Abstract
Phosphorylase kinase (PhK), a regulatory enzyme in the cascade activation of glycogenolysis, is a 1.3-MDa hexadecameric complex, (αβγδ)4. PhK comprises two arched octameric (αβγδ)2 lobes that are oriented back-to-back with overall D2 symmetry and connected by small bridges. These interlobal bridges, arguably the most questionable structural component of PhK, are one of several structural features that potentially are artifactually generated or altered by conventional sample preparation techniques for electron microscopy (EM). To minimize such artifacts, we have solved by cryoEM the first three-dimensional (3D) structure of nonactivated PhK from images of frozen hydrated molecules of the kinase. Minimal dose electron micrographs of PhK in vitreous ice revealed particles in a multitude of orientations. A simple model was used to orient the individual images for 3D reconstruction, followed by multiple rounds of refinement. Three-dimensional reconstruction of nonactivated PhK from approximately 5000 particles revealed a bridged, bilobal molecule with a resolution estimated by Fourier shell correlation analysis at 25 Å. This new structure suggests that several prominent features observed in the structure of PhK derived from negatively stained particles arise as artifacts of specimen preparation. In comparison to the structure from negative staining, the cryoEM structure shows three important differences: (1) a dihedral angle between the two lobes of approximately 90° instead of 68°, (2) a compact rather than extended structure for the lobes, and (3) the presence of four, rather than two, connecting bridges, which provides the first direct evidence for these components as authentic elements of the kinase solution structure.
Keywords: phosphorylase kinase, cryoelectron microscopy, three-dimensional reconstruction, single particle analysis, structural analysis
Phosphorylase kinase (PhK), a regulatory enzyme in the cascade activation of glycogenolysis, is in skeletal muscle a hexadecameric complex composed of four copies of four different subunits: (αβγδ)4 (for review, see Brushia and Walsh 1999). Within the PhK complex, the activity of the catalytic γ subunit is tightly controlled by the regulatory α, β, and δ (endogenous calmodulin) subunits; occupancy of or phosphorylation of allosteric sites on those regulatory subunits leads to PhK’s activation. Because of its large mass, 1.3 MDa, the PhK complex is particularly amenable to analysis by microscopic techniques, and has been studied by transmission (TEM) (Cohen 1974; Schramm and Jennissen 1985; Trempe et al. 1986; Norcum et al. 1994; Wilkinson et al. 1994, 1997), scanning transmission (STEM) (Trempe et al. 1986; Norcum et al. 1994; Wilkinson et al. 1994), and scanning tunneling electron microscopy (STM) (Edstrom et al. 1989, 1990), as well as by atomic force microscopy (AFM) (Edstrom et al. 1990). The two-dimensional images from all these types of microscopy revealed an overall dimeric structure for PhK composed of two large lobes, but of varying dimensions, orientation with respect to each other, and connections, or bridges, between them.
The connecting bridges between the lobes have been one of the most variable structural features observed in PhK using different microscopic techniques. In fact, some techniques (STM and AFM) showed no bridges, but only a large central depression between the lobes (Edstrom et al. 1990). Small discrete bridges between PhK’s lobes were observed in images of both stained and unstained molecules visualized by TEM and STEM, respectively, although their suggested number varied (one, two, or four) (Cohen 1974; Schramm and Jennissen 1985; Trempe et al. 1986; Norcum et al. 1994). Moreover, the possibility that these bridges arise as artifacts of specimen preparation could not be completely ruled out (Norcum et al. 1994), because of potential deformations to PhK’s structure by surface contact interactions, drying, or staining (Norcum et al. 1994; Nadeau et al. 2002).
The first three-dimensional (3D) model proposed for the nonactivated PhK complex was not a reconstruction, but was instead deduced from a comprehensive analysis of the predominant views of the complex in fields of unstained (STEM) and negatively stained (TEM) particles (Norcum et al. 1994). In this model, each lobe was designated as a dimer of αβγδ protomers and the two arched octameric lobes were arranged back-to-back and perpendicularly, i.e., a dihedral angle between them of approximately 90°. The model’s proposed pseudo-tetrahedral arrangement of the protomers with head-to-head packing and D2 symmetry was based on the localization by immunoelectron microscopy of individual subunits within the complex (Wilkinson et al. 1994, 1997; Traxler et al. 2001). Four interlobal connecting bridges were included in that model to coincide with this laboratory’s TEM and STEM results (Norcum et al. 1994). Moreover, a model containing four bridges was used successfully to fit the small-angle X-ray scattering (SAXS) profile of nonactivated PhK in solution (Priddy et al. 2005). The first 3D reconstruction of the nonactivated PhK complex was recently reported (Nadeau et al. 2002). This structure, which was based on negatively stained particles and will henceforth be referred to as the nsEM structure, was solved using a model-based approach, resulting in an estimated resolution of 24 Å (0.5 correlation cutoff). The overall structural features of this 3D reconstruction, which is shown in Figure 2B ▶ (see below), were quite similar to the earlier model, but differed in the dihedral angle between the lobes and the number of interlobal connecting bridges (two instead of four). It should be noted, however, that given the great size differential between PhK’s two large lobes and their small interconnecting bridges, one would predict that both the interlobal bridges and the angle between the lobes would be among the structural features most susceptible to artifactual deformation in any of the above microscopic techniques. In an attempt to gain additional information on PhK’s fine structure and on the bridging and angles between PhK’s two large lobes, a reconstruction of nonactivated PhK has been carried out using frozen hydrated molecules of the kinase in unsupported thin vitrified films (cryoEM structure) to avoid possible artifacts associated with negative staining.
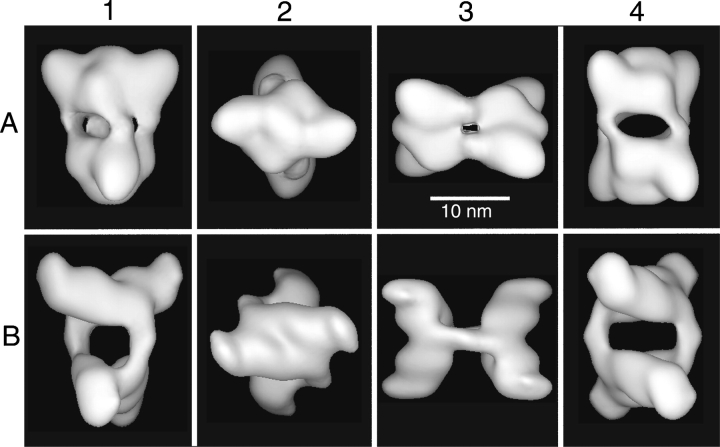
Figure 2.
Surface representations of PhK reconstructed from unstained frozen hydrated (row A) and negatively stained (row B) EM specimens shown in the chalice (1), cross (2), butterfly (3), and cube (4) views. Row B is reprinted from Nadeau et al. (2002), with permission from Elsevier Science © 2002. The surfaces in both reconstructions display a molecular volume consistent with the 1.3-MDa mass of the PhK complex.
Results and Discussion
Electron micrographs of frozen hydrated PhK showed images of the molecule in a wide range of orientations (Fig. 1 ▶), including the four characteristic orientations (chalice, cube, butterfly, and cross) previously identified by visual inspection of fields of the negatively stained kinase (Norcum et al. 1994; Nadeau et al. 2002). Although the bilobate structure commonly observed in previous studies was evident in cryoEM fields of the kinase, several minor differences were readily apparent. For instance, a more compact structure was observed for the PhK complex in ice, where the lobes in several of the common views, including the butterfly (Fig. 2 ▶, column 3), appeared to be foreshortened or less extended. The full butterfly view, previously deduced to potentially be a distorted view of the kinase in negative stain preparations (Norcum et al. 1994; Nadeau et al. 2002), was not observed in any cryoEM fields from over 90 micrographs; thus, this full view likely results from surface contact-induced deformations of the tetrahedral kinase that alter the angle between its lobes, so that each lies in a plane that is approximately parallel to the surface of the grid. Additionally, the molecules in ice were somewhat diffuse, with less distinct outlines than typically observed for more compact structures, such as GroEL (Falke et al. 2001).
Figure 1.

CryoEM field of PhK containing a mixture of orientations of the kinase. The boxes drawn around particles in the field designate examples of single particles.
A model-based approach to 3D reconstruction, used previously to generate the structure of nonactivated PhK from negatively stained molecules (Nadeau et al. 2002), was carried out to generate the structure of the nonactivated kinase from frozen hydrated particles. All images of the approximate dimensions of PhK (~15–25 nm diameter) were selected for analysis, and represented a wide spectrum of viewing directions. Special care was taken during selection to avoid overlapping molecules, resulting in typically 10% of the particles being eliminated after visual inspection, with further losses incurred by correlation cutoff during alignment into each class. A simple spheres model (described in Materials and Methods) was used for initial alignment and orientation of each image with respect to a common reference (Nadeau et al. 2002), and yielded 98 well-represented categories of images from all viewing directions.
Reconstructions from frozen hydrated and negatively stained molecules of the kinase are compared in Figure 2 ▶ (rows A and B, respectively) in the four characteristic orientations first observed in fields of negatively stained PhK (Norcum et al. 1994). The chalice view shows the side view (full length and height) of the upper lobe, which is approximately in the plane of the paper, whereas the bottom lobe projects out from that plane (column 1). The cross view (column 2) is achieved by rotation of the chalice view 90° around the horizontal axis toward the viewer, revealing the complete exterior face for the proximal lobe and a portion of the interior face of the distal lobe. Characteristic topographical features of the lobe faces, which represent the remaining third dimension of the lobes (width), and their symmetrical distribution (also best seen in this view) serve as markers for measuring the dihedral angle between the lobes. A 45° rotation of the chalice (looking down at the upper lobe) around the vertical axis, followed by a second rotation of 90° in the plane of the paper, produces the butterfly view (column 3), which best illustrates the distance between the lobes and the angle of attachment of the connecting bridges to the interior lobe faces. The cube view (column 4), obtained by a 45° rotation of the chalice counterclockwise around the vertical axis, shows the maximum distance separating the bridges. Additionally, a second dimension of the bridges (their thickness) is also revealed in this view, as are the angles of attachment for each bridge pair to the lobes.
Not only has the number of bridges in the PhK complex been previously questioned, but even their very existence, especially since these structural components were not observed by either STM or AFM (Edstrom et al. 1989, 1990). Furthermore, with negatively stained PhK, the question has been raised of whether the formation of pseudo-bridges or the distortion of genuine bridges might occur as artifacts of specimen preparation (Norcum et al. 1994). The presence of connecting bridges in reconstructions from the frozen hydrated molecules of the kinase in this report provides the first direct evidence for these structures as authentic elements of the solution structure of the PhK complex and shows four bridges connecting the four αβγδ protomers across the lobes. As viewed in the cryoEM structure, the bridges are essentially elbow shaped and are arranged in pairs that emanate from the sides of the lobes, as shown in either the whole structure (Fig. 2A ▶, columns 1,4) or in cross sections of the structure in the cross orientation (Fig. 3 ▶, columns 1,2). The points of connection for an individual bridge are structurally different on each of the two lobes it bridges (i.e., the bridges are asymmetric), which is best observed in either the cube view (Fig. 2A ▶, column 4) or the lower-half section of the complex in the cross view (Fig. 3A ▶, column 1). As observed in the cube view, one end of each bridge intersects an interior lobe face approximately perpendicularly, whereas the opposite end of the bridge is oriented 40° incident to the opposing interior lobe face. In the butterfly view (Fig. 2A ▶, column 3), both the proximal and distal bridge pairs, with the former eclipsing the latter, run normal to the interior lobe faces, a previously determined structural characteristic of the nonactivated conformer of PhK (Nadeau et al. 2002). A similar arrangement of the bridges is observed in a structure used to model SAXS of nonactivated PhK in solution (Priddy et al. 2005). In the SAXS study, a model approximating the cryoEM structure, with geometrical shapes corresponding to two lobes and four bridges, generated a theoretical scattering profile almost identical to that observed experimentally for nonactivated PhK in solution.
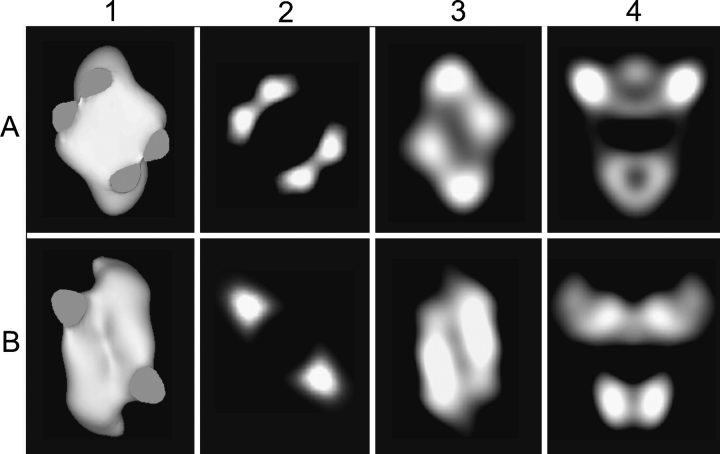
Figure 3.
Sections of reconstructions of PhK from frozen hydrated molecules (row A) and negatively stained molecules (row B) in the cross (columns 1–3) and chalice views (column 4). The points of connection for the bridges on the interior face of the bottom lobe of each structure are revealed by removal of the density corresponding to the upper lobe and bridge halves (column 1). Differences in density between the bridges in each structure are depicted in 5 Å sections that bisect the long axis of each bridge in the plane that runs parallel to the lobe faces in the center of the reconstruction (column 2). Differential density distributions are observed in 5 Å sections bisecting the lobes parallel to the lobe faces (column 3). Vertical slices through the center of the chalice view (normal to the cross view) show different distributions of density in the lobes of each structure, with a region of low density visible at the center of each lobe of the cryoEM reconstruction (column 4).
Besides the differences in bridges, there are other significant differences between the cryoEM and nsEM structures, particularly the shape of the lobes and the dihedral angle between them. Regarding the first of these differences, lobe shape, the lobes of the cryoEM structure are more uniform, with less variation in their height (best viewed in the chalice and cube orientations, Fig. 2 ▶, columns 1,4), progressing from the center of the lobes out toward the lobe tips. The average height of the lobes in the cryo structure (90 Å) exceeds that of the lobes in the nsEM structure by 10 Å. This apparent flattening of the lobes in the nsEM structure is also evident in 5 Å sections bisecting each structure vertically in the chalice view (Fig. 3 ▶, column 4), where the large bulge projecting from the exterior face of the upper lobe in the cryoEM structure is replaced by a concave region of low density in the nsEM reconstruction. Compression of the lobes in other dimensions of the nsEM structure is also apparent from a comparison of the structures in several cross views, including the exterior (Fig. 2 ▶, column 2) and interior (Fig. 3 ▶, column 1) lobe faces and 5 Å sections bisecting the bottom lobe of each molecule (Fig. 3 ▶, column 3). For example, in Figure 2 ▶, column 2, the bridges emanating from the sides of the upper lobe as two large bulges at the 1 o’clock and 7 o’clock positions in the cryoEM structure are barely apparent in the nsEM structure. Sections of the cryoEM structure show a large region of low density in the center of each lobe (Fig. 3 ▶, columns 3,4), which could reflect several internal subunit interfaces; collapse of this region during drying could contribute to the apparent deformations observed in the nsEM structure. A gross flattening of the ryanodine receptor in the structure computed from negatively stained particles (in comparison to the structure from frozen hydrated particles) has been attributed, in part, to deformations imposed by the staining process, i.e., surface interactions of the protein with the carbon surface and the stain, or the effects of drying on the protein, or both (Radermacher et al. 1994). The D2 symmetrical arrangement of the lobes may also promote additional distortions of the nsEM structure through interactions with the carbon surface. Because the lobes have a large axial ratio (length/ average width > 2.3) and are oriented approximately perpendicular to each other, the tips of the lobes necessarily contact the surface, which could account for the upturned tips observed in the nsEM model. This phenomenon would be especially emphasized in the three-point surface landings of the tetrahedral complex that give rise to the chalice and butterfly orientations, which represent the predominant views observed for the negatively stained complex (Norcum et al. 1994; Nadeau et al. 2002).
Another difference between the two structures of the non-activated PhK complex is the orientation of the two lobes with respect to each other, the dihedral angle (assessed using structures in the cross orientation by scribing lines to bisect each lobe along its maximum length, while preserving symmetry). The dihedral angle of ~90° for the cryoEM structure is considerably greater than the average angle of 68° measured for the nsEM structure (Nadeau et al. 2002). The positioning of the two lobes of the complex with respect to each other is likely susceptible to extraneous mechanical forces, especially considering the large size of the lobes (17.6 by 11.5 nm maximum length and width, respectively) versus their narrow connecting bridges (3.0 nm maximum width). Correspondingly, dihedral angles of differing magnitudes have been observed among individual reconstructions of nonactivated conformers computed separately from different preparations of negatively stained kinase (Nadeau et al. 2002), suggesting possible distortions resulting from differential staining or surface contact interactions. Evidence for such distorting forces is further supported by dihedral angles of approximately 90° observed in models that best-fit the SAXS profiles of the nonactivated PhK complex (Priddy et al. 2005). The absence of the full butterfly view in any of the cryoEM fields of the kinase is also consistent with the notion of distortions being present in a significant percentage of the nsEM particles, especially those in the butterfly orientation. Given that different forms of the butterfly view account for approximately 40% of all the images in fields of negatively stained kinase (Norcum et al. 1994; Nadeau et al. 2002), with the full butterfly view (both lobes appearing to be in same plane parallel with grid) accounting for about half of those images (Norcum et al. 1994), incorporation of images of this distorted view in the 3D reconstruction process would undoubtedly influence the positioning of the lobes in the final structure, possibly accounting for the difference in dihedral angles between the cryoEM and nsEM structures. Because the average dihedral angle of 68° measured for the nsEM computed structure differed markedly from the 90° value arbitrarily set in the starting model for its reconstruction, the magnitude of the angle in the computed structure was assumed to be influenced primarily by the data, as opposed to the starting model (Nadeau et al. 2002). To test this assumption with our cryoEM reconstruction, several starting models with lobes of identical dimensions, but with dihedral angles ranging from 50° to 90°, were used to orient and align the images from the same data set used for the reconstruction shown in Figure 2 ▶. Altering the initial dihedral angle in the range specified had little or no effect on the magnitude of the angle realized in the final reconstructions (data not shown); all the final dihedral angles were ~90°, which we thus conclude to be the most appropriate angle for the nonactivated conformer of PhK in solution.
The dihedral angle between the lobes has an intrinsic influence on the orientation of their connecting bridges, because any force that alters the dihedral angle will, by necessity, place mechanical stress on the bridges, altering their juxtaposition. To illustrate, if two rubber bands are attached in parallel at a distance between two cylinders that are oriented perpendicular to each other, and the cylinders are rotated in the planes of their long axes, such as in going from a perpendicular chalice orientation to the distorted full butterfly of the nsEM views, the two strands of each rubber band will approach each other as the cylinders are turned. Correspondingly, in the butterfly orientation the bridge pairs in the cryoEM structure occupy positions that are similar to those occupied by a single bridge in the nsEM structure (Fig. 2 ▶, column 3), with the bridges from each pair in the former structure flanking the position occupied by a single bridge in the latter. The different bridge arrangements in the nsEM and cryoEM structures do not appear to reflect differences in resolution between the reconstructions, as their 0.5 criteria for resolution are essentially the same, but rather reflect distortions imposed on the kinase by the negative staining approach. In agreement, the volume of a single bridge in the nsEM structure is equivalent to the volume of two bridges in the cryoEM structure: The total volume of 5 Å sections, obtained by bisecting all the bridges centrally along their long axes in each structure (Fig. 3 ▶, column 2), was the same for both structures, contoured to a volume of 1600 nm3 (Nadeau et al. 2002). Although the widths of the bridges in both the cryoEM and nsEM structures appear to be more comparable in the chalice and butterfly views, comparison in the cube view clearly shows the bridges in the cryoEM structure to be approximately half the thickness of those in the nsEM structure (Fig. 2A ▶, row 4). The above results are consistent with the collapse of each bridge pair into one bridge during preparation of negatively stained PhK, resulting in four bridges appearing as only two in the nsEM structure. Such a change in the bridges is consistent with differences in the SAXS models of nonactivated versus Ca2+-activated PhK: The scattering models for these two conformers showed four well-resolved bridges in the structure for nonactivated PhK that approached and nearly abutted a counterpart to form two paired bridges in the Ca2+-activated conformer (Priddy et al. 2005).
Further evidence of the interrelationship between the dihedral angle between the lobes and the conformation of the bridges connecting them comes from the effects of Ca2+ on the nsEM structure. Ca2+, which binds to the intrinsic calmodulin subunit (δ) in the PhK complex, caused similar structural changes in independent reconstructions of three different PhK preparations (Nadeau et al. 2002). The changes induced by Ca2+ included, among other things, a decrease in the dihedral angle and a change from parallel to oblique in the relative orientation with respect to each other of the two observed bridges. A similar oblique orientation of the two bridges in an nsEM reconstruction of PhK in the presence of Ca2+ is apparent in a report by Vénien-Bryan et al. (2002), who arrived at their strikingly similar structure using the random conical tilt method, thus confirming the validity of the model-based approach. All of these structural effects of Ca2+ observed by EM, considered together with the SAXS models (Priddy et al. 2005), suggest that this allosteric activator of PhK simultaneously alters the bridges connecting the lobes and the dihedral angle between them, although the effect of Ca2+ on the latter is much smaller in the SAXS models. A Ca2+-induced change in the relative positioning of the bridges is certainly consistent with the location of PhK’s δ (calmodulin) subunits, which were previously identified as being on the interior lobe faces, just distal to the bridges (Traxler et al. 2001).
Although the salient structural differences observed between the current cryoEM and previous nsEM reconstructions have been emphasized, it should be noted that the overall architecture of the PhK complex is essentially preserved in both structures. The similarities between the structures are perhaps most easily seen in the chalice and cube views (Fig. 2 ▶, columns 1,4), with each showing two major lobes associating with overall D2 symmetry, and having similar relative placement of the connecting bridges (albeit four vs. two). An additional important common feature is also evident in the cube view, namely, a large central cavity framed by the opposing lobes and bridges. Comparing the upper exterior lobe faces of the two reconstructions in the cross orientation (Fig. 2 ▶, column 2), it is apparent that both structures also have a ribbed central region from which the bridges and the lobe tips emanate. That major structural features of the nonactivated conformer of the PhK complex are preserved in the nsEM and cryoEM structures argues for the integrity of each reconstruction.
Materials and methods
Sample preparation
PhK was prepared from rabbit skeletal muscle as previously described (King and Carlson 1981). Frozen samples of PhK were thawed, and aggregates of the purified enzyme were removed by size exclusion-HPLC using previously described methods (Traxler et al. 2001). Following HPLC purification, the catalytic activity was determined by measuring the incorporation of 32P from [γ-32P]ATP into phosphorylase-b, measured on phosphocellulose strips (King and Carlson 1981). Final concentrations in the assay mixture at 30°C were PhK (0.7 μg/mL at pH 6.8 or 0.07 μg/mL at pH 8.2), buffer (50 mM Tris/50 mM β-glycerophosphate at pH 6.8 or 8.2), phosphorylase-b (6.0 mg/mL), EGTA (0.1 mM), CaCl2 (0.2 mM), β-mercaptoethanol (13 mM), [γ-32P]ATP (Perkin Elmer/NEN) (1.0 mM, 0.17 Ci/mol), Mg(CH3CO2)2 (10 mM), and sucrose (2%–3%). Only PhK preparations with pH 6.8/pH 8.2 activity ratios of ≤0.05 were used for microscopy. HPLC-purified, nonactivated PhK was diluted to a final concentration of 100 nM in mobile phase HPLC buffer (50 mM HEPES, 0.2 M NaCl, 0.2 mM EDTA at pH 6.8). It was then immediately prepared for cryoelectron microscopy by brief adsorption to plasma-discharged fenestrated carbon films on 300 mesh copper support grids. Preparation of frozen hydrated samples was carried out following previously established procedures (Falke et al. 2001).
Image analysis
Images of PhK were recorded at a magnification of 60,000 using minimal dose protocols with a JEOL 1200EX electron microscope equipped with a Gatan 626-Twin cold stage for cryoEM. Micrographs were digitized with a Eurocore HiScan densitometer using a 30-μm pixel size with 5 Å spacing. Analysis of the cryoEM images was performed as previously described for negatively stained specimens (Nadeau et al. 2002), with only a slight adjustment of the initial model’s dimensions to better match those of the cryoEM reconstruction, yielding the structure shown in Figure 2A ▶. Image analysis was performed on Compaq Alpha workstations with SPIDER and WEB software packages (Frank et al. 1996). The initial model for the cryoEM consisted of two groups of four spheres each, arranged in perpendicular back-to-back arcs, approximating the size and preserving the known D2 symmetry of the PhK complex (Norcum et al. 1994; Wilkinson et al. 1994; Nadeau et al. 2002). To evenly sample all possible viewing directions of the D2 asymmetric unit in the Euler sphere, projections of the model were calculated in angular increments of 5° in the Euler angle φ (from 0° to 90°) and corresponding increments of the Euler angle θ (from 0° to 180°), generating an array of 98 projections. Each image was aligned to each of the projections in the array, and its final alignment parameters and Euler angles were assigned to the projection with the highest correlation. The distribution of particles in each of the 98 classes in the final cryoEM reconstruction was similar to that observed for the nsEM structure (Nadeau et al. 2002). Alternatively, models with dihedral angles varying from 50° to 90°, as well as the nsEM structure itself (Fig. 2B ▶), were also used as starting models for the cryoEM reconstruction and generated results identical to those shown in Figure 2A ▶. Thus, the initial model functions primarily to align and orient the images in the first round of refinement, with pertinent structural details being generated only by the data during successive refinement rounds. The cryoEM reconstruction was based on 4878 molecules of PhK, including 90% of all the identifiable particles selected from 61 minimal-dose (~20 electrons/Å2) cryoelectron micrographs collected using three preparations of the nonactivated enzyme. The resolution of the cryoEM reconstruction is estimated as 25 Å, based on the point at which the Fourier shell correlation drops to a value of 0.5 (Malhotra et al. 1998), compared to a resolution of 24 Å by the same criterion for the structure derived from negatively stained molecules of the kinase (Nadeau et al. 2002).
Acknowledgments
We thank Ms. Jeannette Salazar for her technical contributions. This work was supported by National Institutes of Health Grant DK32953 (G.M.C.).
Abbreviations
AFM, atomic force microscopy
EM, electron microscopy
ns, negative stain
PhK, phosphorylase kinase
SAXS, small-angle X-ray scattering
STEM, scanning transmission EM
STM, scanning tunneling microscopy
TEM, transmission EM
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041123905.
References
- Brushia, R.J. and Walsh, D.A. 1999. Phosphorylase kinase: The complexity of its regulation is reflected in the complexity of its structure. Front. Biosci. 4 D618–D641. [DOI] [PubMed] [Google Scholar]
- Cohen, P. 1974. The role of phosphorylase kinase in the nervous and hormonal control of glycogenolysis in muscle. Biochem. Soc. Symp. 39 51–73. [PubMed] [Google Scholar]
- Edstrom, R.D., Meinke, M.H., Yang, X., Yang, R., and Evans, D.F. 1989. Direct observation of phosphorylase kinase and phosphorylase b by scanning tunneling microscopy. Biochemistry 28 4939–4942. [DOI] [PubMed] [Google Scholar]
- Edstrom, R.D., Meinke, M.H., Yang, X., Yang, R., Elings, V., and Evans, D.F. 1990. Direct visualization of phosphorylase–phosphorylase kinase complexes by scanning tunneling and atomic force microscopy. Biophys. J. 58 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke, S., Fisher, M.T., and Gogol, E.P. 2001. Structural changes in GroEL effected by binding a denatured protein substrate. J. Mol. Biol. 4 569–577. [DOI] [PubMed] [Google Scholar]
- Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M., and Leith, A. 1996. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116 190–199. [DOI] [PubMed] [Google Scholar]
- King, M.M. and Carlson, G.M. 1981. Synergistic activation by Ca2+ and Mg2+ as the primary cause for hysteresis in the phosphorylase kinase reactions. J. Biol. Chem. 256 11058–11064. [PubMed] [Google Scholar]
- Malhotra, A., Penczek, P., Agrawal, R.K., Gabashvili, I.S., Grassucci, R.A., Jünemann, R., Burkhardt, N., Nierhaus, K.H., and Frank, J. 1998. Escherichia coli 70S ribosome at 15 Å resolution by cryo-electron microscopy: Localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280 103–116. [DOI] [PubMed] [Google Scholar]
- Nadeau, O.W., Carlson, G.M., and Gogol, E.P. 2002. A Ca2+-dependent global conformational change in the 3D structure of phosphorylase kinase obtained from electron microscopy. Structure 10 23–32. [DOI] [PubMed] [Google Scholar]
- Norcum, M.T., Wilkinson, D.A., Carlson, M.C., Hainfeld, J.F., and Carlson, G.M. 1994. Structure of phosphorylase kinase: A three-dimensional model derived from stained and unstained electron micrographs. J. Mol. Biol. 241 94–102. [DOI] [PubMed] [Google Scholar]
- Priddy, T.S., MacDonald, B.A., Heller, W.T., Nadeau, O.W., Trewhella, J., and Carlson, G.M. 2005. Ca2+-induced structural changes in phosphorylase kinase detected by small angle X-ray scattering. Protein Sci. (this issue). [DOI] [PMC free article] [PubMed]
- Radermacher, M., Rao, V., Grassucci, R., Frank, J., Timerman, A.P., Fleischer, S., and Wagenknecht, T. 1994. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J. Cell Biol. 127 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, H.J. and Jennissen, H.P. 1985. Two-dimensional electron microscopic analysis of the chalice form of phosphorylase kinase. J. Mol. Biol. 181 503–516. [DOI] [PubMed] [Google Scholar]
- Traxler, K.W., Norcum, M.T., Hainfeld, J.F., and Carlson, G.M. 2001. Direct visualization of the calmodulin subunit of phosphorylase kinase via electron microscopy following subunit exchange. J. Struct. Biol. 135 231–238. [DOI] [PubMed] [Google Scholar]
- Trempe, M.R., Carlson, G.M., Hainfeld, J.F., Furcinitti, P.S., and Wall, J.S. 1986. Analyses of phosphorylase kinase by transmission and scanning transmission electron microscopy. J. Biol. Chem. 261 2882–2889. [PubMed] [Google Scholar]
- Vénien-Bryan, C., Lowe, E.M., Boisset, N., Traxler, K.W., Johnson, L.N., and Carlson, G.M. 2002. Three-dimensional structure of phosphorylase kinase at 22 Å resolution and its complex with glycogen phosphorylase b. Structure 10 33–41. [DOI] [PubMed] [Google Scholar]
- Wilkinson, D.A., Marion, T.N., Tillman, D.M., Norcum, M.T., Hainfeld, J.F., Seyer, J.M., and Carlson, G.M. 1994. An epitope proximal to the carboxyl terminus of the α-subunit is located near the lobe tips of the phosphorylase kinase hexadecamer. J. Mol. Biol. 235 974–982. [DOI] [PubMed] [Google Scholar]
- Wilkinson, D.A., Norcum, M.T., Fitzgerald, T.J., Marion, T.N., Tillman, D.M., and Carlson, G.M. 1997. Proximal regions of the catalytic γ and regulatory β subunits on the interior lobe face of phosphorylase kinase are structurally coupled to each other and with enzyme activation. J. Mol. Biol. 265 319–329. [DOI] [PubMed] [Google Scholar]