Abstract
Immunoconjugates are being explored as novel cancer therapies with the promise of target-specific drug delivery. The immunoconjugate, huN901–DM1, composed of the humanized monoclonal IgG1 antibody, huN901, and the maytansinoid drug, DM1, is being tested in clinical trials to treat small cell lung carcinoma (SCLC). huN901–DM1 contains an average of three to four DM1 drug molecules per huN901 antibody molecule. The drug molecules are linked to huN901 through random modification of huN901 at ɛ-amino groups of lysine residues, thus yielding a heterogeneous population of conjugate species. We studied the drug distribution profile of huN901–DM1 by electrospray time-of-flight mass spectrometry(ESI-TOFMS), which showed that one to six DM1 drug molecules were attached to an antibody molecule. Both light and heavy chains contained linked drugs. The conjugation sites in both chains were determined by peptide mapping using trypsin and Asp-N protease digestion. Trypsin digestion identified modified lysine residues, since these residues were no longer susceptible to enzymatic cleavage after conjugation with the drug. With respect to Asp-N digestion, modified peptides were identified by observing a mass increase corresponding to the modification. The two digestion methods provided consistent results, leading to the identification of 20 modified lysine residues in both light and heavy chains. Each lysine residue was only partially modified. No conjugation sites were found in complementarity determining regions (CDRs). Using structural models of human IgG1, it was found that modified lysine residues were on the surface in areas of structural flexibility and had large solvent accessibility.
Keywords: immunoconjugates, peptide mapping, monoclonal antibody, structural characterization, conjugation site determination
Monoclonal antibody (MAb) therapy has emerged as an important therapeutic modality for the treatment of cancer. One approach is to use naked or unconjugated MAbs as therapeutic agents, such as Rituxan and Herceptin (Grillo-Lopez et al. 2001; Kim 2003; Ross et al. 2003). In another approach tumor-selective MAbs are used to target cytotoxic agents to diseased cells (Liu et al. 1996; Milenic 2002). Typically, several molecules of a potent cytotoxic agent are covalently linked to a MAb, thus forming an immunoconjugate (Blättler and Chari 2001; Milenic 2002; Lo-Coco et al. 2004; Lu et al. 2005). huN901–DM1 is an immunoconjugate composed of the humanized MAb, huN901, and the cytotoxic drug DM1. DM1 is a semisynthetic ansamacrolide, and belongs to the maytansinoid class of anti-microtubular cytotoxic agents originally defined by the natural compound Maytansine (Cassady et al. 2004). HuN901 binds to the CD56 antigen present on cells of small-cell lung carcinomas (SCLC), other neuroendocrine tumors, and a large proportion of multiple myelomas (Zeromski et al. 2001). HuN901–DM1 is currently being evaluated in clinical trials to treat SCLC (Fossella et al. 2002). In addition, it has shown significant anti-myeloma activity in a preclinical study (Tassone et al. 2004).
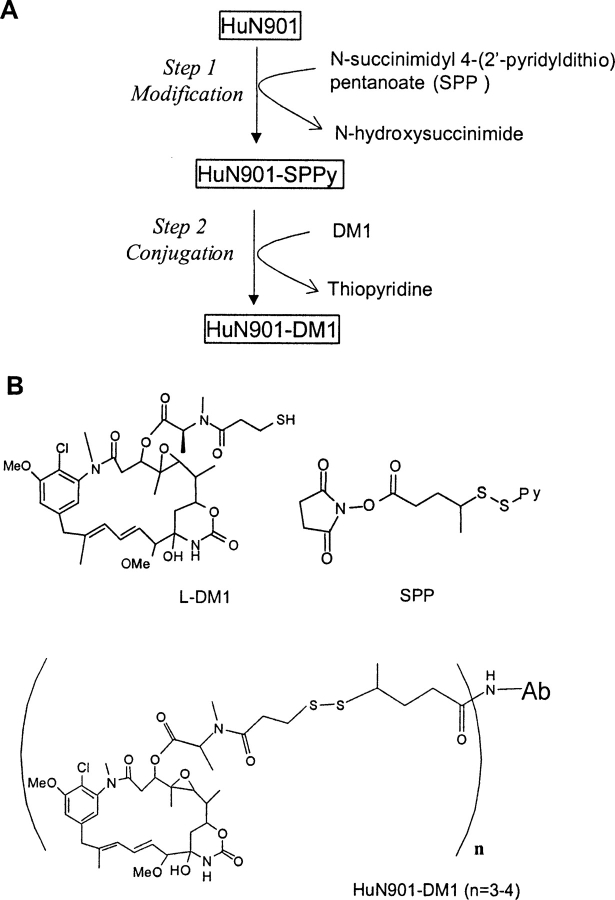
The procedure for the preparation of huN901–DM1 from huN901 and DM1 is illustrated in Figure 1 ▶. The antibody is first reacted with N-succinimidyl 4-(2′-pyridyldithio) pentanoate (SPP), which results in huN901 species with lysine residues that carry a 4-(2′-pyridyldithio)-1-oxopentyl substituent on their ɛ-amino group. The linker-modified antibody is then reacted with DM1, which contains a primary sulfhydryl group that rapidly undergoes a disulfide exchange reaction with the 2′-pyridyldithio moiety of the linker. This conjugation process is well controlled and introduces, on average, three to four DM1 molecules per huN901 molecule.
Figure 1.
Schematic representation of the modification and conjugation reactions in the preparation of huN901–DM1 (A) and illustration of chemical structures of DM1, the SPP linker, and huN901–DM1 conjugate (B).
This method of conjugate preparation will result in a mixture of conjugate species that differ in the number of drugs linked as well as the sites of drug linkage, since huN901 has 86 lysine residues that potentially can be modified. Structural characterization is challenging due to the heterogeneous nature of the conjugate, the large molecular size of antibodies and possible low abundances of modified residues. There are only a few studies in which conjugated antibodies were analyzed by mass spectrometry to give the distribution and load of linked small molecules (Siegel et al. 1997; Adamczyk et al. 2000; Lu et al. 2005). In these cases, however, the exact locations of conjugation sites remained elusive. Determining the conjugation sites is important, since the affinity of MAb conjugates may be affected significantly by modification of residues in complementarity determining regions (CDRs) resulting in non-active conjugate species in the mixture.
Electrospray ionization mass spectrometry (ESIMS) has proven to be a powerful structural characterization technique for large proteins such as MAbs and MAb conjugates (Feng and Konishi 1992; Lewis et al. 1994; Siegel et al. 1997; Bongers et al. 2000). Recently, we published the structural characterization—primary sequence and posttranslational modifications—of huN901, which was performed with tryptic and Asp-N peptide mapping in combination with electrospray ionization time-of-flight mass spectrometry (ESI-TOFMS) (Wang et al. 2005). In the current study, ESI-TOFMS was used to both assess the drug distribution in huN901–DM1 and, for the first time, to determine the conjugation sites. Deglycosylation of huN901–DM1 and tryptic and Asp-N protease digestions were performed to help in the characterization.
Results
Structural analysis of deglycosylated huN901–DM1 by ESI-MS
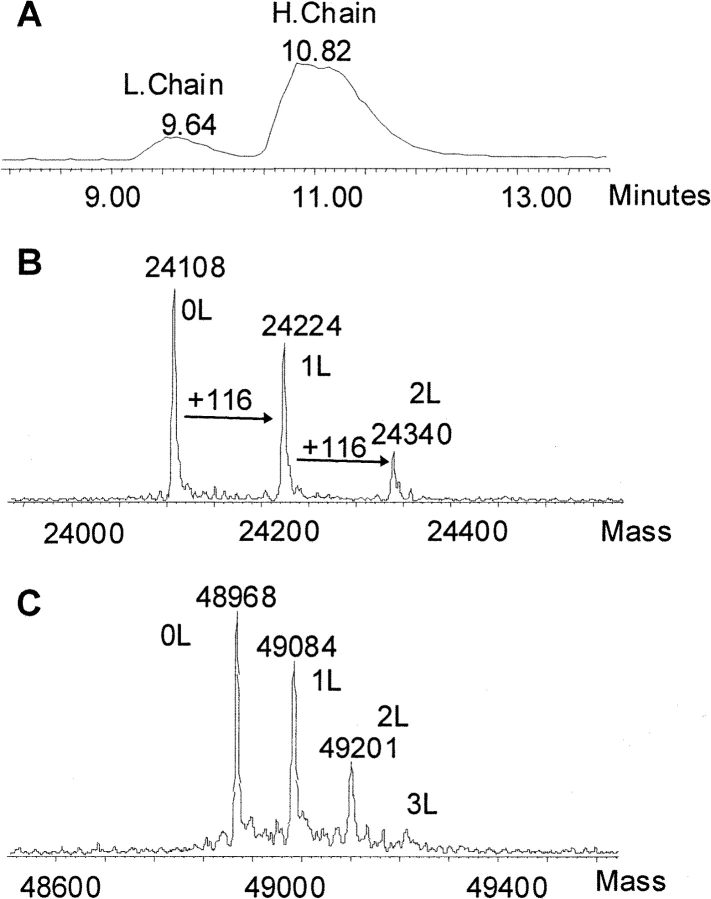
The immunoconjugate, huN901–DM1, is a mixture of conjugate species with varying numbers of covalently linked drugs. The composition of the mixture was analyzed by mass spectrometry of deglycosylated conjugate (dghuN901–DM1), which eliminates the complexity in the MS spectrum caused by the heterogeneity in the glycosylation of the CHO cell-derived huN901 antibody (Wang et al. 2005). Samples were analyzed using size-exclusion chromatography (SEC) coupled online with ESI-TOFMS (Lazar et al. 2005). An organic mobile phase was used in the SEC elution to avoid salt interference with protein ionization. The experimental ESI-MS spectrum of dghuN901–DM1 spanned an m/z range of 2000–4000 (Fig. 2A ▶), which gave a deconvoluted spectrum of seven prominent peaks with the following masses: 146,152 Da, 147,004 Da, 147,858 Da, 148,712 Da, 149,562 Da, 150,416 Da, and 151,268 Da (Fig. 2B ▶). The mass differences between adjacent peaks vary from 850 Da to 854 Da with a mean of 853 Da, which is consistent with the mass of one covalently linked DM1 drug (calculated mass increase 852 Da). The mass of the first peak in the series, 146, 152 Da, is in good agreement with the calculated mass of unconjugated dghuN901 (146,147 Da), given that such absolute mass measurements are typically associated with an error in the range of ±0.01%. The seven major peaks in Figure 2B ▶ can thus be assigned to naked dghuN901 (0D) and dghuN901 with one, two, three, four, five, and six covalently linked DM1 drug molecules (1D–6D), respectively.
Figure 2.
ESI-TOFMS spectra of deglycosylated huN901–DM1. (A) Raw MS data; (B) deconvoluted MS spectrum. “D” in B designates DM1 drug molecule.
In a second step, we analyzed the conjugation profiles of the separated dghuN901 light and heavy chains. The deglycosylated conjugate was denatured and reduced with DTT before the two chains were separated by reverse-phase HPLC and analyzed by online-coupled ESI-TOFMS (Fig. 3 ▶). The deconvoluted MS spectra show three and four prominent peaks for the light chain and the heavy chain, respectively. The mass difference between adjacent peaks varies between 116 Da and 117 Da (Fig. 3B,C ▶), which corresponds to the mass change caused by covalently linking one 4-mercapto-1-oxopentyl moiety (calculated mass increase 116 Da). This moiety is the covalently linked portion of the SPP linker with the free sulfhydryl group, which indicates that DTT reduction of the antibody, as expected, also reduced the disulfide bonds in the drug links. The number of linkers attached to the light and heavy chains can be obtained directly from the deconvoluted MS spectra (Fig. 3B,C ▶); the three prominent peaks in the light-chain spectrum are light chains with zero, one, and two linkers, while the four prominent peaks in the heavy chain spectrum are for species with zero, one, two, and three attached linkers. Therefore, both chains of the antibody are modified and conjugated with DM1 drugs. However, the measured masses of the reduced and alkylated light and heavy chains (24,108 Da and 48,968 Da) differ each from their calculated values (24,113 Da and 48,977 Da) by more than would be expected as the typical error (see above). Based on our previous analysis of the huN901 antibody, we attribute this difference to incomplete reduction of the intrachain disulfide bonds in both chains (Wang et al. 2005).
Figure 3.
LC/MS analysis of deglycosylated and reduced huN901- DM1. (A) Total ion current chromatogram showing the HPLC separation of the light (L) chain and heavy (H) chain; (B) deconvoluted MS spectra of conjugate light chain; (C) deconvoluted MS spectra of conjugate heavy chain. “L” in B and C designates SPP linker moiety.
Determination of conjugation sites by peptide mapping
The first step in conjugate preparation is the modification of huN901 with N-hydroxysuccinimidyl-4-(2′-pyridyldithio)- pentanoate (SPP) (Fig. 1 ▶) in an aqueous buffer solution. Under such conditions, N-hydroxysuccinimide esters preferentially react with primary amino groups on proteins forming stable amide bonds. Classical peptide mapping techniques were used for the determination of drug conjugation sites using either trypsin or Asp-N protease for digestion of the separated heavy and light chains of reduced and alkylated huN901–DM1. Since DTT reduction removes the DM1 drugs from the conjugate, the modification at the conjugation sites will be the alkylated 4-carboxymethylmercapto-1-oxopentyl linker moiety, which adds 174 Da to the peptide mass.
Detecting conjugation sites by trypsin digestion and ESI-TOFMS analysis
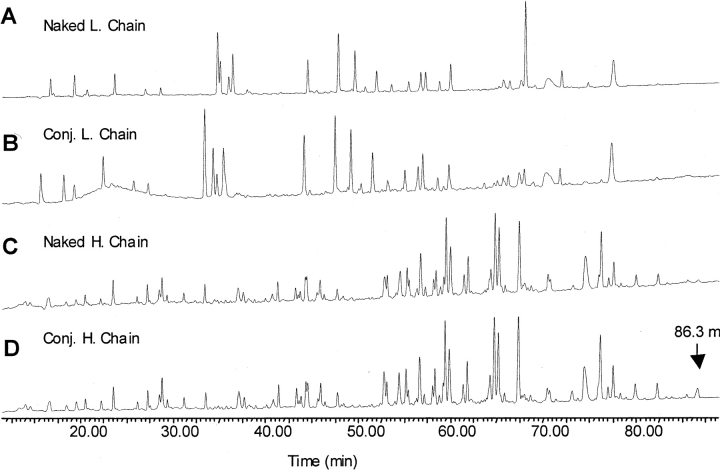
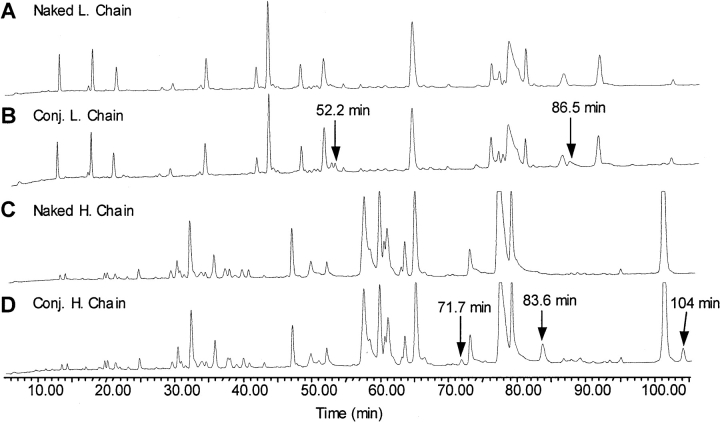
Since modification of a lysine residue will inhibit trypsin cleavage, we compared the tryptic digestion maps of the conjugate heavy and light chains with those from the naked antibody (Fig. 4 ▶). The HPLC UV traces at 214 nm of trypsin digests are presented in Figure 4 ▶, and show that the profiles of the conjugate chains are very similar to those of the naked antibody chains. Some differences in peak intensities and retention times were observed in the light-chain chromatograms from 33.4 to 35.4 min and at 68.0 min (Fig. 4A,B ▶). However, MS data showed that none of these peptides were modified, and the cause of these chromatographic differences is not known. For the heavy chain, a late eluting peak at 86.3 min was found in the conjugate sample only (Fig. 4D ▶). This peak was assigned by MS analysis to be an uncleaved heavy-chain peptide (372GF..K393..LY408) whose mass had increased by 174 Da. Thus, it was identified as a modified peptide and the only internal lysine residue, K393, was designated a conjugation site.
Figure 4.
The UV chromatograms at 214 nm of tryptic digests of light and heavy chains from naked antibody and conjugate. The alkylated light and heavy chains were separated by denaturing size exclusion chromatography prior to the enzymatic digestion. (A) Naked antibody light chain; (B) conjugate light chain; (C) naked antibody heavy chain; (D) conjugate heavy chain.
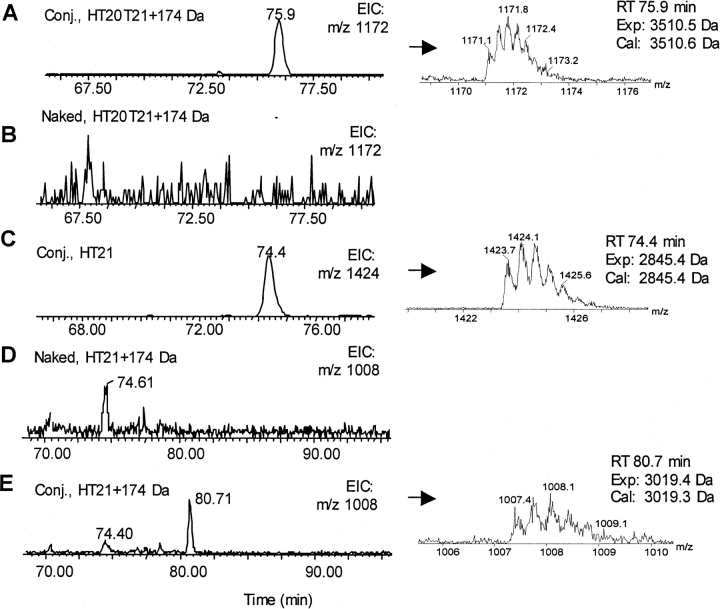
Because of the high similarity between the chromatograms of conjugated and naked huN901 heavy- and light-chain tryptic digests, we could not identify further conjugation sites based on differences in their UV traces. Instead, MS data were used to identify modified peptides. First we assumed that linker modification could occur at any lysine residue and calculated the expected mass of all the putative, uncleaved tryptic peptides with a mass increase of 174 Da. Then we searched in the LC/ MS profile for this mass by plotting extracted ion chromatograms (EICs). The same peptide masses were also extracted from naked-chain peptide mapping profiles. A modified peptide was identified when the corresponding mass was found in the conjugate antibody mapping but not in the same retention time range in the LC/ MS profile of the naked antibody. For example, trypsin digestion of naked huN901 would result in tryptic heavy-chain peptides HT20 (220SCDK223) and HT21 (224TH..PK249). Assuming linker modification at K223 in the heavy chain, an uncleaved peptide HT20T21 (220SC..K223..PK249) would be present with a mass increase of 174 Da (calculated peptide mass 3510.6 Da). As shown in Figure 5, A and B ▶, an EIC peak with a mass of 3510.5 Da was found eluting at 75.9 min in the LC/ MS profile of the conjugated heavy chain but not in that of the naked heavy chain, suggesting that K223 was indeed a modification site. In addition to the modified peptide HT20T21, the unmodified peptide HT21 (m/z 1424) derived from complete trypsin cleavage was also found in the conjugate heavy-chain mapping (Fig. 5C ▶), indicating that K223 was only partially modified.
Figure 5.
The extracted ion chromatograms and ESIMS spectra of tryptic heavy chain peptides HT20T21 (A,B) and HT21 (C–E) derived from the LC/MS analysis of the digests of naked and conjugated huN901. The EIC peaks at 74 min in both D and E do not result from protein conjugation.
Another example of a modified peptide is shown in Figure 5, D and E ▶. Peptide HT21 (224TH..PK247PK249) contains an internal lysine (K247), which is not efficiently cleaved by trypsin due to the proline residue C-terminal to it. This peptide was found to be modified with a linker, since the peptide with an experimental mass of 3019.4 Da, which is 174 Da more than the original mass, was found eluting at 80.7 min in the conjugated heavy-chain map (Fig. 5E ▶), but not seen in the naked-chain map (Fig. 5D ▶). The modification site is likely at residue K247, since it is the only modifiable lysine residue in the peptide.
The results of the complete comparative LC/MS data analysis of tryptic digests of naked and conjugated huN901 light and heavy chains are summarized in Table 1. A total of 18 modified tryptic peptides were identified; six peptides are from the light chain and 12 are from the heavy chain. The experimentally determined mass values of the modified peptides are close to their calculated mass values, with the average deviation being <0.1 Da. The conjugation sites were assigned to the internal lysine residue in each peptide. All of the sites were found only partially modified, since in each case, the uncleaved, modified peptide and the corresponding two peptides generated through cleavage after the internal lysine residue were identified. In fact, the unmodified peptides dominated the chromatographic profiles so that the chromatograms of the trypsin digests of the conjugate huN901 resemble those of the naked antibody (Fig. 4A–D ▶). As a result, most modified peptides could not be observed as new peaks in the UV chromatograms due to their low abundances.
Table 1.
Modification sites in huN901-DM1 detected in peptides from trypsin digestion
| Modified peptidea | Sequence | Cal. massb (Da) | Exp. mass (Da) | Error (Da) | Modified sites |
| LT7T8 | 67 FS. .SR82 | 1832.84 | 1832.88 | 0.04 | K79 |
| LT10T11 | 109 VE. .R113 | 817.41 | 817.43 | 0.02 | K112 |
| LT16T17 | 155VD. .SK188 | 3792.71 | 3792.90 | 0.19 | K174 |
| LT18T19 | 189AD. .HK195 | 1063.44 | 1063.52 | 0.08 | K193 |
| LT19T20 | 194HK. .TK212 | 2315.06 | 2315.13 | 0.07 | K195 |
| LT20T21 | 196VY. .NR216 | 2554.16 | 2554.20 | 0.04 | K212 |
| HT14T15 | 123GP. .VK148 | 2663.29 | 2663.22 | −0.07 | K134 |
| HT18T19 | 215KVEPK219 | 773.38 | 773.38 | 0.00 | K215 |
| HT20T21 | 220 SC. .KPK249 | 3510.60 | 3510.48 | −0.12 | K223 |
| HT21 | 224TH. .KPK249 | 3019.42 | 3019.29 | −0.13 | K247 |
| HT23T24 | 257TP. .AK289 | 3971.79 | 3971.70 | −0.09 | K275 |
| HT24T25 | 276FN. .PR293 | 2333.10 | 2333.08 | −0.02 | K289 |
| HT29T30 | 322CK. .NK327 | 909.37 | 909.35 | −0.02 | K323 |
| HT30T31 | 324VS. . EK335 | 1439.75 | 1439.74 | −0.01 | K327 |
| HT31T32 | 328AL. .SK339 | 1440.77 | 1440.74 | −0.03 | K335 |
| HT36T37 | 357DE. .VK371 | 1921.92 | 1921.86 | −0.06 | K361 |
| HT38T39′ | 372GF. .LY408 | 4359.48 | 4359.59 | 0.11 | K393 |
| HT40T41 | 411LT. .SR417 | 991.47 | 991.50 | 0.03 | K415 |
aL stands for light chain and H for heavy chain; HT38T39′ means the peptide comprised of HT38 and a partial sequence of HT39.
bCalculated mass of the peptides attached with SPP linker.
Detecting conjugation sites by Asp-N protease digestion and ESI-TOFMS analysis
The HPLC UV traces at 214 nm of Asp-N digests of the reduced and alkylated light and heavy chains of naked and conjugate huN901 are presented in Figure 6, A–D ▶. Similar to trypsin digestion, only minor differences were observed in the HPLC profiles of Asp-N digests of light and heavy chains from naked antibody and conjugate. However, since fewer peptides are generated in the Asp- N protease digestion, more chromatographic differences caused by conjugation were evident in the chromatograms than in the tryptic digestion maps. For instance, compared with the naked antibody light-chain profile (Fig. 6A ▶), the chromatogram of the conjugate light chain has two extra peaks eluting at 52.2 min and 86.5 min (Fig. 6B ▶), which were identified by MS as modified light-chain Asp-N peptides LD10 and LD5, respectively. The conjugate heavy-chain chromatogram has three new peaks eluting at 71.7 min, 83.6 min, and 103.9 min, respectively (Fig. 6D ▶). These peaks were identified as modified peptides HD15, HD8, and HD6 (Table 2).
Figure 6.
The UV chromatograms at 214 nm of Asp-N protease digests of the light and heavy chains of naked huN901 (A,C) and huN901-DM1 conjugate (B,D). The observable extra peaks related with antibody conjugation were indicated in the conjugate mapping.
Table 2.
Modification sites in huN901-DM1 detected in peptides from Asp-N digestion
| Modified peptidesa | Sequence | Cal. mass (Da) | Exp. mass (Da) | Error (Da) | Potential mod. sites |
| LD4 | 75DF. .AE86 | 1580.77 | 1580.80 | 0.03 | K79 |
| LD5 | 87DV. .PS126 | 4600.07 | 4599.99 | −0.08 | K108, 112 |
| LD10 | 190DY. .EC219 | 3701.64 | 3701.67 | 0.03 | K193, 195, 212 |
| LD10* | 190DY. .EC219 | 3877.87 | 3877.98 | 0.11 | K193, 195, 212 |
| HD2 | 62DS. .SR72 | 1438.72 | 1438.76 | 0.04 | K65 |
| HD5 | 106DY. .VK148 | 4446.83 | 4446.99 | 0.16 | K122, 134, 148 |
| HD6 | 149DY. .KV212 | 6990.61 | 6991.05 | 0.44 | K206, 211 |
| HD7 | 213DK. .SC221 | 1264.56 | 1264.62 | 0.06 | K214, 215, 219 |
| HD8 | 222DK. .PK249 | 3262.57 | 3262.56 | −0.01 | K223, 247, 249 |
| HD8* | 222DK. .PK249 | 3436.60 | 3436.62 | 0.02 | K223, 247, 249 |
| HD12b | 281DG. .HQ312 | 5317.46 | 5317.64 | 0.18 | K289, 291 |
| HD13 | 313DW. .SR356 | 5199.84 | 5200.56 | 0.72 | 6 K (313–356) |
| HD14 | 357DE. .PS376 | 2473.17 | 2473.24 | 0.07 | K361, 371 |
| HD15 | 377DI. .YL399 | 2774.27 | 2774.30 | 0.03 | K393 |
| HD18′ | 414DK. .MH430 | 2239.94 | 2240.02 | 0.08 | K415 |
aL stands for light chain and H for heavy chain; H18′ means a peptide with partial sequence of Asp-N peptide H18; peptides attached with two linker molecules are indicated by *.
bHD12 is glycosylated at Asn298 as previously reported (Wang et al. 2005).
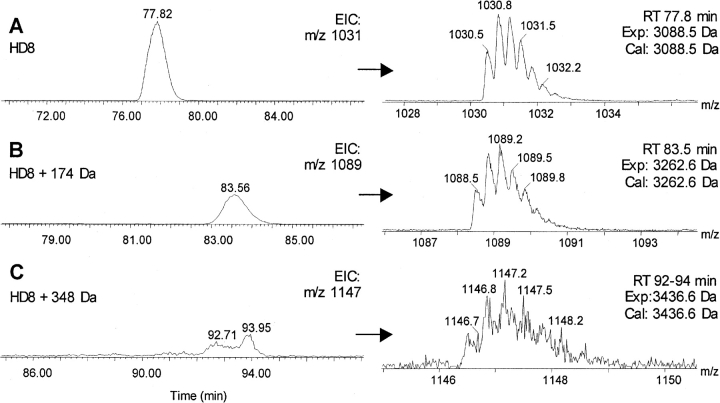
The identification of modified heavy-chain peptide HD8 (222DK..PKPK249) is illustrated in Figure 7 ▶. The unmodified peptide HD8 was found eluting at 77.8 min (Fig. 7A ▶). A new peak was found eluting at 83.6 min, whose experimental mass (3262.6 Da) matches with the calculated mass of HD8 attached with one SPP linker, indicating the peptide is modified (Fig. 7B ▶). The same peptide attached with two linkers at two modification sites was also found, which eluted from 92 to 94 min as a relatively weak EIC peak with a mass increase of 348 Da (Fig. 7C ▶). These results are consistent with trypsin digestion, in which two modification sites, K223 and K247, were identified (Table 1).
Figure 7.
EICs and corresponding ESIMS spectra of the Asp-N heavy chain peptide, HD8, modified with zero (A), one (B), and two (C) linkers derived from the LC/MS analysis of conjugate heavy-chain digest.
To detect additional modified Asp-N peptides, an analogous approach of data processing was adopted as for the trypsin digest described above. Assuming that modification can occur at each lysine residue, the EICs of putative modified Asp-N peptides were plotted for both naked and conjugated chains, and the data were searched for corresponding peaks. A peak was assigned to a modified peptide when the mass of the peptide was increased by 174 Da in the conjugate protein maps but not in the corresponding naked protein maps. A total of 13 modified Asp-N peptides, three from the light chain and 10 from the heavy chain, were thus identified and are listed in Table 2. Two peptides, LD10 (190–219) and HD8 (222–249), were found to contain one or two modifications as measured by mass increases of 174 Da and 348 Da, respectively (Table 2). In Asp-N protease digestion, both modified and unmodified forms of the peptides were found in the conjugate peptide maps, indicating partial modification for all modified peptides. The exact modification sites were generally not assigned for the Asp-N peptides since most of the peptides contain more than one lysine residue. Although further MS/MS analysis may help determine exact modification sites in these peptides, such analysis was not performed since most modified peptides had weak MS signals and are not suitable to be chosen as precursor ions for further fragmentation. Additionally, most modified peptides are not fully resolved from other peptides.
Comparison of the results from trypsin and Asp-N peptide mapping analysis
The results from the trypsin and Asp-N digestion studies are summarized in Figure 8 ▶, which shows the amino acid sequences of huN901 light and heavy chains with all the modified residues labeled that were identified in the trypsin digestion, and all the peptides indicated that were found modified in the Asp-N digestion study. It is apparent that the two sets of results generally agree with each other; i.e., the modifications were detected in either study, with only a few modification sites that were identified in only one of the two peptide mapping studies. For instance, modifications in the three Asp-N peptides, HD2 (62DS..SR72) and HD6 (149DY..KV212) were not detected in the trypsin digestion. The total number of identified modification sites increases, therefore, to 20. In addition, the modification at K174 in the light chain was detected by tryptic mapping but not by the Asp-N mapping.
Figure 8.

The amino acid sequences of huN901 light (top) and heavy (bottom) chains. “<Q” indicates pyroglutamate, and italic residues are CDRs. Conjugation sites detected in trypsin digestion are shown in boxes. Asp-N peptides attached with one or two linker moieties are underlined by one or two arrows.
Discussion
The antibody–drug conjugate, HuN901–DM1, is prepared by first random modification of the antibody with a cross-linking reagent to which the drug is covalently linked in a second step. This method yields a conjugate preparation that is heterogeneous at the molecular level. The current study of the structural composition of the conjugate shows that the heterogeneity is twofold. First, the preparation contains several conjugate species that differ in the number of linked drug molecules per antibody molecule: Species with one to six linked drugs were identified, as well as small amounts of unconjugated (naked) antibody. Second, each species with a certain number of linked drug molecules can have the linked drugs distributed to different sites of modification: Forty different sites of modification were identified considering that there are two copies of light and heavy chains in each antibody molecule. The identification of 40 modification sites in the antibody indicates that the drug molecules distribute over ~47% of the 86 lysine residues in the antibody. This number of conjugation sites is large enough so that each site is only partially modified, which leads to low abundances for many modified peptides in the enzymatic digests. Consequently, such peptides could not be detected in the chromatographic digestion maps. Only careful analysis of the MS data through searching for expected mass peaks allowed the identification of most of the modification sites. Since such an analysis can detect modified peptides present at an amount of as low as a few percent of the total expected amount, one cannot exclude that more than 47% of the lysine residues are modified, albeit at lower levels they are not detectable with the applied technique. It is also worth mentioning that a recent study suggested that tyrosine and histidine residues can also be modified by succinimide esters (Leavell et al. 2004). However, extensive searches for expected masses indicated that modification of tyrosine and histidine residues were not present in huN901–DM1.
The determination of the conjugation sites in huN901–DM1 also showed that the lysine residues in the CDRs of huN901 were not modified (Fig. 8 ▶). This is consistent with the observation that huN901–DM1 had retained the full binding affinity of the naked antibody. Although in this study peptide mapping combined with LC/MS analysis was only performed for one batch of huN901–DM1, the modification sites in different batches of the conjugate are expected to be largely reproducible, assuming that the protein conformation has no change under the same reaction conditions. UV and MS analysis of intact conjugate did show consistent drug load and drug distribution profile from batch to batch (data not shown).
Our results show that not all lysine residues of huN901–DM1 are modified to a degree detectable by ESI-TOFMS. Under the same reaction conditions, whether or not a lysine residue is modified and to what extent it is modified is expected to depend on the location of the residues in the antibody, which determines the local environment. For example, one of the modified Asp-N peptides that were detected in the HPLC peptide mapping, HD8 (heavy-chain residues 222–249), is located in the hinge region. Furthermore, the trypsin digestion analysis (Table 1) showed that both lysine residues in this peptide, K223 and K247, which are the only two lysine residues in the hinge region, were modified. This may be due to the large solvent exposure and the structural flexibility of the hinge region of the antibody.
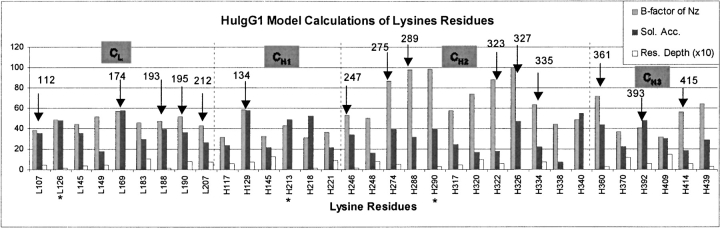
To understand the structural features of the modification sites in huN901–DM1, the crystal structural models of human IgG1 (huIgG1) Fab (PDB code 1UCB; Sheriff et al. 1996), and Fc (PDB code 1H3X; Krapp et al. 2003) were used to mimic the structure of the constant regions of huN901, since huN901 and huIgG1 share identical amino acid sequences in constant regions (Roguska et al. 1994). The solvent accessibility and average depth for the lysine residues in huIgG1 were calculated, and the B-factors of the side-chain nitrogen of lysine residues were also obtained (Fig. 9 ▶). The modified lysine residues in huN901–DM1 were mapped to the corresponding lysine residues in huIgG1. As shown in Figure 9 ▶, most of the modified lysine residues have relatively large solvent- accessible areas, large B-factors, but small values of residue depth, indicating that these residues either are located on the protein surface or have relatively large structural flexibilities. Although this correlation is generally true for most modified lysine residues, exceptions do exist. For example, residues K126 in the light chain, and K213 and K290 in the heavy chain of huIgG1 are surface residues with relatively large solvent-exposure areas or B-factors (Fig. 9 ▶). However, the three lysine residues at similar locations in huN901–DM1 were not modified. These exceptions suggest that other factors, such as effects from neighboring residues or hydrogen bonds, may also contribute to the reactivity of individual lysine residues.
Figure 9.
Plots of the B-factors (Å2) for side-chain nitrogen, solvent accessibility (Å2), and residue depth (Å) of lysine residues in the huIgG1 constant regions, including the light-chain domain CL and the three heavy-chain domains, CH1, CH2, and CH3. Residues marked with arrows correspond to the modified lysine residues detected in huN901–DM1 (labeled with residue numbers in huN901). Solvent accessibility was calculated using the “What_If” Web server (http://swift.cmbi.kun.nl/WIWWWI//) (Rodriguez et al. 1998). The residue depth was calculated using the DPX server (http://hydra.icgeb.trieste.it/dpx/) (Pintar et al. 2003).
The crystal structures of huIgG1 Fab and Fc were also used to examine the locations of the modified lysine residues in the secondary structures of huN901 constant regions. As shown in Figure 10 ▶, most modified lysine residues are located in the surface loops of the antibody, with only two modified lysine residues in the heavy chain (K323 and K393) located in β-sheets. Although they are not located in loop regions, these two residues still have relatively large solvent accessibilities and large B-factors (Fig. 9 ▶).
Figure 10.

Cartoon plots of the crystal structures of human IgG1 Fab (PDB code 1UCB) (A) and Fc (PDB code 1H3X) (B). The two chains are in cyan and yellow and the carbohydrate moieties are in gray. Modified lysine residues in huN901–DM1 are mapped to the corresponding residues in huIgG1 and are shown in red. In the Fc fragment, the modified lysine residues are only labeled in one chain. The N and C termini are labeled as N-and C-, respectively. The protein images were created using Rasmol V2.6.
In summary, the structural heterogeneity caused by random conjugation of lysine residues in huN901– DM1 was characterized by mass spectrometry and peptide mapping. Twofold heterogeneity was revealed: First, the antibody contains species linked with different number of drug molecules; second, antibody with the same number of drug molecules may have different sites of linkage. The conjugation sites were determined by both trypsin and Asp-N protease digestion. Although trypsin digestion led to determination of specific sites of modification, chromatographic differences between maps of naked antibody and conjugate appeared more evident upon Asp-N protease digestion. Modification sites detected by both methods are generally consistent, therefore strengthening the results. All modifications are partial, with about seven lysine residues in the light chain and 13 lysine residues in the heavy chain modified. The detected conjugation sites in huN901–DM1 were mapped in the crystal structural models of huIgG1, and the modified residues were generally found in areas with large solvent accessibility and structural flexibility. Results of this study have enhanced the understanding of the structure of MAb–maytansinoid conjugates and other types of immunoconjugates. Additionally, correlation of residue location and modification is expected to provide useful information to improve conjugation selectivity and to design conjugates with better structural homogeneity and biological activities. The methods used in this study should be applicable to elucidate structural details of other immunoconjugates.
Materials and methods
Materials and equipment
HuN901–DM1 conjugate was manufactured at ImmunoGen, Inc. as an aqueous buffer solution (1 mg/mL antibody in 50 mM potassium phosphate [pH 6.5], 50 mM NaCl). HuN901 was in a solution of 10 mg/mL antibody, 50 mM potassium phosphate buffer [pH 6.5], 50 mM NaCl, 3 mM EDTA. TPCK-treated trypsin was from Worthington Biochemical Co., trifluoroacetic acid (TFA) was from Pierce, and HPLC-grade acetonitrile from Honeywell Burdick & Jackson. All other chemicals and enzymes, unless otherwise stated, were purchased from Sigma. Deionized water used was produced by a MilliQ water purification system. The HPLC system consisted of a Waters Alliance 2695 separation module equipped with a Waters 2478 dual-wavelength UV detector and a column- heating compartment. An LCT ESI-TOF mass spectrometer (Waters Co.) was used for protein mass measurement and peptide mapping with the source temperature in the range of 80°–100°C. The desolvation nitrogen gas flow rate was 400–500 L/h, and desolvation temperature was 150°–200°C. The capillary and cone voltages were optimized for maximum sensitivity. MS data were acquired using Masslynx 4.0. Protein ESIMS spectra were deconvoluted using MaxEnt 1. The mass spectrometer was calibrated using a solution containing 2 mg/mL NaI and 0.05 mg/mL CsI in 50% aqueous isopropanol for the acquiring mass ranges of m/z 200–2000 for peptide mapping and m/z 1000–4000 for protein mass measurement.
SEC/MS analysis of deglycosylated huN901–DM1
Deglycosylation was performed by incubating 100 μg of huN901–DM1 with 4 μL (20 mU) of N-glycanase (Prozym Co.) at 37°C overnight. An TSK gel Super 3000 SWXL SEC column (4.6 mm × 30 cm, 4-μm particle size, 250 Å pore size; Tosoh Biosciences) was online coupled with ESIMS for protein desalting and MS analysis. About 50 μg of deglycosylated huN901–DM1 was loaded onto the column and eluted with a mobile phase of 50% (v/v) aqueous acetonitrile containing 0.1% (v/v) TFA in 20 min at 60°C with a flow rate of 0.25 mL/min. The eluate passed through a UV detector (280 nm) and was split 1:5 before it was directed toward the mass spectrometer. The conjugated antibody eluted at about 7.5 min and the flow was diverted to waste at about 14 min to the end of the run to avoid salt contamination in the MS source.
LC/MS analysis of deglycosylated and reduced huN901–DM1
Deglycosylated huN901–DM1 from above was reduced with 50 mM DTT in 50 mM potassium phosphate buffer [pH 6.5], at 37°C for 60 min. Approximately 25 μg reduced, deglycosylated huN901–DM1 was loaded onto a PLRP-S reverse-phase HPLC column (2.1 × 50 mm, 8 μm, 4000 Å ; Polymer Laboratories) equilibrated at 60°C, and coupled with ESI-TOFMS. The column was eluted at a flow rate of 0.2 mL/min using a gradient of acetonitrile in water as follows: 5% acetonitrile for 2 min, then from 5%–30% acetonitrile in 0.5 min, from 30%– 50% in 10 min, from 50%–98% in 1 min, and back to 5% acetonitrile in 0.5 min and equilibration at 5% for 6 min. The HPLC flow was split 1:5 before ESIMS and diverted to waste in the first 5 min of the gradient to avoid salt interference.
Reduction and alkylation of huN901 and huN901–DM1 for peptide mapping
HuN901–DM1 was concentrated to the same concentration of naked huN901 (10 mg/mL) using an Amicon 10-kDa cutoff filter (Millipore). Naked and conjugated huN901 was reduced by mixing 2 mg of antibody (200 μL) with 1 mL of denaturing buffer (6 M guanidine hydrochloride, 1 mM EDTA, 0.5 M Tris buffer [pH 8.3]) and 60 μL of 1MDTT and incubating at 37°C for 1 h. Then 120 μL of 1 M iodoacetic acid in 2 M NaOH solution was added and the solution was kept in the dark at room temperature for 30 min. Finally, 120 μL of 1MDTT was added to quench the reaction.
Separation of heavy and light chains of naked and conjugated huN901
Separation of the antibody light and heavy chains was performed using a Hitachi HPLC system equipped with a Superdex 200 HR SEC column (Pharmacia) and a mobile phase of 3 M guanidine hydrochloride at a flow rate of 1 mL/min. About 700–800 μL of denatured, reduced huN901–DM1 was injected each time and baseline separation of light and heavy chains was obtained. The elution was monitored by UV absorbance at 280 nm and fractions were collected from 9.2 to 12.2 min for the heavy chain and from 12.4 to 14.5 min for the light chain. Fractions were concentrated to 200–300 μL using an Amicon 10-kDa cutoff filter (Millipore) and subjected to a fast desalting gel filtration column (Pharmacia Biotech, flow rate 1.5 mL/min) to change the buffer to 50 mM Tris.HCl (pH 8.2), containing 1 mM calcium chloride for trypsin digestion or 100 mM Tris.HCl (pH 8.2), containing 10 mM calcium chloride for Asp-N digestion.
Enzyme digestions
The separated light and heavy chains of naked and conjugated huN901 were digested with TPCK treated trypsin at an enzyme: substrate (E:S) ratio (w/w) of 1:50 at 37°C overnight. The reaction was then quenched by adding 30 μL of 1 M HCl to 1 mL of reaction solution. Similarly, Asp-N protease digestion of naked and conjugated light and heavy chains was performed at 37°C overnight. The E:S ratio (w/w) was 1:100. About 55 μL of 1 M HCl was added to 1 mL of reaction solution to quench the digestion.
LC/MS analysis of trypsin digests
Tryptic digests of naked and conjugated huN901 light and heavy chains were analyzed with a Jupiter Proteo C18 column (4.6 × 250 mm, 4 μm, 90 Å; Phenomenex) connected to an ESI-TOFMS. The HPLC column was kept at 37°C and eluted at a flow rate of 1.0 mL/min with 40 μL/min sent to the MS probe. Peptides were separated using a gradient of acetonitrile containing 0.1% TFA (mobile phase B) in water with 0.1% TFA (mobile phase A). Gradient conditions were as follows: 0–5 min, 0% B; 5–95 min, 0%–40% B; 95–96 min, 40%–100% B; 96–105 min, 100% B; 105–106 min, 100%–0% B; 106–120 min, 0% B. The HPLC flow in the first 5 min after a sample application was diverted to waste.
LC/MS analysis of Asp-N protease digests
Asp-N protease digests were analyzed using a narrow-bore C18 HPLC column (2.1 × 150 mm, 5 μm, 300 Å; Vydac) eluting at 30°C with a flow rate of 0.2 mL/min. The column was coupled with an ESI-TOFMS machine. A flow of 40 μL/min was directed to the MS probe using a T splitter after the column. Peptides were separated by gradient elution using water as mobile phase A and acetonitrile as mobile phase B, both with 0.05% (w/v) TFA. The gradient used was as follows: 0–10 min, 2%–10% B; 10–100 min, 10%–32% B; 100–105 min, 32%– 90% B; 105–110 min, 90%–98% B; 110–111 min, 98%–2% B; 111–120 min, 2% B. The HPLC flow was diverted to waste in the first 5 min after sample application.
Acknowledgments
We thank Drs. Alex Lazar and John Thomas for helpful discussions.
Abbreviations
CDR, complementarity determining region
DTT, dithiothreitol
EIC, extracted ion chromatogram
ESI-TOFMS, electrospray ionization time-of-flight mass spectrometry
HPLC, high-performance liquid chromatography
LC/MS, liquid chromatography/mass spectrometry
MAb, monoclonal antibody
PDB, Protein Data Bank
SCLC, small cell lung carcinoma
SEC, size-exclusion chromatograph
TIC, total ion current
TFA, trifluoroacetic acid.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051478705.
References
- Adamczyk, M., Gebler, J., Shreder, K., and Wu, J. 2000. Region-selective labeling of antibodies as determined by electrospray ionization-mass spectrometry (ESI-MS). Bioconjug. Chem. 11 557–563. [DOI] [PubMed] [Google Scholar]
- Blättler, W.A. and Chari, R.V.J. 2001. Drugs to enhance the therapeutic potency of anti-cancer antibodies: Antibody-drug conjugates as tumor-activated prodrugs. In Anticancer agents: Frontiers in cancer chemotherapy (eds. OjimaI. et al.), pp. 317–338. American Chemical Society, Washington, DC.
- Bongers, J., Cummings, J.J., Ebert, M.B., Federici, M.M., Gledhill, L., Gulati, D., Hilliard, G.M., Jones, B.H., Lee, K.R., Mozdzanowski, J., et al. 2000. Validation of a peptide mapping method for a therapeutic monoclonal antibody: What could we possibly learn about a method we have run 100 times? J. Pharm. Biomed. Anal. 21 1099–1128. [DOI] [PubMed] [Google Scholar]
- Cassady, J.M., Chan, K.K., Floss, H.G., and Leistner, E. 2004. Recent developments in the maytansinoid antitumor agents. Chem. Pharm. Bull. (Tokyo) 52 1–26. [DOI] [PubMed] [Google Scholar]
- Feng, R. and Konishi, Y. 1992. Analysis of antibodies and other large glycoproteins in the mass range of 150,000–200,000 Da by electrospray ionization mass spectrometry. Anal. Chem. 64 2090–2095. [DOI] [PubMed] [Google Scholar]
- Fossella, F.V., Tolcher, A., Elliott, M., Lambert, J.M., Lu, R., Zinner, R., Lu, C., Oh, Y., Forouzesh, B., McCreary, H., et al. 2002. Phase I trial of the monoclonal antibody conjugate, BB-10901, for relapsed/refractory small cell lung cancer (SCLC) and other neuroendocrine (NE) tumors. In American Society of Clinical Oncology Annual Meeting, Orlando, FL.
- Grillo-Lopez, A.J., Dallaire, B.K., McClure, A., Weaver, R., Varns, C., Wei, A., Allen, R., Lee, D., Shen, D., Leonard, J., et al. 2001. Monoclonal antibodies: A new era in the treatment of non-Hodgkin’s lymphoma. Curr. Pharm. Biotechnol. 2 301–311. [DOI] [PubMed] [Google Scholar]
- Kim, J.A. 2003. Targeted therapies for the treatment of cancer. Am. J. Surg. 186 264–268. [DOI] [PubMed] [Google Scholar]
- Krapp, S., Mimura, Y., Jefferis, R., Huber, R., and Sondermann, P. 2003. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 325 979–989. [DOI] [PubMed] [Google Scholar]
- Lazar, A.C., Wang, L., Blättler, W.A., Amphlett, G., Lambert, J.M., and Zhang, W. 2005. Analysis of the composition of immunoconjugates using size-exclusion chromatography coupled to mass spectrometry. Rapid Commun. Mass Spectrom. 19 1806–1814. [DOI] [PubMed] [Google Scholar]
- Leavell, M.D., Novak, P., Behrens, C.R., Schoeniger, J.S., and Kruppa, G.H. 2004. Strategy for selective chemical cross-linking of tyrosine and lysine residues. J. Am. Soc. Mass Spectrom. 15 1604–1611. [DOI] [PubMed] [Google Scholar]
- Lewis, D.A., Guzzetta, A.W., Hancock, W.S., and Costello, M. 1994. Characterization of humanized anti-TAC, an antibody directed against the interleukin 2 receptor, using electrospray ionization mass spectrometry by direct infusion, LC/MS, and MS/MS. Anal. Chem. 66 585–595. [DOI] [PubMed] [Google Scholar]
- Liu, C., Tadayoni, B.M., Bourret, L.A., Mattocks, K.M., Derr, S.M., Widdison, W.C., Kedersha, N.L., Ariniello, P.D., Goldmacher, V.S., Lambert, J.M., et al. 1996. Eradication of large colon tumor xenografts by targeted delivery of maytansinoids. Proc. Natl. Acad. Sci. 93 8618–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco, F., Cimino, G., Breccia, M., Noguera, N.I., Diverio, D., Finolezzi, E., Pogliani, E.M., Di Bona, E., Micalizzi, C., Kropp, M., et al. 2004. Gemtuzumab ozogamicin (Mylotarg) as a single agent for molecularly relapsed acute promyelocytic leukemia. Blood 104 1995–1999. [DOI] [PubMed] [Google Scholar]
- Lu, S.X., Takach, E.J., Solomon, M., Zhu, Q., Law, S.J., and Hsieh, F.Y. 2005. Mass spectral analyses of labile DOTA-NHS and heterogeneity determination of DOTA or DM1 conjugated anti-PSMA antibody for prostate cancer therapy. J. Pharm. Sci. 94 788–797. [DOI] [PubMed] [Google Scholar]
- Milenic, D.E. 2002. Monoclonal antibody-based therapy strategies: Providing options for the cancer patient. Curr. Pharm. Des. 8 1749–1764. [DOI] [PubMed] [Google Scholar]
- Pintar, A., Carugo, O., and Pongor, S. 2003. DPX: For the analysis of the protein core. Bioinformatics 19 313–314. [DOI] [PubMed] [Google Scholar]
- Rodriguez, R., Chinea, G., Lopez, N., Pons, T., and Vriend, G. 1998. Homology modeling, model and software evaluation: Three related resources. CABIOS 14 523–528. [DOI] [PubMed] [Google Scholar]
- Roguska, M.A., Pedersen, J.T., Keddy, C.A., Henry, A.H., Searle, S.J., Lambert, J.M., Goldmacher, V.S., Blättler, W.A., Rees, A.R., and Guild, B.C. 1994. Humanization of murine monoclonal antibodies through variable domain resurfacing. Proc. Natl. Acad. Sci. 91 969–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J., Gray, K., Schenkein, D., Greene, B., Gray, G.S., Shulok, J., Worland, P.J., Celniker, A., and Rolfe, M. 2003. Antibody-based therapeutics in oncology. Expert Rev. Anticancer Ther. 3 107–121. [DOI] [PubMed] [Google Scholar]
- Sheriff, S., Chang, C.Y., Jeffrey, P.D., and Bajorath, J. 1996. X-ray structure of the uncomplexed anti-tumor antibody BR96 and comparison with its antigen-bound form. J. Mol. Biol. 259 938–946. [DOI] [PubMed] [Google Scholar]
- Siegel, M. M., Tabei, K., Kunz, A., Hollander, I.J., Hamann, R.R., Bell, D.H., Berkenkamp, S., and Hillenkamp, F. 1997. Calicheamicin derivatives conjugated to monoclonal antibodies: Determination of loading values and distributions by infrared and UV matrix-assisted laser desorption/ ionization mass spectrometry and electrospray ionization mass spectrometry. Anal. Chem. 69 2716–2726. [DOI] [PubMed] [Google Scholar]
- Tassone, P., Gozzini, A., Goldmacher, V., Shammas, M.A., Whiteman, K.R., Carrasco, D.R., Li, C., Allam, C.K., Venuta, S., Anderson, K.C., et al. 2004. In vitro and in vivo activity of the maytansinoid immunoconjugate huN901-N2′-deacetyl-N2′-(3-mercapto-1-oxopropyl)-maytansine against CD56+ multiple myeloma cells. Cancer Res. 64 4629–4636. [DOI] [PubMed] [Google Scholar]
- Wang, L., Amphlett, G., Lambert, J.M., Blättler, W.A., and Zhang, W. 2005. Structural characterization of a recombinant monoclonal antibody by electrospray time-of-flight mass spectrometry. Pharm. Res. (in press). [DOI] [PubMed]
- Zeromski, J., Nyczak, E., and Dyszkiewicz, W. 2001. Significance of cell adhesion molecules, CD56/NCAM in particular, in human tumor growth and spreading. Folia Histochem. Cytobiol. 39 (Suppl. 2): 36–37. [PubMed] [Google Scholar]