Abstract
The filamentous bacteriophage Pf1 undergoes a reversible temperature-dependent transition that is also influenced by salt concentrations. This structural responsiveness may be a manifestation of the important biological property of flexibility, which is necessary for long, thin filamentous assemblies as a protection against shear forces. To investigate structural changes in the major coat protein, one- and two-dimensional solid-state NMR spectra of concentrated solutions of Pf1 bacteriophage were acquired, and the structure of the coat protein determined at 0°C was compared with the structure previously determined at 30°C. Despite dramatic differences in the NMR spectra, the overall change in the coat protein structure is small. Changes in the orientation of the C-terminal helical segment and the conformation of the first five residues at the N-terminus are apparent. These results are consistent with prior studies by X-ray fiber diffraction and other biophysical methods.
Keywords: NMR, filamentous bacteriophage, temperature transition, protein structure
Pf1 is a filamentous bacteriophage that infects Pseudomonas aeruginosa. Each bacteriophage particle consists of ~7000 symmetrically arranged copies of its 46-residue major coat protein, a single-stranded circular DNA genome stretched lengthwise on the interior, and a few copies of “minor” coat proteins at each end. The structure of the major coat protein has been determined in bacteriophage particles, where it has been found to have a remarkably complex structure for such a small helical protein (Nambudripad et al. 1991; Welsh et al. 2000; Thiriot et al. 2004). Further, the X-ray fiber diffraction studies show that the bacteriophage can exist in two forms with slightly different symmetries, and that it can undergo a reversible temperature-dependent transition between them (Wachtel et al. 1976; Nave et al. 1979; Specthrie et al. 1987; Marvin et al. 1992; Welsh et al. 2000). This macromolecular transition has been investigated previously by calorimetry, density, and circular dichroism (Hinz et al. 1980) and Raman spectroscopies (Thomas et al. 1983). The fiber diffraction results predict that lowering the temperature below 8°C, the midpoint of the temperature transition, results in a 1000 Å increase in the length of the particle. Thus, this transition provides an opportunity to examine how the cumulative effects of small changes in the structure of the protein subunits results in a large net effect on a supramolecular assembly.
In order to understand this transition and its effects on the bacteriophage particles at the molecular level, it is essential to compare the three-dimensional structures of the low- and high-temperature forms of Pf1 coat protein. We recently reported the structure of the high-temperature form of Pf1 coat protein as determined by solid-state NMR spectroscopy using samples of concentrated solutions of bacteriophage particles at 30°C (Thiriot et al. 2004). The structure determined by NMR spectroscopy complemented those previously obtained from X-ray fiber (Welsh et al. 2000) and neutron diffraction (Nambudribad et al. 1991). We have now determined the structure of the low-temperature form of the protein, also by solid-state NMR spectroscopy, enabling us to compare the low- and high-temperature forms of the coat protein with atomic resolution.
Results
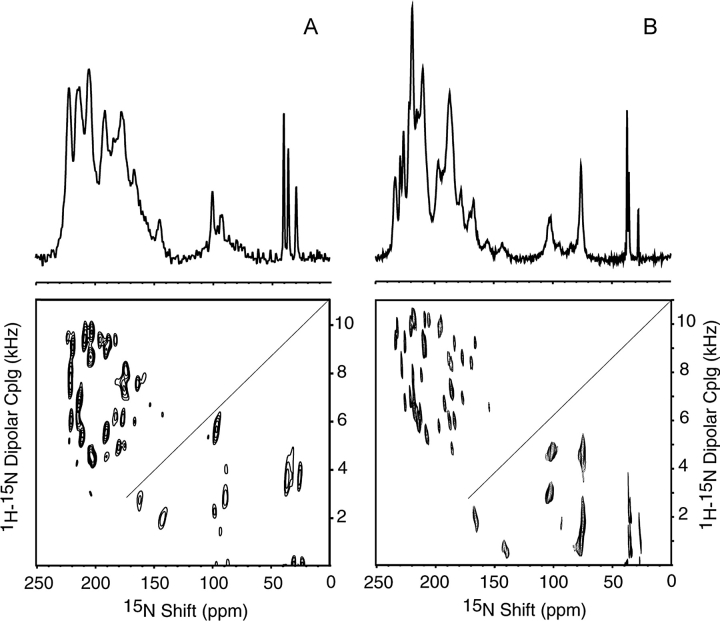
NMR spectra of uniformly and selectively 15N-labeled samples of Pf1 bacteriophage were obtained at 0°C, which was chosen to be well below the transition temperature but comfortably above the freezing point of the samples. The spectra obtained from concentrated solutions of magnetically aligned Pf1 bacteriophage particles have narrower line widths and hence higher resolution at 0°C than at 30°C. This is apparent in the comparisons of both the one- and two-dimensional spectra of uniformly 15N-labeled Pf1 bacteriophage shown in Figure 1A ▶ (30°C) and Figure 1B ▶ (0°C). The line widths are similar to those observed in single crystals of model peptides, and indicate that all of the protein subunits have the same unique structure, and that the particles are nearly perfectly aligned parallel to the direction of the applied magnetic field. The two-dimensional spectra in Figure 1 ▶ are composites; the diagonal line separates the upper part acquired using the PISEMA pulse sequence (Wu et al. 1994) and the lower part acquired using the SAMMY pulse sequence (Nevzorov and Opella 2003a), which is better suited for measuring smaller dipolar couplings over a broad chemical shift range.
Figure 1.
Comparison of one- and two-dimensional solid-state NMR spectra of uniformly 15N labeled Pf1 bacteriophage at 30°C (A) and 0°C (B). The upper diagonal part of each spectrum was acquired using PISEMA; the lower part was acquired using SAMMY. Experimental NMR parameters for (A) are described in Thiriot et al. (2004); for (B): 4.18 μsec 90° pulse length; 10-sec recycle delay, 128 scans.
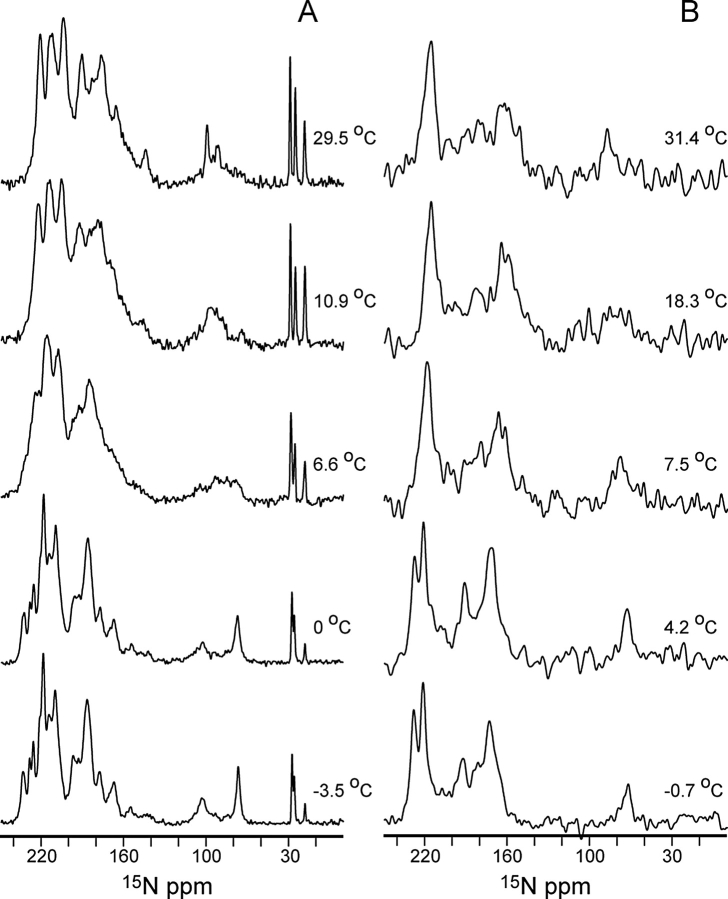
One-dimensional NMR spectra of uniformly 15N and selectively 15N Ile-labeled samples of Pf1 bacteriophage obtained over a range of temperatures are shown in Figure 2 ▶. There are dramatic and fully reversible changes in the appearance of these spectra that occur between ~6°C and 8°C, which corresponds, within the accuracy of the calibrations of the apparatus, to the temperature at which the fiber diffraction symmetry changes were found to occur (Marvin et al. 1992). The effects of the narrower line widths observed at low temperatures can be seen in the spectra of the selectively labeled sample (Fig. 2B ▶) where the resonances from Ile 32 (233 ppm) and Ile 39 (226 ppm) are clearly resolved in the low-temperature form, but overlap in the spectra obtained at the higher temperatures.
Figure 2.
One-dimensional 15N NMR spectra of uniformly 15N-labeled (A); and selectively 15N-Ile-labeled Pf1 bacteriophage (B), recorded as a function of sample temperature (indicated to the right of each spectrum).
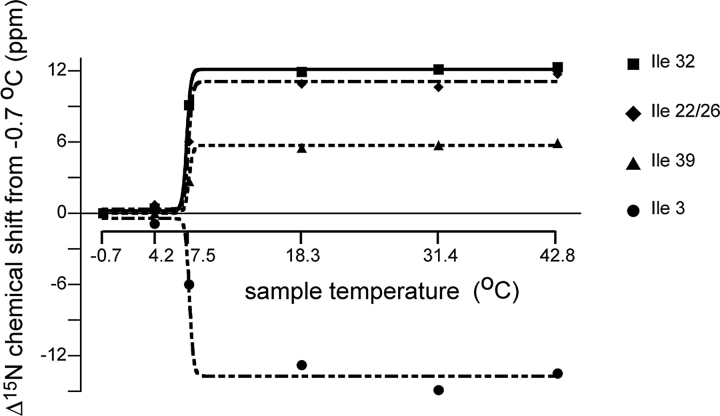
The one-dimensional spectra of the selectively 15N Ile-labeled sample are sufficiently well resolved to enable the measurement of individual chemical shift frequencies as a function of temperature. In Figure 3 ▶, the frequencies of the Ile resonances are plotted as a function of temperature, and are fit to a two-state transition curve with a midpoint of 7.5°C. All of the resonances appear to have the same transition temperature, and since the Ile residues are distributed throughout the sequence, this indicates that the entire protein is affected.
Figure 3.
Chemical shift changes for Ile residues as a function of temperature. Each data set is fit with a sigmoidal two-state transition curve, indicating that for all of these isoleucine residues (distributed widely throughout the protein sequence) a single transition occurs at 7.5°C.
The chemical shift frequencies in solid-state NMR spectra of aligned samples reflect the orientations of the amide nitrogens with respect to the direction of the filament axis and magnetic field (Cross et al. 1983). The data in Figures 2 ▶ and 3 ▶ clearly show that all of the residues in the coat protein change orientation as a function of temperature, but they do not differentiate between changes in protein conformation and rearrangements of the protein subunits. We determined the structure of the coat protein in Pf1 bacteriophage at 0°C in order to make structural, not just spectroscopic, comparisons with the results obtained at 30 °C.
Spectral expansions containing the resonances from the helical residues are presented in Figure 4 ▶. Spectra were obtained from nine selectively 15N-labeled samples, which account for most of the residues in the coat protein. (In the Arg sample, but not the Lys sample, the side-chain nitrogens are also labeled.) The assignments marked in Figure 4A ▶ were obtained by comparison to the previously assigned two-dimensional spectrum of the high-temperature form of Pf1 coat protein (Thiriot et al. 2004), as well as by an independent application of the “shot gun” approach (Marassi and Opella 2003) that utilizes simultaneous analysis of Pisa Wheel (Marassi and Opella 2000, 2002; Wang et al. 2000) and Dipolar Wave (Mesleh et al. 2002, 2003; Mesleh and Opella 2003) patterns. These analysis methods rely on the periodicity and orientational patterns in the spectra that result from the secondary structure of the protein. Although ambiguous assignments are a possibility for overlapping or nearby resonances, the actual structural consequences of switching a small number of such assignments are minimal.
Figure 4.
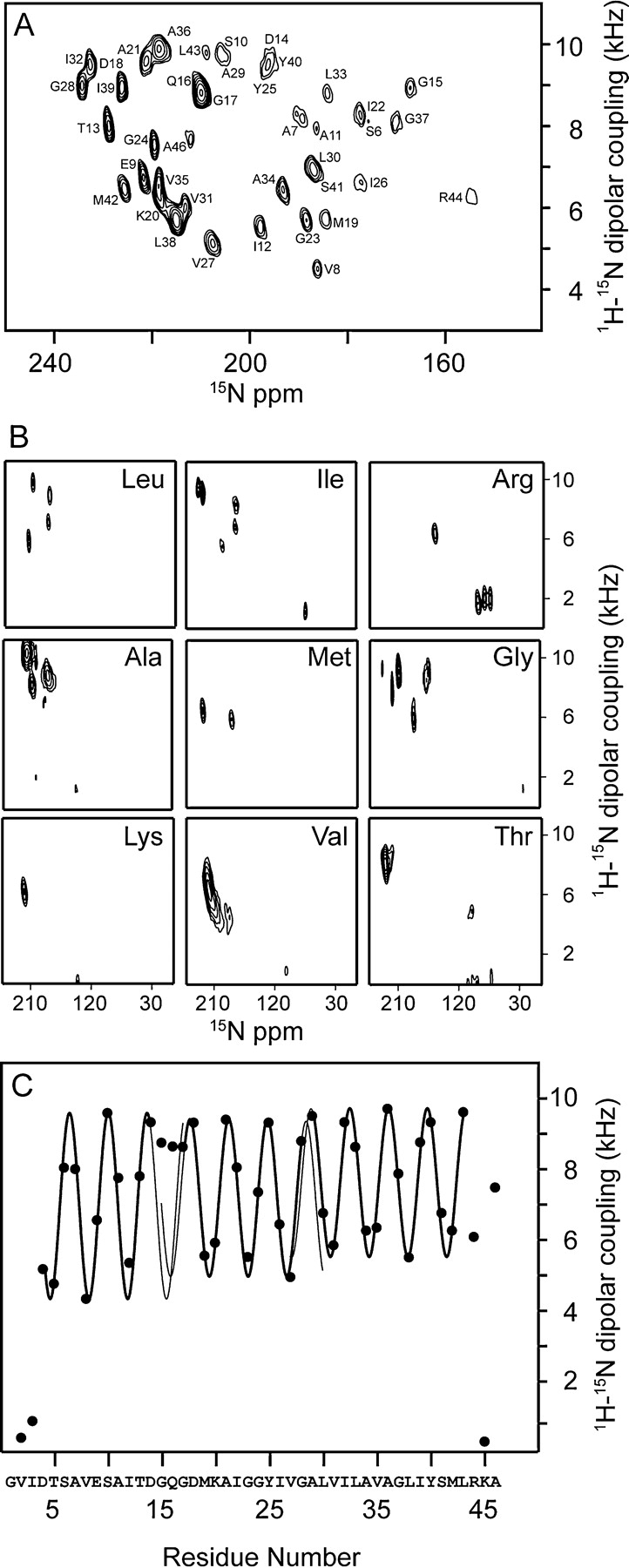
Assigned spectra and dipolar waves for Pf1 coat protein at 0°C. (A) Two-dimensional PISEMA spectrum of uniformly 15N-labeled Pf1 at 0°C. (B) Two-dimensional PISEMA spectra of nine different selectively 15N-labeled samples of Pf1. These represent 36 out of 46 residues in the protein. (C) Dipolar wave of Pf1 coat protein at 0°C indicating the three helical segments with slightly different tilt angles. The thick colored lines indicate the predicted extent of the helical segments, and are continued as thin lines to demonstrate discontinuities (amplitude and phase differences in the waves) near Q16 and A29.
Dipolar waves describe the periodic variation in the magnitudes of the dipolar couplings in the backbone of a protein as a function of residue number; for residues in an α-helix, these waves have a periodicity near 3.6. As a result, they are useful not only as part of the assignment process, but also for the analysis of protein structure. The dipolar couplings plotted as a function of residue number for the low-temperature form of the protein are measured from the experimental spectrum in Figure 4C ▶. Although the spectra obtained at 0°C and 30°C appear to be quite different, the resulting dipolar waves are similar, describing the secondary structure as three helical segments separated by small kinks. An overlay of the dipolar waves for the two forms of the protein in Figure 5 ▶ indicates that the structures and orientations of the protein subunits are indeed very similar, except for a difference in the tilt angles of the C-terminal segment.
Figure 5.
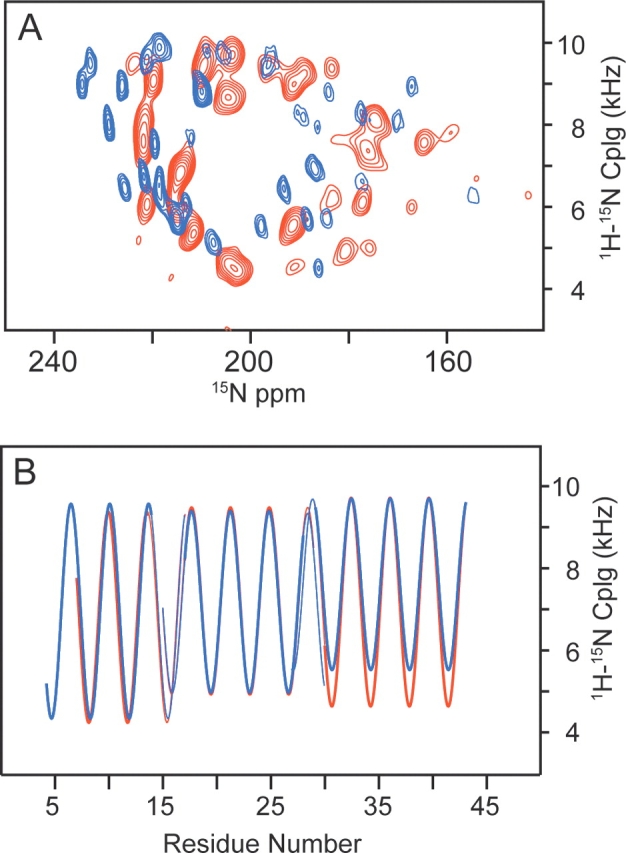
Comparison of the (A) two-dimensional PISEMA spectra and (B) dipolar waves for Pf1 at 0°C and 30°C. Data for 0°C are plotted in blue, and data for 30°C are plotted in red. These data indicate that while essentially all of the resonance frequencies change, the secondary structure as predicted from the dipolar wave is essentially the same, except for a small change in the tilt angle of the C-terminal segment.
The backbone structure of the low-temperature form of Pf1 coat protein was calculated using the 1H-15N dipolar couplings and 15N chemical shift frequencies in Table 1 as orientational constraints, as previously described for the high-temperature form of Pf1 coat protein (Thiriot et al. 2004) and fd coat protein (Zeri et al. 2003). Dipolar waves are extremely sensitive indicators of the tilt angles and polarity of helices and, as noted above, they do show differences in the C-terminal segment. However, the atomic details of such differences can only be revealed by the structural fitting using solid-state NMR frequencies as orientational constraints (Nevzorov and Opella 2003b). As can be seen from Figure 6 ▶, the differences between the low- and high-temperature forms of the coat protein are relatively subtle with an RMSD between them of <2 Å. When the central helical segments of the two structures are overlaid using only uniaxial rotations and translations consistent with solid-state NMR of aligned samples the C-terminal segments have slightly different tilt angles. There are also pronounced differences in the first five N-terminal residues that appear to have “double hook” conformations at both temperatures. These structural difference are consistent with those previously identified by X-ray fiber diffraction (Fig. 3 ▶ of Welsh et al. 2000) and particularly with the finding of “. . . some change in the structure of the NH2-terminal . . .” (Specthrie et al. 1987).
Table 1.
Values used to calculate the 0°C solid-state NMR structure of Pf1 major coat protein in virions
| Residue | 15N ppm | 3H-15N dipolar couping (kHz) |
| G 1 | ||
| V 2 | 106 | 0.60 |
| I 3 | 77 | 1.07 |
| D 4 | 78 | 5.36 |
| T 5 | 102 | 4.95 |
| S 6 | 177 | 8.24 |
| A 7 | 189 | 8.19 |
| V 8 | 186 | 4.52 |
| E 9 | 222 | 6.75 |
| S 10 | 206 | 9.78 |
| A 11 | 186 | 7.94 |
| I 12 | 198 | 5.54 |
| T 13 | 229 | 8.00 |
| D 14 | 196 | 9.51 |
| G 15 | 167 | 8.94 |
| Q 16 | 210 | 8.82 |
| G 17 | 210 | 8.82 |
| D 18 | 233 | 9.52 |
| M 19 | 184 | 5.74 |
| K 20 | 218 | 6.11 |
| A 21 | 221 | 9.59 |
| I 22 | 177 | 8.24 |
| G 23 | 188 | 5.71 |
| G 24 | 219 | 7.55 |
| Y 25 | 196 | 9.51 |
| I 26 | 177 | 6.63 |
| V 27 | 207 | 5.14 |
| G 28 | 234 | 8.99 |
| A 29 | 205 | 9.70 |
| L 30 | 187 | 6.95 |
| V 31 | 213 | 6.03 |
| I 32 | 233 | 9.52 |
| L 33 | 184 | 8.81 |
| A 34 | 193 | 6.45 |
| V 35 | 219 | 6.54 |
| A 36 | 219 | 9.90 |
| G 37 | 170 | 8.06 |
| L 38 | 215 | 5.70 |
| I 39 | 226 | 8.95 |
| Y 40 | 196 | 9.51 |
| S 41 | 187 | 6.95 |
| M 42 | 225 | 6.46 |
| L 43 | 209 | 9.80 |
| R 44 | 154 | 6.27 |
| K 45 | 140 | 0.48 |
| A 46 | 212 | 7.68 |
Figure 6.
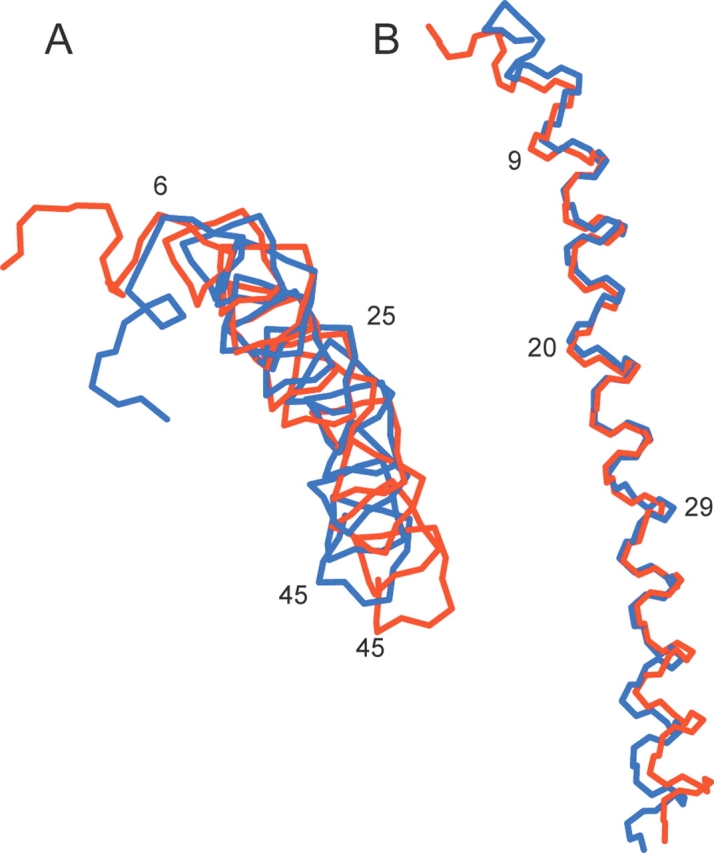
Comparison of the 0°C (blue) and 30°C (red) backbone structures of the coat protein in Pf1 bacteriophage. (A) Top view, looking down the filament axis. (B) Side view.
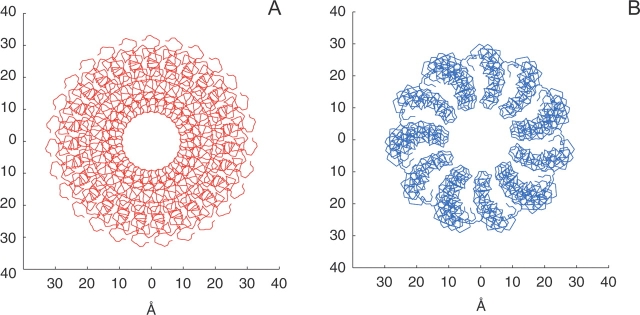
The structures of the 0°C and 30°C forms of the virions are shown in Figure 7 ▶. The 30°C form of the virion has been described previously (Thiriot et al. 2004); for the low-temperature forms, the arrangements of the Pf1 subunits were taken from Specthrie et al. (1987) and Welsh et al. (2000) with subunit packing in a right-handed sense (Bryan et al. 1983). The parameters for the low-temperature virion are: 65.9° rotation per Pf1 monomer subunit followed by a 3.05 Å vertical rise. The pseudoatom distances relative to the virion axis were assumed to be the same as at 30°C since no neutron diffraction data are available for the low-temperature form. The main difference found in the low-temperature virion is a smaller diameter and broader inner pore, which results in the lengthening of the whole virion. This is a consequence of the slightly smaller tilt of the helical subunits in the low-temperature form. The spacings between the individual subunits are also different as a result of the different rotation angle per coat protein monomer in the low- and high-temperature forms of the virion.
Figure 7.
Comparison of Pf1 virion structures at 30°C (A) and 0°C (B). The slightly smaller tilt angle of the monomer subunits in the cold-temperature form gives rise to a smaller diameter and a larger inner pore.
Discussion
The one-dimensional 15N NMR spectra of magnetically aligned solutions of bacteriophage particles obtained as a function of temperature have sufficient resolution to enable the transition to be monitored at individual residues throughout the sequence. Two-dimensional PISEMA spectra provide the input for structure determination of the low-temperature (0°C) form of the coat protein, enabling direct comparisons to the previously determined high-temperature (30°C) form (Thiriot et al. 2004). At both 0°C and 30°C, the coat protein has the same secondary structure, which consists predominantly of three distinct helical segments. Each segment has a slightly different tilt angle relative to the filament axis delimited by kinks near Q16 and A29. Solid-state NMR spectra of aligned samples are exquisitely sensitive to structure because the resonance frequencies reflect orientations with respect to the filament axis and magnetic field; therefore, the rather dramatic spectroscopic changes that occur as a function of temperature reflect small changes in structure. The ~5% increase in the length of the lower temperature coat protein monomer is mirrored by the ~5% increase in the length of the whole phage particle. Thus, the cumulative effects of the small structural changes in the protein monomers lead to a substantial (1000 Å) change in the length of the bacteriophage particles.
It has been estimated, based on calorimetric and isopotential specific volume measurements (Hinz et al. 1980) that the energy difference associated with the Pf1 bacteriophage temperature transition is on the order of the energy involved in altering the interactions of one or two hydrophobic side chains with water (Specthrie et al. 1987). This is consistent with the structural information obtained from the solid-state NMR studies described here, and previous studies by other biophysical methods. The energetically balanced packing interactions present in the Pf1 virion may be biologically relevant, since the size and shape of the bacteriophage particles present a difficult task for packaging by the coat protein. Pf1 is an unusually long (~20,000 Å) and narrow (~60 Å) virus particle, and therefore is highly susceptible to damage by shear forces. To protect its DNA genome, the bacteriophage particles must have substantial structural flexibility. The temperature-sensitive structural transition of the coat proteins may reflect the careful balancing of packing interactions, and it may also be advantageous for survival under the wide range of temperatures encountered in the wild. Another indication of the small energy barriers to structural responsiveness in Pf1 is the finding that at 30°C, the “hinge” region around Q16 displays evidence of internal dynamics (Tobias et al. 1993; Thiriot et al. 2004).
These results demonstrate that solid-state NMR of aligned samples is capable of probing structural changes with high precision. The orientationally dependent 1H-15N dipolar coupling and 15N chemical shift frequencies provide input for calculation of three-dimensional protein structures. The direct comparisons of the structures determined at 0°C and 30°C provide an atomic resolution description of the temperature transition of this supramolecular assembly.
Materials and methods
Sample preparation
For uniformly 15N-labeled Pf1 bacteriophage, the growth media was comprised of the following: 11 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L MgSO4−7H2O, 0.025 g/L CaCl2−2H2O, 10 g/L dextrose, 0.1 g/L thiamine hydrochloride, 10 mL/L 100X MEM vitamin solution (Sigma #M6895; www.sigma-aldrich.com), and 1 g/L 15N ammonium sulfate (Cambridge Isotope Laboratories; www.isotope.com). For the selectively labeled samples, one 15N labeled amino acid (Cambridge) was added at a concentration of 0.1 g/L, and the other 19 unlabeled amino acids were added at higher (>0.2 g/L) concentrations to the growth media. The final pH of the media was adjusted to ~7.4. Pseudomonas aeruginosa (ATCC# 25102; www.atcc.org) was grown in 2 L baffled flasks to O.D. 0.3, and then infected with Pf1 phage (ATCC# 25102-B1) at a high multiplicity. Growth was allowed to continue from 4 h to overnight, after which the cultures were centrifuged at low speed, the supernatant retained, and the bacteriophage particles precipitated by addition of PEG-8000 (40 g/L) and NaCl (29.2 g/L) at 4°C. The precipitated bacteriophage particles were collected by centrifugation, resuspended in tris-EDTA buffer (pH 8), centrifuged to remove any remaining bacteria, reprecipitated with 4% PEG-8000/0.5 M NaCl, and resuspended overnight in 5 mM sodium borate buffer (pH 8). The concentration was adjusted to ~1 mg/mL and then mixed with solid CsCl (0.4 g per mL of 1 mg/mL phage solution) and prepared in OptiSeal tubes (Beckman #362185; www.beckman.com) for density gradient ultracentrifugation. Centrifugation was performed in a Beckman NVT90 rotor from 4 h to overnight at 70,000 rpm and 10°C. The phage particles form a pale blue band near the center of the tube, and are removed with a short Pasteur pipette and placed into 10-kDa cutoff dialysis tubing. Following dialysis, the phage solution was centrifuged at 50,000 rpm for 2 h to obtain a pellet of viscous phage gel (~90 mg/mL), which was diluted with sodium borate/sodium azide buffer to ~50 mg/mL. A final volume of 100 μL to 200 μ(L was placed in 15–16-mm-long, 5-mm outer diameter thin-wall glass tubes for the NMR experiments.
NMR spectroscopy
The NMR experiments were performed on a spectrometer with a Bruker Avance console, a Magnex 750/54 magnet, and a home-built probe with single horizontal solenoid coil double tuned to the resonance frequencies of 15N and 1H (750 MHz). The temperature of the sample was controlled by directing precooled nitrogen gas through a dewared glass tube past a heater element, and the temperature reading was calibrated using a sample tube at the same position in the probe coil, but containing a thermocouple. Chemical shift frequencies are referenced relative to external solid, powdered 15N ammonium sulfate, which is defined as 26.8 ppm.
The one-dimensional 15N NMR spectra were obtained using single-contact spin-lock cross-polarization (Pines et al. 1973) on magnetically aligned solutions of virus particles (Cross et al. 1983). The two-dimensional 1H-15N heteronuclear dipolar coupling/15N chemical shift spectra separated local field spectra were obtained using PISEMA (polarization inversion spin exchange at the magic angle) sequence (Wu et al. 1994; Ramamoorthy et al. 1999) and SAMMY (Nevzorov and Opella 2003a) as previously described (Thiriot et al. 2004). In a typical experiment, the 1H carrier frequency was set on the water resonance. The 15N carrier frequency was set at 200 ppm, or +173.2 ppm relative to the signal from powdered 15N ammonium sulfate. The 1H 90° pulse lengths were 4.15–4.8 μsec, and the initial spin lock cross polarization time was 1 msec. During the frequency switched Lee-Goldburg period of the PISEMA, the power on the 15N channel was increased to match the power level on the 1H channel. Typically a recycle delay of 8–10 sec was used to avoid sample heating by high-power RF irradiation.
Data processing
The experimental data were processed using the program Felix (www.accelrys.com). The data from selectively 15N labeled samples were typically apodized with parameters that improved the signal to noise ratio, at the expense of some resolution; sine bells and/or exponential line broadening was applied in both the 15N chemical shift and 1H-15N dipolar coupling dimensions prior to Fourier transformation. In the spectra of uniformly 15N-labeled samples, apodization with phase-shifted sine bells in both dimensions was used to optimize resolution.
Structure calculations
Fitting of dipolar waves to the experimental dipolar couplings as a function of residue number was performed using MATLAB scripts (Mesleh et al. 2002, 2003; Mesleh and Opella 2003). Structural fitting was performed using a modified version of a MATLAB script and algorithm previously described (Nevzorov and Opella 2003b). Briefly, the fitting algorithm uses both the dipolar coupling and chemical shift data together with restraints on the allowed Ramachandran Φ and Ψ backbone angles and an estimate of the errors in the resonance peak positions to limit the number of structures consistent with the NMR data to a unique family, or small set of related structural families. The peptide plane geometry is assumed to be constant with the values listed below, which provide chemical bond constraints for the fitting. The fitting algorithm was modified to iteratively adjust the allowed Φ and Ψ values independently for each amino acid position to maintain the searched Ramachandran space as near as possible to the ideal helical values (Φ = −65°, Ψ = −40°). The tolerance, or peak position error, used in the calculation of the structure was ± 76 Hz (1 ppm) for all residues except near the N-terminus, which fall outside the optimal chemical shift range for the carrier frequencies used in the experiments, and were allowed up to ± 2000 Hz. The bond angles used were the following: 〈HNσ33 = 18.5°; 〈HNCα = 118.2°; 〈NCoCα = 115.6°; 〈NCαCo = 110.5°; 〈CαCoO = 121.1°; 〈NCoO = 123.2°. The bond lengths used were the following: rCαCo = 1.52 Å; rCoN = 1.33 Å; rNCα = 1.46 Å; rNH = 1.07 Å. The 15N chemical shift tensors were the following: For all residues except glycine, σ11 = 64 ± 7 ppm; σ22 = 77 ± 7 ppm; σ33 = 222 ± 7 ppm. For glycine residues, σ11 = 41 ± 7 ppm; σ22 = 64 ± 7 ppm; σ33 = 215 ± 7 ppm. To account for the small number of peaks that fell above this range, the five peaks with chemical shifts higher than 224 ppm were fit by using a value of σ33 = 234 ± 7 ppm.
Structure coordinates
The coordinates of the low-temperature form of Pf1 coat protein in bacteriophage particles have been deposited at the RCSB Protein Database (www.rcsb.org) and BioMagRes Bank (www.bmrb.wisc.edu) for immediate release upon publication.
Acknowledgments
This research was supported the National Institutes of Health through grants RO1EB001966, by RO1EB002169, and P41EB002031, which supports the Biomedical Technology Resource for NMR Molecular Imaging of Proteins, and postdoctoral fellowship F32GM63300 to D.S.T.
Abbreviations
NMR, nuclear magnetic resonance
DNA, deoxyribonucleic acid
RMSD, root-mean-square deviation
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041220305.
References
- Bryan, R.K., Bansal, M., Folkhard, W., Nave, C., and Marvin, D.A. 1983. Maximum-entropy calculation of the electron density at 4 angstrom resolution of Pf1 filamentous bacteriophage. Proc. Natl. Acad. Sci. 80 4728–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, T.A., Tsang, P., and Opella, S.J. 1983. Comparison of protein and DNA structure in fd and Pf1 bacteriophages. Biochemistry 22 721–726 [DOI] [PubMed] [Google Scholar]
- Hinz, H.J., Greulich, K.O., Ludwig, H., and Marvin, D.A. 1980. Calorimetric, density and circular dichroism studies of the reversible structural transition in Pf1 filamentous bacterial virus. J. Mol. Biol. 144 281–289. [DOI] [PubMed] [Google Scholar]
- Marassi, F.M. and Opella, S.J. 2000. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 144 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. Using pisa pies to resolve ambiguities in angular constraints from PISEMA spectra of aligned proteins. J. Biomol. NMR 23 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 12 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin, D.A., Nave, C., Bansal, M., Hale, R.D., and Salje, E.K.H. 1992. Two forms of Pf1 Inovirus: X-ray diffraction studies on a structural phase transition and a calculated libration normal mode of the asymmetric unit. Phase Transitions 39 45–80. [Google Scholar]
- Mesleh, M.F. and Opella, S.J. 2003. Dipolar waves as NMR maps of helices in proteins. J. Magn. Reson. 163 288–299. [DOI] [PubMed] [Google Scholar]
- Mesleh, M.F., Veglia, G., DeSilva, T.M., Marassi, F.M., and Opella, S.J. 2002. Dipolar waves as NMR maps of protein structure. J. Am. Chem. Soc. 124 4206–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesleh, M.F., Lee, S., Veglia, G., Thiriot, D.S., Marassi, F.M., and Opella, S.J. 2003. Dipolar waves map the structure and topology of helices in membrane proteins. J. Am. Chem. Soc. 125 8928–8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambudripad, R., Stark, W., and Makowski, L. 1991. Neutron diffraction studies of the structure of filamentous bacteriophage Pf1. Demonstration that the coat protein consists of a pair of alpha-helices with an intervening, non-helical surface loop. J. Mol. Biol. 220 359–379. [DOI] [PubMed] [Google Scholar]
- Nevzorov, A.A. and Opella, S.J. 2003a. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J. Magn. Reson. 164 182–186. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Structural fitting of PISEMA spectra of aligned proteins. J. Magn. Reson. 160 33–39. [DOI] [PubMed] [Google Scholar]
- Pines, A., Gibby, M.G., and Waugh, J.S. 1973. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 59 569–590. [Google Scholar]
- Ramamoorthy, A., Wu, C.H., and Opella, S. J. 1999. Experimental aspects of multidimensional solid-state NMR correlation spectroscopy. J. Magn. Reson. 140 131–140. [DOI] [PubMed] [Google Scholar]
- Specthrie, L., Greenberg, J., Glucksman, M.J., Diaz, J., and Makowski, L. 1987. Structural responsiveness of filamentous bacteriophage Pf1: Comparison of virion structure in fibers and solution. The effect of temperature and ionic strength. Biophys. J. 52 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriot, D.S., Nevzorov, A.A., Zagyanskiy, L., Wu, C.H., and Opella, S.J. 2004. Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. J. Mol. Biol. 341 869–879. [DOI] [PubMed] [Google Scholar]
- Thomas Jr., G.J., Prescott, B., and Day, L.A. 1983. Structure similarity, difference and variability in the filamentous viruses fd, If1, IKe, Pf1 and Xf. Investigation by laser Raman spectroscopy. J. Mol. Biol. 165 321–356. [DOI] [PubMed] [Google Scholar]
- Tobias, D.J., Klein, M.L., and Opella, S.J. 1993. Molecular dynamics simulation of Pf1 coat protein. Biophys. J. 64 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtel, E.J., Marvin, R.J., and Marvin, D.A. 1976. Structural transition in a filamentous protein. J. Mol. Biol. 107 379–383. [DOI] [PubMed] [Google Scholar]
- Wang, J., Denny, J., Tian, C., Kim, S., Mo, Y., Kovacs, F., Song, Z., Nishimura, K., Gau, Z., Fu, R., et al. 2000. Imaging membrane protein helical wheels. J. Magn. Reson. 144 162–167. [DOI] [PubMed] [Google Scholar]
- Welsh, L.C., Symmons, M.F., and Marvin, D.A. 2000. The molecular structure and structural transition of the alpha-helical capsid in filamentous bacteriophage Pf1. Acta Crystallogr. D Biol. Crystallogr. 56 137–150. [DOI] [PubMed] [Google Scholar]
- Wu, C.H., Ramamoorthy, A., and Opella, S.J. 1994. High-resolution hetero-nuclear dipolar solid-state NMR spectroscopy. J. Magn. Reson. A 109 270–272. [Google Scholar]
- Zeri, A.C., Mesleh, M.F., Nevzorov, A.A., and Opella, S.J. 2003. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. 100 6458–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]