly processed to yield MBP-TEVP-rsTEV and EGFP-His6, respectively (Fig. 1B ▶, lane 3, below). The resulting EGFP-His6 protein was confirmed first by Western blot using the anti-His6 antibody (Fig. 3C ▶, lane 3, below). We noted that the yield of intracellular processing is nearly 100%, since almost no signal of the unprocessed MBP-TEVP-rsTEV-EGFP-His6 was detected by Western blot using the anti-His6 antibody (Fig. 3C ▶, lane 3). Extracts containing EGFP-His6 were also subjected to purification on Ni2+-containing resins that selectively retain His6-tagged polypeptides (data not shown). Peptide sequencing of the purified protein showed that the NH2-terminal pentamer GEFGL matched the first five amino acid residues of EGFP-His6. Finally, when both E. coli cells were examined by a fluorescence microscope, only cells expressing the MBP-TEVP-rsTEV-EGFP-His6 fusion protein emitted green fluorescence upon UV light illumination (Fig. 1D ▶). Taken together, we concluded that the MBP-TEVP-rsTEV-EGFP-His6 fusion protein is able to carry out near 100% autonomous site specific processing in vivo.
Figure 3.
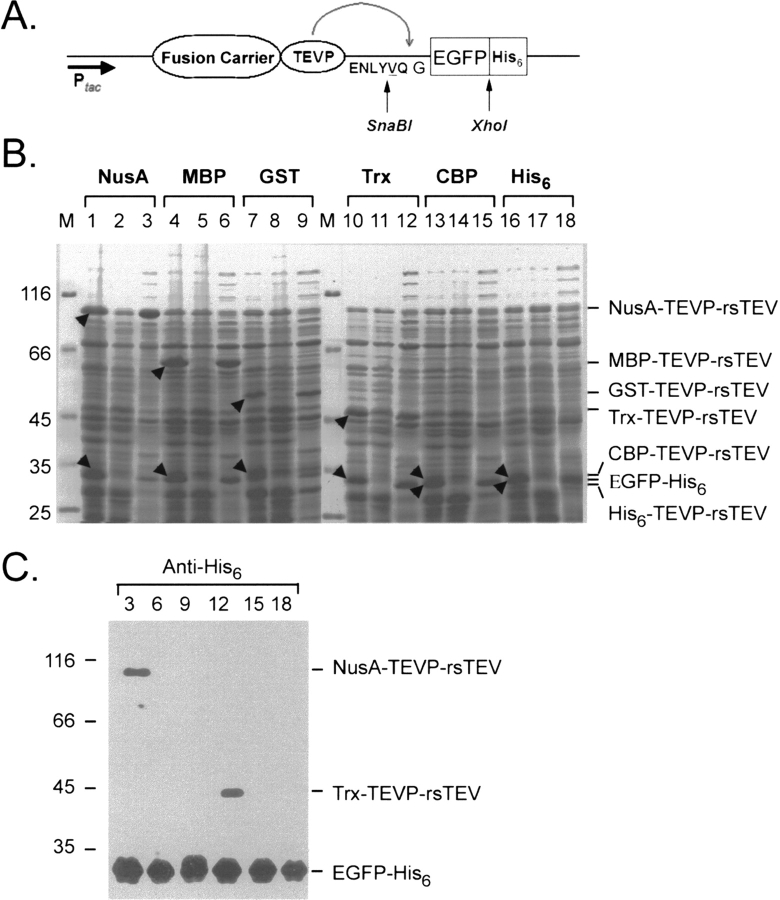
Intracellular processing of multiple FC-TEVP-rsTEV-EGFP-His6 fusion protein expression vectors; each scenario contains a different fusion carrier (FC). (A) Schematic map of the FC-TEVP-rsTEV-EGFP-His6 construct. (B) Protein solubility tests were carried out as described in Figure 1B ▶. Protein samples were separated by SDS-PAGE and analyzed by Coomassie blue staining: lanes 1, 4, 7, 10, 13, and 16. Total cell lysates of E. coli cells induced with IPTG; lanes 2, 5, 8, 11, 14, and 17. Total cell lysates of E. coli cells without IPTG induction; lanes 3, 6, 9, 12, 15, and 18. Soluble fractions of E. coli cells induced with IPTG. The positions of cleaved products were marked by arrowheads and also indicated on the left. (C) Western blot analysis of the soluble fractions of E. coli cells induced with IPTG using anti-His6 antibody. Note that NusA-TEVP-rsTEV and Trx-TEVP-rsTEV were also recognized by the anti-His6 antibody, because both NusA and Trx contain an additional His6 tag. The molecular weight standards are shown on the left.
This self-processing fusion protein strategy also has been successfully applied to a different passenger target protein, i.e., the Sulfolobus solfataricus (Sso) 1889 protein (as a different model system). Here, EGFP-His6 was replaced by Sso1889-His6, and a solubility test was carried out as described above. SDS-PAGE stained with Coomassie blue indicated that MBP-TEVP-rsTEV-Sso1889-His6 indeed self-cleaved into MBP-TEVP-rsTEV and Sso1889-His6 (Fig. 1E ▶). Like EGFP-His6, Sso1889-His6 is completely cleaved off since MBP-TEVP-rsTEV-Sso1889-His6 could not be detected by Western blotting using the anti-His6 antibody (Fig. 1F ▶).
Production of recombinant proteins with a native amino acid sequence
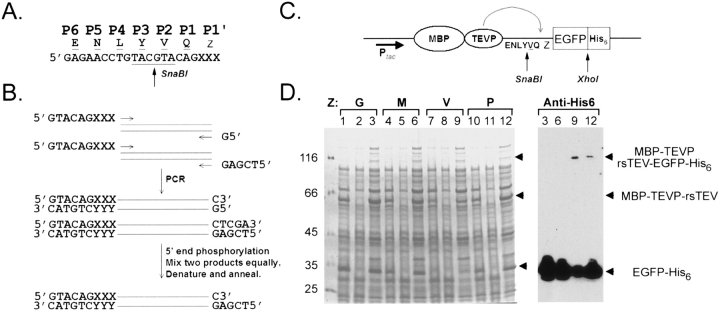
Owing to the presence of aminopeptidase (and also endopeptidase) activities in both eukaryotic and prokaryotic cells, the N-terminal fMet or Met amino acid is often split off, leaving the other amino acid residues as the N terminus in processed native proteins. It is often desirable to carry out site-specific cleavage to yield native N termini, since they may play an essential structural or functional role. Here we design a general approach that is more effective in PCR cloning and is able to autonomously produce recombination proteins with native amino termini. First of all, an SnaBI restriction enzyme site (5′-TACGTA-3′) was created as described in Figure 2A ▶, so as that the amino acid residue in the P2 position will be replaced from Phe (Fig. 1A ▶) to Val (Fig. 2A ▶). This modification allows cloning of any target protein gene into the MBP-TEV expression vector between the 5′ end SnaBI and the 3′ end XhoI sites (with or without the stop codon) by the sticky-end PCR method (Fig. 2B ▶). The method requires three PCR primers (one forward and two reverse) and reactions in two separate tubes. Both PCR products were purified and mixed equally. After denaturation and renaturation, ∼50% of the final products carry one SnaBI blunt end and one XhoI cohesive end, and are ready for ligation even without restriction digestion of PCR products. This method is suitable for cloning any gene, even genes with internal SnaBI or XhoI restriction sites. To optimize cloning efficiency, sticky-end PCR products were 5′ phosphorylated with T4 poly-nucleotide kinase and the vectors were dephosphorylated by calf intestine alkaline phosphatase. Finally, this cloning strategy allows one to express proteins with native amino termini, because all 20 amino acid residues can be chosen at the P1′ position. The resulting fusion protein construct was illustrated as Figure 2C ▶, where Z represents the P1′ amino acid.
Figure 2.
Cloning design and intracellular cleavage of otherwise identical MBP-TEVP-rsTEV-EGFP-His6 fusion proteins with different amino acid residues in the P1′ position. (A) Introduction of an SnaBI site at the coding sequence of the TEVP recognition site. (B) Sticky-end PCR cloning strategy. One forward and two reverse PCR primers as well as PCR reactions were used in two separate tubes. An equal amount of the two PCR products were mixed, and then 5′ ends were phosphorylated with T4 polynucleotide kinase. After denaturing (95°C for 5 min) and renaturing (65°C for 5 min), ∼50% of the final products carry SnaBI (5′) and XhoI (3′) ends and are ready for ligation into the vector. The codon and anticodon of the amino acid residues in the P1′ position is indicated as “XXX” or “YYY,” respectively. (C) Schematic representation of the new MBP-TEVP-rsTEV-EGFP-His6 fusion protein vector. The amino acid residue in the P1′ position is indicated as “Z.” (D) Samples of soluble protein lysates from IPTG-induced E. coli cells producing MBP-TEVP-rsTEV-EGFP-His6 with different amino acid residues in the P1′ position (indicated in a single-letter code) were separated by SDS-PAGE and analyzed either by Coomassie blue staining or by Western blot using anti-His6 antibody. The positions of MBP-TEVP-rsTEV and EGFP-His6 protein bands are marked. The molecular weight standards are shown on the left.
To further validate the efficacy of this new design, we constructed and expressed four different MBP-TEVP-rsTEV-EGFP-His6 fusion proteins in the JM109(DE3) strain. Each one of them has different amino acid residues at the P1′ position, i.e., Met, Gly, Pro, and Val, respectively. Host cells were harvested and lysed, and then subjected to a protein solubility test in parallel. All of these four fusion proteins were effectively processed into MBP-TEVP-rsTEV and EGFP-His6 in vivo, as revealed by both SDS-PGE and Western blot using anti-GFP antibody (Fig. 2D ▶). It is surprising to find a near complete processing of the fusion protein with Pro in the P1′ position. A previous study by Kapust et al. (2002) indicated that MBP-rsTEV-NusG with Pro in the P1′ position exhibited no processing in E. coli cells coexpressing TEVP. One possibility is that the GFP-His6 fusion protein used in this study is simply a better TEVP substrate than NusG. Alternatively, self-cleavage was carried out in MBP-TEVP-rsTEV-GFP-His6, whereas a separate TEVP molecule processes MBP-rsTEV-NusG. An intramolecular catalysis is more effective than an intermolecular enzymatic reaction. However, we could not rule out the possibility that trans proteolytic cleavage reaction may still occur in our cis fusion protein construct. Finally, we are not certain if the cleavage reaction occurs during or before the GFP-His6 is completely folded.
Parallel cloning and screening of multiple self-cleavage fusion protein vectors
This self-processing strategy was further expanded to several other fusion carrier or affinity tag expression systems, including NusA, thioredoxin (Trx), glutathione S-transferase (GST), calmodulin binding protein (CBP), His6 tag, etc. We constructed five additional TEVP fusion vectors, including GST-TEVP, Trx-TEVP, NusA-TEVP, CBP-TEVP, and His6-TEVP. All these vectors shares the same TEV recognition site as well as the SnaBI and XhoI restriction sites (Fig. 3A ▶), so that one could carry out parallel cloning of sticky-end PCR products as described in Figure 2C ▶. As indicated by SDS-PAGE (Fig. 3B ▶) and Western blot using anti-GFP antibody (Fig. 3C ▶), all six vectors successfully carried out intracellular cleavage and produced EGFP-His6 proteins (Fig. 3 ▶).
Discussion
In the postgenomic era, high-throughput protein expression technologies are essential tools. Conceivably, two greatest technical obstacles to the production of recombinant proteins for functional and structural analysis are solubility and yield. The fusion protein approach offers a means of circumventing these two problems, and therefore has become a cornerstone of modern biological research. However, due to concerns about the deleterious effects of fusion carrier on the structure and activity of a passenger, it is often desirable to obtain the native protein free from its fusion carrier partner.
Here we have developed an intracellular self-processing fusion protein system for producing soluble native protein in E. coli. This same strategy can also be applied to facilitate native protein production in other parkaryotic and eukaryotic heterologous expression systems. Our new design avoids not only the use of expensive proteases for fusion protein cleavage but also the tedious cloning efforts into different expression vectors. Parallel cloning was achieved here by the sticky-end PCR method in conjunction with two unique cloning sites: SnaBI and XhoI. It can be applied to clone any gene, including those with internal SnaBI and XhoI.
The choice of an SnaBI site is also very intriguing, because it greatly improves vectors for the expression of fusion proteins with a TEV protease cleavage site. Following proteolytic cleavage with TEV protease, a passenger protein with the desired N terminus can be obtained. This design is not only feasible in cis, as described in this study (e.g., the MBP-TEVP-rsTEV-passenger protein), it is also useful for common trans approaches (e.g., the MBP-rsTEV-passenger protein). Therefore, we believe this approach will greatly help in screening the expression of a large number of native proteins for functional and structural studies.
Most protein carriers used in the fusion protein approach also are affinity tags. Admittedly, the in vivo cleavage approach described in the present study may deprive the advantage of affinity tags in protein purification. Nevertheless, we still find this approach very useful, particularly in those cases that functionality and solubility of proteins must be taken as priorities. After all, there are many methods available for protein purification. For example, our new design would significantly augment the practicability of fusion protein approach in protein drug research.
Another technical concern of our cis approach is the potential interference with folding of the passenger protein caused by upstream TEV protease. It had been reported that one partner of a hybrid protein can be destabilized by the other partner while maintaining its structural and functional characteristics (Blondel et al. 1996). Intriguingly, a similar model has also been proposed to explain why MBP is uncommonly effective at promoting the solubility and folding of its fusion partners (Kapust and Waugh 1999). Therefore, it would be interesting to find out if the TEV protein could interfere (either negatively or positively) with foldability or stability of the C-terminal passenger proteins. However, in the present study, we did not observe any apparent folding problem with MBP-TEVP-rsTEV-GFP-His6 and MBP-TEVP-rsTEV-Sso1889-His6. Finally, we suggest that the folding interference problem may likely be overcome by a proper design of the “linker sequence” between the TEV protease and passenger protein.
Materials and methods
Molecular cloning and protein analysis
The cDNA of EGFP was amplified by PCR from the pEGFP-N2 vector (Clontech). Six fusion protein vectors used in this study were described previously (Shih et al. 2002; Wang and Wang 2004), including MBP, NusA, Trx, GST, CBP, and His6 tag. Parallel sticky-end PCR cloning, protein induction, and solubility testing were also carried out as previous described (Shih et al. 2002; Wang and Wang 2004). For protein induction, bacterial cultures in the log phase (OD600 ∼0.6) were induced with 0.1 mM IPTG at 18°–20°C for 24 h. We found that low temperature and long induction time greatly facilitate correct protein folding (Shih et al. 2002). Anti-His6 antibody (Clontech) and anti-GFP antibody (Molecular Probes) were used for Western blot analysis.
Living cell microscopy
The EGFP fusion proteins were visualized in living cells. After IPTG induction, E. coli cells were harvested by centrifugation, washed once, and then resuspend with the same volume of phosphate-base saline. About 2 μL was applied to a microscope slide, excess liquid was aspirated, and a glass coverslip was placed on the slide. The cell outlines were visualized simultaneously with the GFP signal using Chroma filter set no. 86002v1. Images were captured with a Leica DMR microscopy plus a cooled charge-coupled device (CCD) camera (Roper Scientific) and MetaVue software (Universal Imaging Corporation).
Acknowledgments
This study was supported by grants from the National Science Council and Academia Sinica (AS92IBC3) of Taiwan to T.F.W. and A.H.J.W. The National Genomic Medicine Project funded the National Core Facility of High Throughput Protein Production to A.H.J.W. from the National Science Council, Taiwan. We thank the National Core Facility of Proteomic Research for N-terminal peptide analysis.
Abbreviations
TEVP, tobacco etch virus protease
rsTEV, TEV recognition site
Sso, Sulfolobus solfataricus
MBP, maltose binding protein
Trx, thioredoxin
GST, glutathione S-transferase
CBP, calmodulin binding protein
His6, hexahistidine tag
FC, fusion carrier
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041129605.
References
- Blondel, A., Nageotte, R., and Bedouelle, H. 1996. Destablizing interactions between the partners of a bifunctional fusion protein. Protein Eng. 9 231–238. [DOI] [PubMed] [Google Scholar]
- Dougherty, W.G., Cary, S.M. and Parks, T.D. 1989. Molecular genetic analysis of a plant virus polyprotein cleavage site: A model. Virology 171 356–364. [DOI] [PubMed] [Google Scholar]
- Gruber, S., Haering, C.H., and Nasmyth, K. 2003. Chromosomal cohesin forms a ring. Cell 112 765–777. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust, R.B. and Waugh, D.S. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr. Purif. 19 312–318. [DOI] [PubMed] [Google Scholar]
- Kapust, R.B., Tozser, J., Copeland, T.D., and Waugh, D.S. 2002. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 294 949–955. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8 1300–1309. [DOI] [PubMed] [Google Scholar]
- Phan, J., Zdanov, A., Evdokimov, A.G., Tropea, J.E., Peters 3rd, H.K., Kapust, R.B., Li, M., Wlodawer, A., and Waugh, D.S. 2002. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277 50564–50572. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2000. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shih, Y.P., Kung, W.-M., Chen, J.C., Yeh, C.H., Wang, A.H.-J., and Wang, T.F. 2002. High-throughput screening of soluble recombinant proteins. Protein Sci. 11 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.F. and Wang, A.H.-J. 2004. High-throughput screening of soluble recombinant proteins. In Purifying proteins for proteomics: A laboratory manual (ed. R.J. Simpson), pp. 111–119. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.