Abstract
The flavoprotein component (AhpF) of Salmonella typhimurium alkyl hydroperoxide reductase contains an N-terminal domain (NTD) with two contiguous thioredoxin folds but only one redox-active disulfide (within the sequence -Cys129-His-Asn-Cys132-). This active site is responsible for mediating the transfer of electrons from the thioredoxin reductase-like segment of AhpF to AhpC, the peroxiredoxin component of the two-protein peroxidase system. The previously reported crystal structure of AhpF possessed a reduced NTD active site, although fully oxidized protein was used for crystallization. To further investigate this active site, we crystallized an isolated recombinant NTD (rNTD); using diffraction data sets collected first at our in-house X-ray source and subsequently at a synchrotron, we showed that the active site disulfide bond (Cys129–Cys132) is oxidized in the native crystals but becomes reduced during synchrotron data collection. The NTD disulfide bond is apparently particularly sensitive to radiation cleavage compared with other protein disulfides. The two data sets provide the first view of an oxidized (disulfide) form of NTD and show that the changes in conformation upon reduction of the disulfide are localized and small. Furthermore, we report the apparent pKa of the active site thiol to be ~5.1, a relatively low pKa given its redox potential (~265 mV) compared with most members of the thioredoxin family.
Keywords: X-ray crystallography, thioredoxin, disulfide, peroxiredoxin, thiolate, alkyl hydroperoxide reductase, synchrotron radiation, pKa, radiation damage
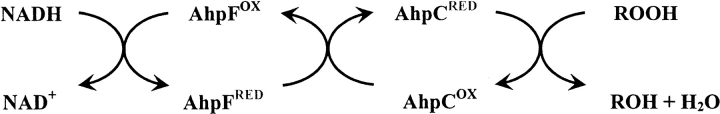
The alkyl hydroperoxide reductase system found in many eubacteria is responsible for catalyzing the reduction of hydrogen peroxide and organic hydroperoxides to their corresponding alcohols and water (Jacobson et al. 1989; Niimura et al. 1995; Poole and Ellis 1996). This peroxide-inducible system helps to protect cells against oxidative and nitrosative damage to DNA and other cellular macromolecules (Chen et al. 1998). The two protein components, AhpF and AhpC, work together to use NADH to reduce the hydroperoxide substrates as seen in Scheme 1 ▶:
Scheme 1.
The AhpC component of the Salmonella typhimurium system is a member of the peroxiredoxin family and has two redox-active cysteinyl residues involved in the reduction of hydroperoxides (Poole 2005). The structure of AhpC has been solved in both the oxidized (Wood et al. 2002) and reduced (Wood et al. 2003) forms, and it is proposed to go through a dimer–decamer transition during its catalytic cycle (Wood et al. 2002). The AhpF component is a homodimeric flavoenzyme (2 × 57 kDa) composed of three domains, a 196-residue N-terminal redox domain (NTD), and a portion similar to E. coli thioredoxin reductase (TrxR), which includes an FAD binding domain and an NADH/redox-active disulfide domain (NADH/SS) (Poole et al. 2000b).
The NTD domain of AhpF, the focus of this work, is the direct electron donor to AhpC, and it effectively acts as an appended substrate for the TrxR-like portion of AhpF (Poole and Ellis 1996; Poole et al. 2000b). Indeed, both soluble recombinant NTD (rNTD) mixed with TrxR, and NTD fused to TrxR, endowed TrxR with AhpC reducing activity (Poole et al. 2000a; Reynolds and Poole 2000). In the crystal structure of AhpF (Wood et al. 2001), the NTD was seen to be attached to the FAD domain of AhpF by a flexible 14-residue linker consistent with its role as an appended substrate. In addition, the NTD was seen to be representative of a new class of thioredoxin-related proteins that contain a single redox-active disulfide in the context of two fused thoredoxin folds, the active site itself having a mirror image relationship to that of thioredoxin. A surprising result from the crystal structure of AhpF was that, although the oxidized protein was used for crystallization, the active site cysteines of the NTD (Cys129 and Cys132) were found to be in the reduced form. They demonstrated a very close sulfur–sulfur distance of ~3 Å, and this interaction was proposed to be a thiol–thiolate hydrogen bond (Wood et al. 2001).
To further characterize this interesting variation on the thioredoxin motif, we are pursuing biochemical and structural (NMR and X-ray) investigations of the isolated rNTD, which is smaller and more amenable to analysis than is intact AhpF. Here, we characterize the rNTD crystal structure and the pKa of the active site Cys. We show that the rNTD active site disulfide is exquisitely sensitive to radiation-induced cleavage, and using laboratory-based diffraction data, we describe the structure of the disulfide form of the rNTD active site.
Results
Background
In preliminary work, two data sets from crystals of rNTD were collected at synchrotron sources. These data sets, at 2.1 and 2.8 Å resolution, were easily solved by molecular replacement. One crystal was in space group P212121 and the other was space group P41212, but both showed a similar active site structure as was seen in the intact AhpF structure (Wood et al. 2001); that is, the active site Cys residues did not form a disulfide but appeared to be in a close, nonbonded interaction (~3 Å separation). As we were surprised again that the disulfide was reduced despite the lack of reducing agent in the purification and crystallization buffers, we biochemically assessed the oxidation state of purified NTD using a thiol-sensitive assay. Quantification of free thiol levels with DTNB demonstrated that the protein used for crystallization began in the oxidized state (0.05 mol of reactive thiol per mole of rNTD as purified vs. 2.21 thiols per mole of DTT-reduced rNTD). This led us to conjecture that the disulfide was being reduced during synchrotron data collection, as has been seen for some other proteins (Weik et al. 2000; Alphey et al. 2003). To test this hypothesis, and if possible, determine a structure of the unperturbed oxidized NTD, we selected a single large crystal with the P41212 space group (two molecules per asymmetric unit) and collected two data sets: one from our laboratory X-ray source and then one at a synchrotron.
Structure determinations and overall structure
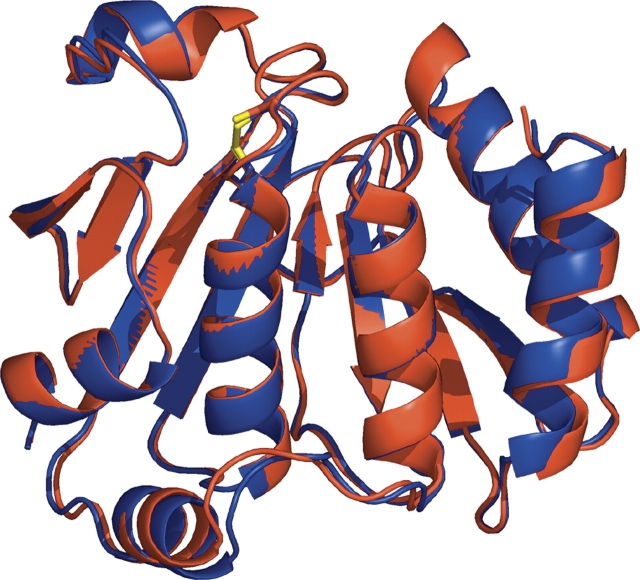
The refinements against both the LAB (2.3 Å resolution) and SYNC (2.4 Å resolution) data behaved well and led to structures with an expected accuracy for well-ordered parts of the protein of ~0.3 Å (Read 1986). Consistent with this expected coordinate accuracy, the LAB and SYNC structures are very similar to one another, with Cα root mean square deviations of 0.2 Å for chains ALAB versus ASYNC and BLAB versus BSYNC. In contrast, comparisons of NTD chains in different crystal environments, chain A versus chain B and chains A and B versus the NTD as seen in intact AhpF, differed by 0.5 Å . Overall, no significant differences are observed between the isolated rNTD and intact AhpF crystal structures (Fig. 1 ▶) so the structural descriptions of Wood et al. (2001) are relevant. For simplicity, the discussion and the figures refer to molecule A from the LAB and SYNC data sets, but the results hold for molecule B also.
Figure 1.
Comparison of rNTD with the N-terminal domain of AhpF. The backbones of rNTD (blue) and the N-terminal domain of AhpF (red) are represented as a ribbon; the bonds of the redox center (Cys129–Cys132) are depicted as sticks. The root mean square deviation for the overlay is 0.5 Å for 196 Cα atoms. The figure was generated by using Pymol (DeLano Scientific).
Oxidation state of active site cysteines in the crystal structures
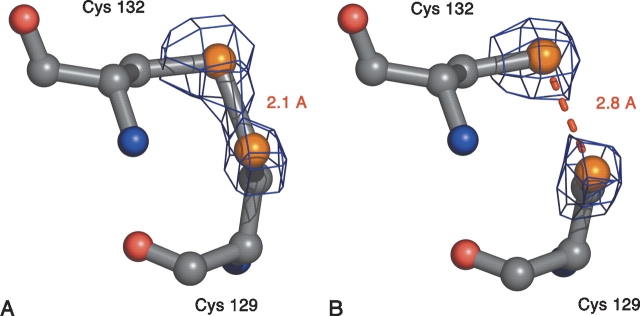
The structure determined from the LAB data set unambiguously revealed cysteines 129 and 132 to be in a disulfide bond (Fig. 2 ▶). Confirmation of the oxidized state came from two trial refinements; in the first case, cysteines 129 and 132 were mutated to alanine, and in the second case, the Sγ sulfurs of cysteines 129 and 132 were refined without van der Waals interactions. The results of both experiments were in agreement with the sulfurs being at a normal covalent disulfide bonding distance (~2.1 Å ).
Figure 2.
Reduction of the NTD redox-active disulfide center by syn-chrotron radiation. (A) The active site Cys-X-X-Cys of rNTD in its electron density based on the LAB data set, and (B) the equivalent image based on the SYNC data set. The electron density in both panels is contoured at 3.5 σ, and the Sγ–Sγ distances are given. The figure was generated by using Pymol (DeLano Scientific).
The structure deduced from the SYNC data set, again refined without van der Waals constraints on the active site sulfur atoms, resulted in an electron density distribution and a model that were not consistent with a disulfide bond. The refined Sγ–Sγ distance of 2.8 Å (Fig. 2 ▶) was very similar to that reported for intact AhpF (Wood et al. 2001).
Determination of the pKa for the active site Cys
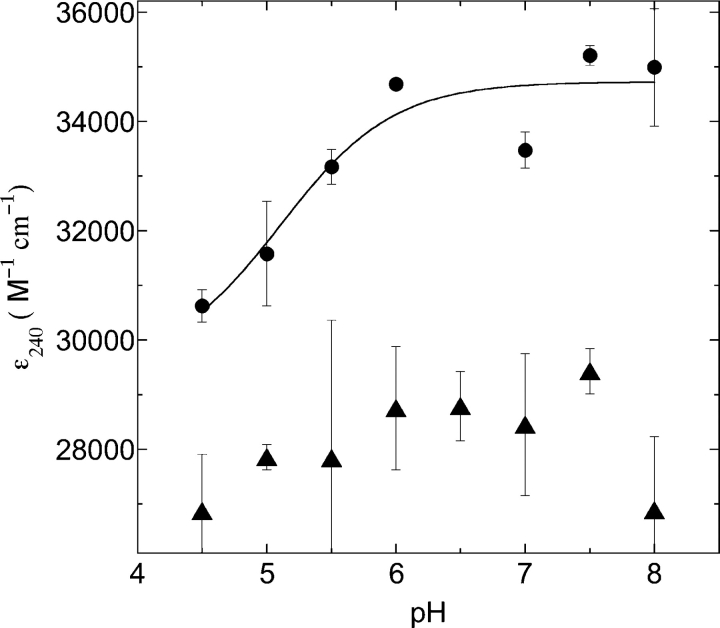
To confirm whether the short Sγ–Sγ distances observed in the synchrotron-exposed crystals of AhpF (Wood et al. 2001) and the rNTD were due to a thiol–thiolate interaction, we determined the pKa of the reduced “dithiol” form of the active site. Following the formation of the thiolate ion by absorbance at 240 nm (Benesch et al. 1955; Polgar 1974; Graminski et al. 1989; Kortemme et al. 1996), we obtained an apparent pKa of 5.1 ± 0.4 (Fig. 3 ▶). Two observations suggested that indeed the absorbance change is due to thiolate formation rather than another change in structure: First, the observation that the absorbance of oxidized (disulfide) rNTD remained constant over the pH range studied (Nelson and Creighton 1994) and, second, that the change in extinction coefficient of ~6000 M−1cm−1 (Fig. 3 ▶) matches that expected for a single thiolate (Benesch et al. 1955).
Figure 3.
Apparent pKa determination of the active site Cys129 thiol(ate). Absorbance values at 240 nm of reduced (circles) and oxidized (triangles) rNTD proteins were determined over the pH range of 4.5–8 as described in Materials and Methods. The difference in ɛ240nm between the reduced and the oxidized (average oxidized ɛ240nm 28,100 M−1cm−1) protein was ~6000M−1cm−1, which is in the range of 4000– 6000 M−1cm−1 as is expected for the titration of a single thiol group (Benesch et al. 1955). Data points were fit to the following equation:
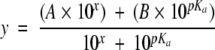 |
In this equation, x is the pH value and y the corresponding absorbance value, and A and B are allowed to vary and represent the upper and lower absorbances of the titration data, respectively. Final values from the fit using Kaleidagraph (Synergy Software) were 5.1 ± 0.4, 34,700 ± 400, and 29,500 ± 1500 for pKa, A, and B, respectively.
Discussion
Disulfide cleavage from synchrotron radiation
The two crystal structures of rNTD reveal an interesting story of radiation-generated cleavage of a disulfide bond. Cleavage of disulfide bonds has been reported to selectively occur when disulfide-containing proteins are exposed to intense radiation (Weik et al. 2000). In fact, this phenomenon of disulfide cleavage has been used to purposely generate structures of reduced tryparedoxin, when other attempts had proven unsuccessful (Alphey et al. 2003). The Sγ–Sγ bond distance increased from 2.05 Å in the oxidized state to 2.8 Å and 3.0 Å in the radiation-generated reduced state. In that case, reduction required prolonged X-ray exposures; accompanying the reduction of the disulfide bond, there were also decreases in diffraction strength, indicating that general radiation damage to the crystal had also occurred (Weik et al. 2000; Alphey et al. 2003). In contrast, the reduction of the rNTD disulfide was much more rapid and without evidence of general radiation damage. It occurred so rapidly, that a single 15-min data set behaves as if the protein was fully reduced. These observations indicate that the disulfide bond of rNTD is exquisitely sensitive to radiation cleavage and will be a useful model system for studying this process. Although we cannot recreate the radiation doses used for the structure solution of AhpF, this disulfide cleavage also occurred during the collection of a single data set, consistent with it being similarly sensitive (Wood et al. 2001).
The oxidized structure of the NTD
The structure solved from the rNTD LAB data set represents the first crystal structure of the oxidized form of this unusual domain. The AhpF crystals analyzed by Wood et al. (2001) were not large enough to be analyzed by using a laboratory source, but it was our good fortune that the size and order of the rNTD crystals did support such an analysis. Compared with the reduced form (as described by Wood et al. 2001 and as seen here in the SYNC structure), the oxidized rNTD redox center shows only minor structural changes confined to the immediate vicinity of the disulfide bond. The only consistent and significant movement is the small rotation of the Cys129-χ torsion angle needed to form the disulfide. In one of the two monomers (chain A), there is also a shift of a His130-χ to a different rotamer, but this side chain is very disordered already. The small differences between the reduced and the oxidized rNTD structures are consistent with what has been seen for thioredoxin (Weichsel et al. 1996).
The oxidized rNTD is the redox form that is attacked by one of the Cys residues in the thioredoxin reductase-like portion of AhpF, which initiates the thiol-disulfide interchange reaction, resulting in NTD reduction (Li Calzi and Poole 1997; Wood et al. 2001). Two further structures needed to complete a structural view of the catalytic cycle of the NTD are the mixed disulfides it forms when it is being reduced by the thioredoxin reductase-like part of AhpF and when it is passing reducing equivalents to AhpC. We have been able to covalently trap both of these mixed disulfide complexes, one via the generation of a C132S/C348S double mutant of AhpF (Jönsson and Poole 2002) and the second with two single mutants, C132S AhpF and C46S of AhpC (Poole 1999). We are working to crystallize these constructs.
The NTD pKa and thiol–thiolate hydrogen bonding
As seen in the full-length AhpF structure (Wood et al. 2001), the two cysteinyl residues (Cys129, Cys132) are involved in a close, ~3 Å noncovalent interaction. Wood et al. (2001) presented compelling arguments that this is a thiol–thiolate interaction, and now the measurement of a pKa of 5.1 ± 0.4 for one of the active-site Cys residues confirms that one Cys would be expected to be mostly deprotonated (i.e., in a thiolate form) both in the rNTD crystals studied here, which are at pH 8.4, and in the original AhpF crystals, which were at pH 5.6. Although we have no direct evidence as to which Cys is being titrated, the geometry of the active site (see discussion in Wood et al. 2001) and analogy with thioredoxin and related proteins containing the Cys-X-X-Cys motif both lead to the expectation that the low pKa belongs to the Cys129, the more N-terminal and exposed Cys that attacks disulfide substrates (Jönsson and Poole 2002).
This low pKa is not unprecedented for thioredoxin-fold proteins, as the pKa of E. coli DsbA is as low as ~3.2 (Nelson and Creighton 1994; Grauschopf et al. 1995), but it is lower than the pKas of other Prx-reducing domains/proteins. Other reductants of Prxs such as E. coli Trx and Trypanosoma brucei tryparedoxin (Txn) have reported pKas ~7.0 (Kallis and Holmgren 1980; Jeng et al. 1995; Dyson et al. 1997; Reckenfelderbaumer and Krauth-Siegel 2002). Thus, for all of these Trx-like - Cys-X-X-Cys- containing proteins, the pKa of the more N-terminal Cys thiol is shifted significantly below the value of ~8.3 for an unperturbed Cys thiol in solution, but the specific values cover a wide range between 3.2 and 7.2. While the thiol–thiolate hydrogen bond exerts an important influence on this residue’s pKa (see Jeng et al. 1995; Mössner et al. 2000), most or all proteins in this group are thought to have such an interaction (Weichsel et al. 1996; Guddat et al. 1998; Nordstrand et al. 1999), so this cannot be the distinctive feature. Additional contributing factors documented in the literature are structure-related effects, including hydrogen bonding and electrostatic interactions (Mössner et al. 1998, 2000; Foloppe and Nilsson 2004). For Trx-fold proteins, numerous studies have used mutations to study the role of the residues present between the two Cys residues in the -Cys-X-X-Cys- motif (Krause et al. 1991; Grauschopf et al. 1995; Kortemme et al. 1996; Chivers et al. 1997; Guddat et al. 1997; Huber-Wunderlich and Glockshuber 1998; Mössner et al. 1998). An important insight synthesizing many observations was that the pKa of the N-terminal Cys appears largely determined by the number of hydrogen bonds that the -X-X-Cys residues provide to stabilize the thiolate (Foloppe et al. 2001; Foloppe and Nilsson 2004), with proteins having four, three, and two hydrogen bonds to the thiolate showing pKas of ~3 (e.g., DsbA), ~5 (e.g., Grx3), and ~7 (e.g., Trx). The NTD fits this generalization as it has three hydrogen bonds (from the Cys132 thiol, and the backbone amides of residues 131 and 132). In the NTD, the nearest positively charged side chains, His130 and Arg185, cannot approach Cys129 to form a hydrogen bond without rearrangements in backbone structure. As has been done for other Trx-like proteins (Grauschopf et al. 1995; Kortemme et al. 1996; Huber-Wunderlich and Glockshuber 1998; Mössner et al. 1998), this explanation for modulation of the pKa could be tested by mutation of position 131 to His to add a fourth hydrogen bond and match the DsbA sequence or to Pro to remove a hydrogen bond and match the Trx sequence. Still, additional factors such as protein dynamics (Foloppe and Nilsson 2004) and global electrostatics (Gane et al. 1995) also contribute to the active site properties, as some Trx family members with common dipeptide sequences but from different organisms exhibit considerably different pKa values (see Nelson and Creighton 1994; Ruddock et al. 1996).
Finally, a surprising result is that the pKa of ~5.1 for the NTD does not fit with the free energy correlation between the N-terminal Cys pKa values and the redox potentials that has been demonstrated empirically for a number of Trx family members (Szajewski and Whitesides 1980; Grauschopf et al. 1995; Huber-Wunderlich and Glockshuber 1998; Mössner et al. 2000). Trx and Txn exhibit quite low redox potentials at pH 7.0 (Eo′) of −270 and −249 mV, consistent with their highly reducing nature and relatively high pKas of ~7 (Mössner et al. 2000; Reckenfelderbaumer and Krauth-Siegel 2002). The redox potential Eo ′ of ~264 ± 8 mV (Reynolds and Poole 2000) and function of the NTD are quite similar to these other redox donors, but the pKa for Cys129 in the NTD, at ~5.1, is not. In this regard, characterization of the mutants of the -Cys-X-X-Cys- motif suggested above will be a useful first step to probe this unusual property of the NTD and will reveal whether a group of NTD mutants will have a pKa-to-redox potential relationship that is shifted but with the same slope as seen for other Trx proteins, or whether it has a different slope. Either way, because of its unusual properties, further work on the NTD promises to yield new insights into the factors governing the relationship between the pKas and redox potentials of the Trx family proteins.
Materials and methods
Purification of rNTD
The separately expressed rNTD of AhpF, also known as F[1–202], was purified as previously described (Poole et al. 2000a) from a 10 L fermentor growth by sequential chromatography on Q Sepharose HP and Superose 12 columns using an Äkta Explorer 10S FPLC instrument (Amersham Biosciences). For crystallization, rNTD was stored at 22.7 mg/mL in 25 mM potassium phosphate buffer (pH 7.0), containing 1 mM EDTA. rNTD concentration was assessed using an ɛ279= 15,100 M−1 cm−1 (Poole et al. 2000a).
Thiol quantification
Thiols were quantified via reaction with DTNB (5,5′-dithio-bis-( 2-nitrobenzoic acid)) (Ellman’s reagent) as described previously (Poole 1996). Briefly, the free thiol groups within the oxidized and dithiothreitol(DTT)-reduced rNTD protein were measured at pH 7.0 by the addition of 150 μM DTNB in the presence or the absence of 4M guanidine hydrochloride (denaturing or nondenaturing conditions, respectively) followed by detection of the TNB produced at 412 nm (ɛ142=14,150 M−1 cm−1) (Riddles et al. 1979) on a thermostated Milton Roy Spectronic 3000 diode array spectrophotometer.
Crystallization
Rod-like crystals (~l × 0.2 × 0.2 mm3) were grown at room temperature in ~3 wk using a reservoir of 0.2 M ammonium acetate, 0.1 M Tris (pH 8.4), 30%–35% polyethylene glycol 4000, and a drop made from 2 μL of rNTD storage solution mixed with 2 μL of the reservoir solution. Most crystals had splayed ends, but a few were single. For data collection, crystals were scooped into cryoloops through a mineral oil monolayer and immediately flash-frozen by plunging into liquid nitrogen.
Data collection and refinement
Two data sets were collected at −170°C from a single rNTD crystal. Various parts of the crystal had variable diffraction quality, and for each data set, we attempted to find an optimal volume of the crystal through trial exposures. The first data set (LAB) was collected on an R-axis IV image plate detector (Molecular Structure Corp.) with a Rigaku RUH3R rotating anode (CuKα) operating at 50 kV and 100 mA with a 0.3-mm collimator. The crystal was recovered and stored in liquid nitrogen until 8 wk later when the second data set (SYNC) was collected at beamline 5.0.3 of the Advanced Light Source (Lawrence Berkeley National Laboratory) in two passes. The first pass included 50° of data (15 sec per 1° frame; λ =1.01 Å ), and to avoid saturation of the lower resolution reflections, a second 45° of data were collected more rapidly (3 sec per 1° frame). X-ray data were processed using programs DENZO and SCALEPACK (Otwinowski and Minor 1997), and the LAB and SYNC data each have reasonable statistics, extending to 2.3 Å and 2.4 Å , respectively (Table 1). We speculate that the slightly poorer resolution limit of the synchrotron data was limited by the intrinsic order of the volume of the crystal used. The crystals belong to the tetragonal space group P41212 with two molecules in the asymmetric unit and 56% solvent. A random 10% of the data were selected for cross validation.
Table 1.
Data collection and refinement statistics
| LAB | SYNC | |
| Resolution limits (Å) | 50–2.3 (2.38–2.3) | 50–2.4 (2.49–2.4) |
| Space group | P41212 | P41212 |
| Unit cell (Å) | a = 85.41 | a = 85.27 |
| b = c = 108.60 | b = c = 107.94 | |
| Data collection | ||
| Unique observations | 22,423 | 19,094 |
| Multiplicity | 6.5 (6.5) | 5.7 (3.2) |
| Average I/σ | 16.2 (2.7) | 12.2 (1.6) |
| Rmeas (%) | 7.2 (46.1) | 9.2 (51.9) |
| Rmergd-F (%) | 7.7 (35.3) | 9.6 (53.7) |
| Completeness (%) | 99.7 (99.5) | 98.7 (97.5) |
| Refinement | ||
| No. of protein molecules | 2 | 2 |
| No. of amino acid residues | 392 | 392 |
| Average B-factor (Å2) | 45 | 44 |
| Non-hydrogen protein atoms | 3042 | 3042 |
| R-factor | 0.199 (0.262) | 0.206 (0.298) |
| Free R-factor (10% of data) | 0.256 (0.325) | 0.261 (0.343) |
| RMSD bond lengths (Å) | 0.010 | 0.006 |
| RMSD bond angles (°) | 1.41 | 1.20 |
Numbers in parentheses correspond to values in the highest resolution shell. RMSD, rootmean square deviation.
The structure of NTD was solved by molecular replacement using the CNS suite of programs (v1.1) (Brunger et al. 1998) and a search model based on the N-terminal domain (residues 1–196) from the structure of AhpF (Protein Data Bank code 1HYU; Wood et al. 2001). Two molecules were unambiguously placed using data from 50.0 to 4.0 Å resolution, and rigid body refinement led to R and Rfree values of 0.330 and 0.329, respectively. Positional and B-factor refinement from 50.0 to 2.3 Å resolution rapidly dropped the R-factors to R=0.267 and Rfree=0.299. Manual rebuilding of the model into 2FO–FC and FO–FC electron density maps was done using the program O (Jones et al. 1991). Only small modifications of side chains were necessary. Waters were added both manually and using the CNS utility Water-Pick with the following criteria: (1) a minimum 3 σ peak in 2FO–FC maps and (2) a minimum distance of 2.6 Å and a maximum distance of 3.5 Å to potential hydrogen-bond donor or acceptor. To allow for unbiased determination of the Cys129 and Cys132 positions, the van der Waals interactions for the sulfurs were turned off by use of an “IGROUP” statement in the minimization input file for CNS. In a separate control refinement, Cys129 and Cys132 were mutated to alanine to remove model bias. Refinements were considered complete when the largest difference map peaks were not interpretable and R and Rfree had converged. Refinement using the SYNC data set was done starting with the final model from the LAB data set, including waters. After one round of positional and B-factor refinement from 50.0 to 2.4 Å , R and Rfree were 0.221 and 0.267, respectively. Two additional rounds of manual rebuilding and positional and B-factor refinement completed the refinement. Final statistics of both models are reported in Table 1. The coordinates of the new structures have been deposited with the RCSB Protein Data Bank (http://www.rcsb.org/pdb) with PDB codes 1ZYN (LAB data set, oxidized NTD) and 1ZYP (SYNC data set, reduced NTD).
Cysteine pKa determination
The thiolate anion was directly monitored at 240 nm (using a Shimadzu UV-2401PC UV-Vis spectrophotometer) as previously described by Kortemme et al. (1996). All measurements were carried out at 25°C in buffer A: 1 mM each sodium citrate (pKa=3.13, 4.76, and 6.40), sodium borate (pKa=9.24), and sodium phosphate (pKa=2.15, 7.20, and 12.38), with 200 mM NaCl. Reduced NTD was prepared by incubation with 0.1 M Tris-HCl and 5 mM DTT (pH 7.4) for 15 min at room temperature. After incubation, the DTT was removed by a 1000-fold dilution into buffer-A at the appropriate pH by three successive 10-fold dilutions, and concentration using Centricon concentrators (Millipore, 5 kDa cutoff); buffer without protein was used to control for residual DTT, which had an absorbance of <0.001 at the wavelengths measured. Protein concentrations were between 10 and 15 μM. As a control, the absorbance at 240 nm for the NTD in the disulfide form was measured over the same pH range. All measurements were carried out in triplicate.
Acknowledgments
This publication was made possible in part by the Environmental Health Sciences Center’s Molecular Structure and Interactions Facilities core supported by grant number P30 ES00210 from the National Institute of Environmental Health Sciences, NIH. We thank Rick Faber and Ganapathy Sarma for all their help and support. This research was supported by a grant (RO1 GM50389) from the NIH to L.B.P. and P.A.K., and grant ES00040 from the National Institute of Environmental Health Sciences to Joe S. Beckman.
Abbreviations
AhpF, alkyl hydroperoxide reductase flavoprotein or NADH:peroxiredoxin oxidoreductase
NTD, N-terminal domain of AhpF
rNTD, recombinant NTD
DTT, 1,4-dithiothreitol
DTNB, 5,5′-dithio-bis-(2-nitrobenzoic acid)
TNB, 2-nitro-5-thiobenzoate.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051459705.
References
- Alphey, M.S., Gabrielsen, M., Micossi, E., Leonard, G.A., McSweeney, S.M., Ravelli, R.B., Tetaud, E., Fairlamb, A.H., Bond, C.S., and Hunter, W.N. 2003. Tryparedoxins from Crithidia fasciculata and Trypanosoma brucei: Photoreduction of the redox disulfide using synchrotron radiation and evidence for a conformational switch implicated in function. J. Biol. Chem. 278 25919–25925. [DOI] [PubMed] [Google Scholar]
- Benesch, R.E., Lardy, H.A., and Benesch, R. 1955. The sulfhydryl groups of crystalline proteins. I. Some albumins, enzymes, and hemoglobins. J. Biol. Chem. 216 663–676. [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J.,Nilges, M., Pannu,N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt. 5): 905–921. [DOI] [PubMed] [Google Scholar]
- Chen, L., Xie, Q.W., and Nathan, C. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1 795–805. [DOI] [PubMed] [Google Scholar]
- Chivers, P.T., Prehoda, K.E., and Raines, R.T. 1997. The CXXC motif: A rheostat in the active site. Biochemistry 36 4061–4066. [DOI] [PubMed] [Google Scholar]
- Dyson, H.J., Jeng, M.F., Tennant, L.L., Slaby, I., Lindell, M., Cui, D.S., Kuprin, S., and Holmgren, A. 1997. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: Structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry 36 2622–2636. [DOI] [PubMed] [Google Scholar]
- Foloppe, N. and Nilsson, L. 2004. The glutaredoxin -C-P-Y-C- motif: Influence of peripheral residues. Structure (Camb) 12 289–300. [DOI] [PubMed] [Google Scholar]
- Foloppe, N., Sagemark, J., Nordstrand, K., Berndt, K.D., and Nilsson, L. 2001. Structure, dynamics and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: Comparison with functionally related proteins. J. Mol. Biol. 310 449–470. [DOI] [PubMed] [Google Scholar]
- Gane, P.J., Freedman, R.B., and Warwicker, J. 1995. A molecular model for the redox potential difference between thioredoxin and DsbA, based on electrostatics calculations. J. Mol. Biol. 249 376–387. [DOI] [PubMed] [Google Scholar]
- Graminski, G.F., Kubo, Y., and Armstrong, R.N. 1989. Spectroscopic and kinetic evidence for the thiolate anion of glutathione at the active site of glutathione S-transferase. Biochemistry 28 3562–3568. [DOI] [PubMed] [Google Scholar]
- Grauschopf, U., Winther, J.R., Korber, P., Zander, T.P., Dallinger, P., and Bardwell, J.C.A. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83 947–955. [DOI] [PubMed] [Google Scholar]
- Guddat, L.W., Bardwell, J.C., Glockshuber, R., Huber-Wunderlich, M., Zander, T., and Martin, J.L. 1997. Structural analysis of three His32 mutants of DsbA: Support for an electrostatic role of His32 in DsbA stability. Protein Sci. 6 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guddat, L.W., Bardwell, J.C., and Martin, J.L. 1998. Crystal structures of reduced and oxidized DsbA: Investigation of domain motion and thiolate stabilization. Structure 6 757–767. [DOI] [PubMed] [Google Scholar]
- Huber-Wunderlich, M. and Glockshuber, R. 1998. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold. Des. 3 161–171. [DOI] [PubMed] [Google Scholar]
- Jacobson, F.S., Morgan, R.W., Christman, M.F., and Ames, B.N. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: Purification and properties. J. Biol. Chem. 264 1488–1496. [PubMed] [Google Scholar]
- Jeng, M.F., Holmgren, A., and Dyson, H.J. 1995. Proton sharing between cysteine thiols in Escherichia coli thioredoxin: Implications for the mechanism of protein disulfide reduction. Biochemistry 34 10101–10105. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 (Pt. 2) 110–119. [DOI] [PubMed] [Google Scholar]
- Jönsson, T. and Poole, L.B. 2002. Intramolecular electron transfer pathway in mutants of AhpF, a flavoprotein disulfide reductase. In Flavins and flavoproteins 2002 (eds. S. Chapman et al.), pp. 691–696. Agency for Scientific Publications, Berlin.
- Kallis, G.B. and Holmgren, A. 1980. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J. Biol. Chem. 255 10261–10265. [PubMed] [Google Scholar]
- Kortemme, T., Darby, N.J., and Creighton, T.E. 1996. Electrostatic inter-actions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry 35 14503–14511. [DOI] [PubMed] [Google Scholar]
- Krause, G., Lundstrom, J., Barea, J.L., Pueyo de la Cuesta, C., and Holmgren, A. 1991. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J. Biol. Chem. 266 9494–9500. [PubMed] [Google Scholar]
- Li Calzi, M. and Poole, L.B. 1997. Requirement for the two AhpF cystine disulfide centers in catalysis of peroxide reduction by alkyl hydroperoxide reductase. Biochemistry 36 13357–13364. [DOI] [PubMed] [Google Scholar]
- Mössner, E., Huber-Wunderlich, M., and Glockshuber, R. 1998. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol-disulfide oxidoreductases. Protein Sci. 7 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mössner, E., Iwai, H., and Glockshuber, R. 2000. Influence of the pKa value of the buried, active-site cysteine on the redox properties of thioredoxin-like oxidoreductases. FEBS Lett. 477 21–26. [DOI] [PubMed] [Google Scholar]
- Nelson, J.W. and Creighton, T.E. 1994. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33 5974–5983. [DOI] [PubMed] [Google Scholar]
- Niimura, Y., Poole, L.B., and Massey, V. 1995. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 270 25645–25650. [DOI] [PubMed] [Google Scholar]
- Nordstrand, K., Aslund, F., Meunier, S., Holmgren, A., Otting, G., and Berndt, K.D. 1999. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 449 196–200. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Polgar, L. 1974. Mercaptide-imidazolium ion-pair: The reactive nucleophile in papain catalysis. FEBS Lett. 47 15–18. [DOI] [PubMed] [Google Scholar]
- Poole, L.B. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 2. Cystine disulfides involved in catalysis of peroxide reduction. Biochemistry 35 65–75. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Flavin-linked redox components required for AhpC reduction in alkyl hydroperoxide reductase systems. In Flavins and flavoproteins (eds. S. Ghisla et al.), pp. 691–694. Agency for Scientific Publications, Berlin.
- ———. 2005. Bacterial defenses against oxidants: Mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433 240–254. [DOI] [PubMed] [Google Scholar]
- Poole, L.B. and Ellis, H.R. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35 56–64. [DOI] [PubMed] [Google Scholar]
- Poole, L.B., Godzik, A., Nayeem, A., and Schmitt, J.D. 2000a. AhpF can be dissected into two functional units: Tandem repeats of two thioredoxinlike folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39 6602–6615. [DOI] [PubMed] [Google Scholar]
- Poole, L.B., Reynolds, C.M., Wood, Z.A., Karplus, P.A., Ellis, H.R., and Li Calzi, M. 2000b. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267 6126–6133. [DOI] [PubMed] [Google Scholar]
- Read, R.J. 1986. Improbed Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42 140–149. [Google Scholar]
- Reckenfelderbaumer, N. and Krauth-Siegel, R.L. 2002. Catalytic properties, thiol pK value, and redox potential of Trypanosoma brucei tryparedoxin. J. Biol. Chem. 277 17548–17555. [DOI] [PubMed] [Google Scholar]
- Reynolds, C.M. and Poole, L.B. 2000. Attachment of the N-terminal domain of Salmonella typhimurium AhpF to Escherichia coli thioredoxin reductase confers AhpC reductase activity but does not affect thioredoxin reductase activity. Biochemistry 39 8859–8869. [DOI] [PubMed] [Google Scholar]
- Riddles, P.W., Blakeley, R.L., and Zerner, B. 1979. Ellman’s reagent: 5,5′-dithiobis(2-nitrobenzoic acid):A reexamination. Anal. Biochem. 94 75–81. [DOI] [PubMed] [Google Scholar]
- Ruddock, L.W., Hirst, T.R., and Freedman, R.B. 1996. pH-dependence of the dithiol-oxidizing activity of DsbA (a periplasmic protein thiol:disulphide oxidoreductase) and protein disulphide-isomerase: Studies with a novel simple peptide substrate. Biochem. J. 315 (Pt. 3): 1001– 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewski, R.P. and Whitesides, G.M. 1980. Rate constants and equilibrium constants for thiol-disulfide interchange reactions involving oxidized glutathione. J. Am. Chem. Soc. 102 2011–2026. [Google Scholar]
- Weichsel, A., Gasdaska, J.R., Powis, G., and Montfort, W.R. 1996. Crystal structures of reduced, oxidized, and mutated human thioredoxins: Evidence for a regulatory homodimer. Structure 4 735–751. [DOI] [PubMed] [Google Scholar]
- Weik, M., Ravelli, R.B., Kryger, G., McSweeney, S., Raves, M.L., Harel, M., Gros, P., Silman, I., Kroon, J., and Sussman, J.L. 2000. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. 97 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, Z.A., Poole, L.B., and Karplus, P.A. 2001. Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis. Biochemistry 40 3900–3911. [DOI] [PubMed] [Google Scholar]
- Wood, Z.A., Poole, L.B., Hantgan, R.R., and Karplus, P.A. 2002. Dimers to doughnuts: Redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry 41 5493–5504. [DOI] [PubMed] [Google Scholar]
- Wood, Z.A., Poole, L.B., and Karplus, P.A. 2003. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300 650–653. [DOI] [PubMed] [Google Scholar]