Abstract
Previous reports detailing mutational effects within the hydrophobic core of human acidic fibroblast growth factor (FGF-1) have shown that a symmetric primary structure constraint is compatible with a stably folded protein. In the present report, we investigate symmetrically related pairs of buried hydrophobic residues in FGF-1 (termed “mini-cores”) that are not part of the central core. The effect upon the stability and function of FGF-1 mutations designed to increase primary structure symmetry within these “mini-core” regions was evaluated. At symmetry-related positions 22, 64, and 108, the wild-type protein contains either Tyr or Phe side chains. The results show that either residue can be readily accommodated at these positions. At symmetry-related positions 42, 83, and 130, the wild-type protein contains either Cys or Ile side chains. While positions 42 and 130 can readily accommodate either Cys or Ile side chains, position 83 is substantially destabilized by substitution by Ile. Tertiary structure asymmetry in the vicinity of position 83 appears responsible for the inability to accommodate an Ile side chain at this position, and is known to contribute to functional half-life. A mutant form of FGF-1 with enforced primary structure symmetry at positions 22, 64, and 108 (all Tyr) and 42, 83, and 130 (all Cys) is shown to be more stable than the reference FGF-1 protein. The results support the hypothesis that a symmetric primary structure within a symmetric protein superfold represents a solution to achieving a foldable, stable polypeptide, and highlight the role that function may play in the evolution of asymmetry within symmetric superfolds.
Keywords: fibroblast growth factor, β-trefoil, mini-core, protein engineering, de novo design, folding kinetics, protein evolution
Human acidic fibroblast growth factor (FGF-1) is a member of the fibroblast growth factor family that comprises at least 23 structurally related polypeptides (Strewler 2001). The FGFs are potent modulators of cell proliferation, motility, differentiation, and survival. FGF-1 triggers mitogenic activity by binding and activating dimerized tyrosine kinase receptors (FGFR1– FGFR4) at the cell surface (Pellegrini et al. 2000; Plotnikov et al. 2000). As a member of the β-trefoil superfold (Murzin et al. 1992), FGF-1 exhibits a characteristic pseudo-threefold symmetry when viewed down the β-barrel axis (Fig. 1 ▶). The repeating structural unit of this threefold symmetry comprises of a pair of anti-parallel β-sheets, referred to as a “β-trefoil fold.” The tertiary structure can be described as a six-stranded β-barrel closed off at one end by a β-hairpin triplet. The β-trefoil structure is hypothesized to have evolved by successive gene duplication and fusion events, and the β-trefoil has been identified as a monomeric structural element in epidermal growth factor, as a dimeric element in the structure of the protease inhibitor ecotine and as a trimeric arrangement in the β-trefoil superfold (Mukhopadhyay 2000; Ponting and Russell 2000).
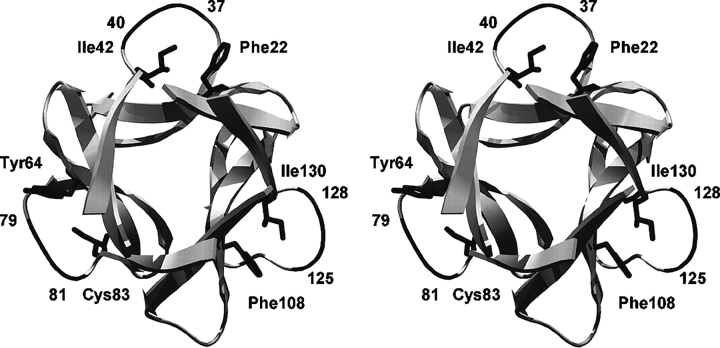
Figure 1.
Ribbon drawing of the FGF-1 structure (PDB accession no. 1JQZ) oriented so as to view down the threefold axis of symmetry intrinsic to the β-trefoil superfold. Also indicated in wireframe representation are the symmetry-related pairs of residue positions that comprise the “mini-core” regions in the structure Phe22/Ile42, Tyr64/ Cys83, and Phe108/Ile130 (the view has been sectioned in the Z-axis so as to make these residues more visible). Also indicated in dark shading are the adjacent main-chain regions that pack against residues Ile42, Cys83, and Ile130.
The threefold tertiary structure symmetry present in FGF-1 is largely absent in the primary structure (Fig. 2 ▶). This raises intriguing questions regarding the relationship of tertiary and primary structure symmetry to protein stability, function, folding, evolution, and de novo design (Richardson and Richardson 1989; Wolynes 1996; Lang et al. 2000; Heidary and Jennings 2002; Brych et al. 2004). Prior studies from our laboratory have shown that it is possible to successfully redesign the central core packing group of FGF-1 using a symmetric primary structure constraint (Brych et al. 2001, 2003, 2004). Optimal symmetric packing interactions within the core region of FGF-1 were achieved by including several loop deletion mutations designed to increase the tertiary structure symmetry. This highly symmetric form of FGF-1, referred to as “SYM6ΔΔ” (Table 1) was shown to be substantially more stable than the wild-type protein (Brych et al. 2004), albeit at the expense of certain functionality. These results led to the hypothesis that a primary structure symmetric constraint, within a symmetric protein superfold, is a solution to, rather than a constraint upon the stability and foldability of the polypeptide (Brych et al. 2004) and supports the “stability/function tradeoff” hypothesis of protein evolution (Schreiber et al. 1994; Shoichet et al. 1995; Wang et al. 2002; Bloom et al. 2004).
Figure 2.
Primary structure of human FGF-1 aligned to show the relationship between the three trefoil fold subdomains. The shaded regions indicate the residue positions that comprise the three “mini-core” regions in the structure. Beneath each of these positions is listed the frequency of occurrence of amino acids for the 23-member FGF family of proteins. Residue positions 79 and 80 are in parentheses to indicate that they do not align structurally with positions 37–39 or 125–127.
Table 1.
Nomenclature of mutant forms of FGF-1
| Acronym | Mutations |
| WT* | Wild-type (His tagged) |
| SYM6Δ Δa | Leu44→Phe/Met67→I1e/Leu73→Val/Ala103→Gly/Δ104–106/Val109→Leu/Leu111→I1e/Cys117→Val/ Arg119→Gly/Δ120–122 |
| SYM7Δ Δ | SYM6Δ Δ/Phe22→Tyr/Phe108→Tyr |
| SYM8Δ Δ | SYM7Δ Δ/Ile42→Cys/Ile130→Cys |
In addition to the central core region, FGF-1 contains an outer “mini-core” within each of the three β-trefoil folds. Each “mini-core” comprises a pair of buried residues (Phe22/Ile42, Tyr64/Cys83, and Phe108/Ile130) related by the threefold symmetry of the tertiary structure (Fig. 1 ▶). The Phe/Ile pairs are symmetric at positions 22/42 and 108/130, however, the 64/83 pair is a uniquely different Tyr/Cys pair (Fig. 2 ▶). In a continuation of our efforts toward understanding the consequences of introducing primary structure symmetry within a symmetric superfold, we have studied effects of mutations within the “mini-core” regions of FGF-1. Positions 22, 64, and 108 are observed to tolerate either Phe or Tyr mutations, although a symmetric Tyr mutant form of FGF-1 is observed to be moderately more stable than the reference protein. Positions 42 and 130 are similarly observed to tolerate either Ile or Cys mutations; however, position 83 is unable to accommodate an Ilemutation without substantial destabilization. A symmetric combination mutation, involving Tyr/Cys pairs at each “mini-core” region is observed to be more stable than the asymmetric parental sequence. The inability of position 83 to accommodate an Ile side chain is found to be due to asymmetry within the tertiary structure forming the packing environment. Thus, the ability of position 83 to accommodate an Ile residue is postulated to require further mutations to improve the tertiary structure symmetry. The results support the hypothesis that a primary structure symmetric constraint, within a symmetric protein superfold, is a solution to, rather than a constraint upon the stability and foldability of the polypeptide, and highlight the role that functionality may play in the observed asymmetry of proteins belonging to a symmetric superfold.
Results
Mutant protein purification
All mutant proteins, except those containing the Cys83→Ile mutation, were purified with high yield (~50 mg/L). Mutants containing the Cys83→Ile mutation were observed to be expressed in the form of inclusion bodies. For these mutants, only the soluble fraction was used for purification of the mutant protein. Thus, a typical yield for the purification of a mutant containing the Cys83→Ile mutation was ~8–10 mg/L.
Isothermal equilibrium denaturation
The derived thermodynamic parameters for the mutant proteins, in comparison to wild type and SYM6ΔΔ are summarized in Table 2. The thermodynamic stabilities for wild type and SYM6ΔΔ are somewhat higher than previously reported values determined in 20 mM N- (2-acetamido)iminodiacetic acid (ADA), 100 mM NaCl, 2 mM DTT (pH 6.60) (ADA buffer) (Brych et al. 2004; Kim et al. 2004). The buffering system used in the present study contains phosphate and sulfate ions, which permit the higher protein concentrations necessary for CD studies. These ions are known to interact with FGF-1 within the heparin-binding site (Blaber et al. 1996; Romero et al. 1996) and have also been shown thermodynamically to have a stabilizing effect (Mach and Middaugh 1994; Adamek et al. 1998).
Table 2.
Thermodynamic parameters for FGF-1 mutants as determined by isothermal equilibrium denaturation using GuHC1 and monitored using CD spectroscopy
| Mutant | ΔG (kJ/mol) | μ-value (kJ/mol M) | Cm (M) | aΔΔG (kJ/mol) |
| SYM6ΔΔ | 37.7 ± 2.8 | 19.1 ± 1.4 | 1.97 ± 0.01 | _ |
| Group A: | ||||
| I42→C | 35.2 ± 1.3 | 18.0 ± 0.6 | 1.95 ± 0.01 | 0.7 |
| C83→I | 30.3 ± 2.1 | 20.5 ± 1.4 | 1.47 ± 0.07 | 9.9 |
| I130→C | 35.2 ± 1.2 | 17.4 ± 0.6 | 2.02 ± 0.01 | −0.9 |
| F22→Y | 39.1 ± 1.6 | 19.5 ± 0.8 | 2.00 ± 0.01 | −0.6 |
| Y64→F | 32.6 ± 2.6 | 16.8 ± 1.4 | 1.94 ± 0.01 | 0.5 |
| F108→Y | 40.5 ± 1.1 | 18.9 ± 0.5 | 2.14 ± 0.01 | −3.2 |
| Group B: | ||||
| F22→Y/F108→Y (SYM7ΔΔ) | 41.2 ± 1.8 | 19.2 ± 0.8 | 2.14 ± 0.01 | −3.3 |
| F22→Y/I42→C/F108→Y/I130→C (SYM8ΔΔ) | 41.1 ± 0.9 | 19.0 ± 0.5 | 2.16 ± 0.01 | −3.7 |
| Group C: | ||||
| I42→V | 31.1 ± 1.7 | 18.7 ± 1.0 | 1.66 ± 0.01 | 5.8 |
| C83→V | 31.2 ± 1.1 | 21.1 ± 0.8 | 1.47 ± 0.01 | 10.1 |
| I130→V | 32.3 ± 1.0 | 19.0 ± 0.6 | 1.70 ± 0.01 | 5.1 |
| Y55→L/C83→I | 32.3 ± 2.4 | 25.7 ± 1.9 | 1.25 ± 0.01 | 16.1 |
| P79→G/C83→I | 21.0 ± 1.3 | 18.7 ± 1.1 | 1.12 ± 0.01 | 16.1 |
| T78[G]/P79→G/C83→I | 20.2 ± 1.7 | 18.3 ± 1.5 | 1.10 ± 0.01 | 16.3 |
| WTb | 26.6 ± 0.9 | 20.3 ± 0.7 | 1.29 ± 0.01 | 13.4 |
All mutants were constructed in the SYM6 Δ Δ FGF-1 background protein (Table 1).
a Δ ΔG = (CmREF − Cmmutant )(mREF + mmutant )/2 as described by Pace and Scholtz (1997), and where the reference protein is SYM6Δ Δ. A negative value of Δ ΔG indicates a more stable mutation, and error is stated as the standard error from multiple data sets.
b His-tagged wild-type FGF-1 in the same buffer conditions is included for reference.
The mutations within “GroupA” of Table 2 are intended to evaluate the effects of sequence-swapping point mutations within the sets of symmetry-related positions at (42, 83, 130) and (22, 64, 108). The wild-type Ile side chains at symmetry-related positions 42 and 130 can be substituted by Cys with little detectable effect upon stability (i.e., |ΔΔG| values<1.0 kJ/mol). In contrast, the Cys at position 83 exhibits a dramatic destabilization of 9.9 kJ/mol when substituted by Ile. Tyr substitutions at symmetry-related positions 22 and 108 are accommodated with a slight increase in stability (position 22), or with a 3.2 kJ/mol increase in stability (position 108). A Phe substitution at position 64 exhibits essentially no effect upon stability.
The mutations within “Group B” of Table 2 are designed to probe the effects of combining appropriate point mutations so as to constrain the primary structure at symmetry-related positions (42, 83, 130) and (22, 64, 108) to the threefold symmetry inherent in the tertiary structure. The double mutant Phe22→Tyr/Phe108→Tyr (“SYM7ΔΔ” mutant), constraining symmetry-related positions 22, 64, and 108 to Tyr, is accommodated with a 3.3 kJ/mol increase in stability (essentially, the additive contribution of the two Tyr point mutations). With this as a background form of FGF-1, the double-mutant Ile42→Cys/Ile130→Cys, constraining symmetry-related positions (42, 83 130) to Cys, is accommodated with a 0.4 kJ/mol increase in stability (essentially, the additive contribution of the two Cys point mutations).
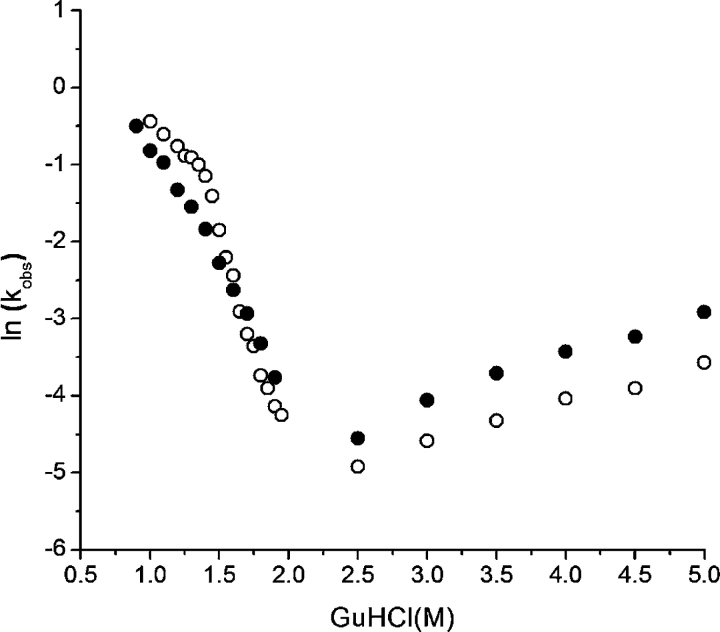
The mutations within “Group C” of Table 2 are an attempt to elucidate the basis for the inability of position 83 to accommodate an Ile substitution. The Ile42→Val and Ile130→Val point mutations probe the consequences of the loss of the side-chain Cδ1 group. The effects upon stability are essentially identical for these symmetry related positions, with a net decrease in stability of 5.1–5.8 kJ/mol. The Cys83→Val point mutation probes the effective addition of a side-chain Cγ1 group (forming a β-branched residue). The protein is destabilized by a substantial 10.1 kJ/mol in response to this mutation, almost exactly the same amount as the Cys83→Ile mutation (Table 2). The remaining “Group C” mutations were designed to compensate for postulated structural strain in response to a Cys83→Ile mutation. The Tyr55→Leu/ Cys83→Ile and Pro79→Gly/Cys83→Ile mutations were designed to eliminate bad contacts with the Ile side chain at position 83 and to permit greater flexibility of this region. The Thr78[Gly]/Pro79→Gly/Cys83→Ile mutation includes an inserted Gly residue after position 78 in an attempt to increase the length of this adjacent structure to relieve postulated structural strain due to the Ile side chain at position 83. All of these mutations were, however, substantially destabilizing (ΔΔG ~6 kJ/mol) in relationship to the Cys83→Ile point mutant. Folding and unfolding kinetic studies were performed on the SYM7ΔΔ mutant and showed that the increase in stability is accomplished primarily through a modest decrease in the rate of unfolding and that there are minimal overall effects upon the folding transition state intermediate (Fig. 3 ▶).
Figure 3.
Folding and unfolding kinetic data (“chevron plot”) for the SYM7ΔΔ FGF-1 mutant (○) and the SYM6ΔΔ FGF-1 reference protein (•).
Mitogenic activity of His-tagged wild type and mutants of FGF-1
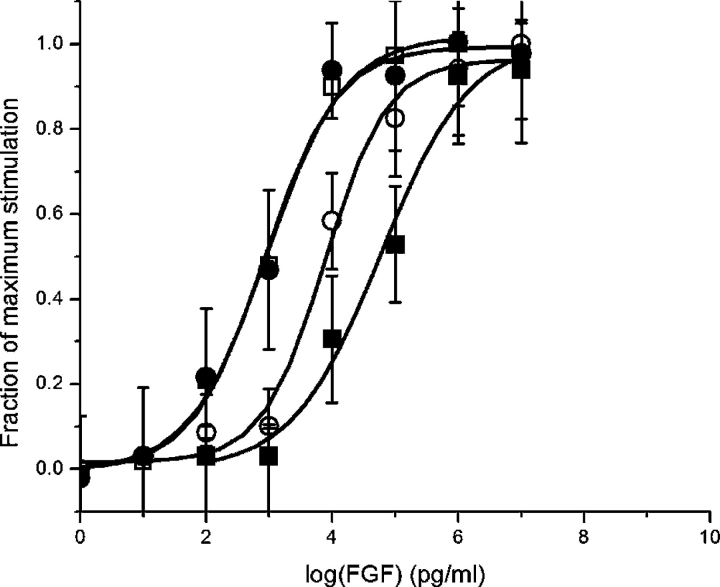
Mitogenic activity was determined for His-tagged wild-type FGF-1 (EC50=60 ng/mL), SYM6ΔΔ (EC50=0.84 ng/ mL), Phe22→Tyr/SYM6ΔΔ (EC50=7.6 ng/mL), and Tyr64→Phe/SYM6ΔΔ (EC50=0.92 ng/mL) mutants (Fig. 4 ▶). As previously described, the SYM6ΔΔ mutant is approximately two orders of magnitude more potent in mitogenic activity than His-tagged wild-type FGF-1 (Brych et al. 2004). The Tyr64→Phe/SYM6ΔΔ mutant is essentially indistinguishable from the SYM6ΔΔ mutant with regard to mitogenic activity, reflecting the minimally destabilizing effect of this mutation (Table 2). Conversely, although the Phe22→Tyr/ SYM6ΔΔ mutation is associated with a slight increase in stability, the mitogenic assay indicates that it exhibits only 10% of the mitogenic activity of the SYM6ΔΔ protein (Fig. 4 ▶).
Figure 4.
Mitogenic activity assay of WT (•), Phe22→Tyr SYM6ΔΔ (○), Tyr64→Phe SYM6ΔΔ (□), and SYM6ΔΔ (▪) proteins toward NIH 3T3 fibroblasts.
Discussion
Phe vs. Tyr residues at symmetry-related positions 22, 64, and 108
A sequence analysis derived from a comparison of 23 members of the FGF family indicates that a Tyr residue at position 64 is present with a frequency of occurrence of 82.6% (Fig. 2 ▶). The only other residue found at this position (17.4% occurrence) is the related aromatic Phe side chain. A similar trend is observed at symmetry-related position 108, where Tyr occurs with 47.8% frequency and Phe follows with 34.7% frequency (with His, Ile, and Pro residues being present at single-digit frequencies). In contrast, at symmetry-related position 22, Phe occurs with a higher frequency (30.4%) than Tyr (13.0%), and His is actually present at a higher frequency (56.5%) than Phe. Thus, when comparing symmetry-related positions 22, 64, and 108, position 22 stands out as having an unusually high occurrence of Phe in comparison to Tyr (in addition to the high prevalence of His). A Phe residue is observed at position 22 in FGF-1, and while there is no apparent basis for the selection of Phe over Tyr in terms of protein stability (Table 2), the mitogenic assay indicates a substantial functional preference for Phe at position 22 (Fig 3 ▶).
Position 22 has been described as forming part of an interface between FGF-1 and domain D2 of FGF receptor 2 ectodomain from the X-ray structure of an FGF-1/FGFR2/heparin ternary complex (PDB accession no. 1E0O) (Pellegrini et al. 2000). This interface includes residue positions Tyr15, Gly20, Phe22, Tyr94, Leu133, and Leu135 of FGF-1 with residues Lys164, Leu166, Ala168, Val169, and Pro170 of FGFR2 and is, therefore, largely hydrophobic. Model building of a Phe22→Tyr mutant into the 1E0O FGF-1/FGFR2/ heparin structure identifies a potential close contact (2.2–2.7 Å) between the Tyr Oη and the Nɛ moiety of residue Arg37 within the same FGF-1 molecule. However, the B factors of the Arg37 Nɛ moiety are in excess of 100 Å2, suggesting the potential for adjustment to accommodate the introduced Tyr Oη; furthermore, this interaction has the potential of satisfying the hydrogen-bonding requirement of the introduced Tyr side chain. Despite these structural considerations, it is clear from the mitogenic assay that maintaining the hydrophobic nature of position 22 is critical for efficient receptor complex formation.
As with position 22, position 64 can accommodate either a Phe or Tyr residue with essentially no effect upon overall stability (with Tyr being the naturally occurring residue at this position). This position in FGF-1 is distal to any receptor or heparin contacts in the FGF-1/FGFR/heparin complex structure (Pellegrini et al. 2000), and the mitogenic assay indicates that there is no effect upon functional activity for the Tyr64→Phe mutation (Fig. 4 ▶). Symmetry-related position Phe108 exhibits a 3.2 kJ/mol increase in stability. Analysis of the wild-type structure (PDB accession no. 1JQZ) indicates that the aromatic ring of the Phe108 side chain is pointed toward an adjacent Tyr125 side chain. The aromatic rings of these two side chains are essentially orthogonal, with the Phe108 Cζ positioned near the center of the Tyr125 aromatic ring. However, in the wild-type structure, the Phe108 Cζ and the Tyr125 aromatic ring are ~3.7 Å distal; thus, a direct interaction of the π electron orbital for these rings is unsupported. However, the Phe108→Tyr mutation would position the introduced side chain Oη group within appropriate hydrogen-bonding distance of the Tyr125 aromatic ring π electrons, and is therefore postulated as the basis for the observed increase in stability for the Phe108→Tyr mutation. In this regard, positions Arg37 and Thr78 are the symmetry-related positions to Tyr125, and therefore, similar stabilizing π electron interactions are not possible with Tyr residues at positions 22 and 64.
Ile vs. Cys residues at symmetry-related positions 42, 83, and 130
Analysis of sequence data for the FGF family of proteins indicates that a Cys residue is present at position 83 with a 100% frequency of occurrence (Fig. 2 ▶). The other two symmetry-related positions (42 and 130) exhibit a variety of side chains, with the most commonly observed residues being small (Ala, Ser, and Thr); however, there is no example of Cys at either of these positions in any of the 23 members of the FGF family. Positions 42 and 130 in FGF-1 are both Ile residues that also share identical rotamer orientations (χ1=gauche-, χ2=trans); thus, they exhibit the threefold symmetric relationship inherent in the β-trefoil architecture. Positions Ile42 and Ile130 are highly tolerant to substitution by Cys, exhibiting essentially no effect upon stability. However, the converse is not true at symmetry-related position Cys83, and the Cys83→Ile mutation is substantially destabilizing (Table 2). Thus, despite their symmetric relationship within the tertiary structure of FGF-1, there is an extreme disparity of preferred amino acids at positions 42, 83, and 130, both in terms of statistical frequency and stability effects upon mutation. How are positions 42 and 130 able to structurally accommodate either Cys or Ile residues, but position 83 cannot?
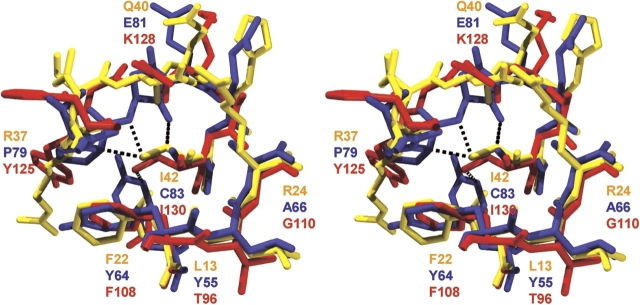
The structural environment surrounding Cys83 is defined by the side chains of residue positions Tyr55, Tyr64, Ala66, and the main-chain atoms of the region Pro79–Asn80 (Fig. 5 ▶). Symmetrically equivalent sidechain interactions are conserved in the packing environment of Ile42 (residue positions Leu13, Phe22, and Arg24, respectively), but the adjacent main-chain interactions involves the more extended region of Asp36– Asp39. Likewise, symmetrically equivalent side-chain interactions are conserved in the packing environment of Ile130 (residue positions Thr96, Phe108, and Gly110, respectively), but the adjacent main-chain interaction also involves a more extended region, involving His124–Gln127. Structurally, therefore, a significant difference in the packing environment of Cys83 involves main-chain region Pro79–Asn80. An overlay of these symmetry-related main-chain regions indicates that the Cα of residue positions Asp36, Thr78, and His124 are essentially juxtaposed, as are the Cα of residue positions Gln40, Glu81, and Lys128. Thus, the main-chain region packing against Cys83 is structurally “one residue shorter” than the symmetry-related regions. When evaluating the consequences of the substitution of Cys83 by Ile (and assuming that the adopted rotamer is identical to that observed for Ile42 and Ile130), a series of close contacts are apparent between the introduced Ile83 Cγ1 and Cδ1 moieties. These close contacts include 2.3 Å between position 79Cα and an introduced Ile Cδ1, 2.3 Å between position 80N and an introduced Ile Cδ1, 2.0 Å between 80O and an introduced Ile Cγ1, and 2.8 Å between 55Cɛ1 and an introduced Ile Cδ1 (Fig. 5 ▶).
Figure 5.
Relaxed stereo diagram of an overlay of the packing environments surrounding symmetry-related positions Ile42 (yellow), Cys83 (blue), and Ile130 (red). Close contacts between the mainchain region 79–80 and Ile residues that would occupy the symmetry related positions to residue 83 are indicated by broken lines. Fig. 5 live 4/c
The inferred flexibility of the main chain regions that pack against positions Ile42, Cys83, and Ile130 is another structural difference. An Ile→Cys mutation within a buried region is expected to destabilize the structure due to the effective loss of van der Waals interactions involving the side-chain Cδ1 and Cγ1 atoms. However, the effects upon stability can be minimized if the structure is able to collapse to fill the resulting void caused by the mutation (Eriksson et al. 1992). The ability of positions 42 and 130 to accommodate Ile→Cys mutations with essentially no effect upon stability indicates that main-chain regions 36–39 and 124–127 exhibit a certain flexibility and are able to adjust to avoid a resulting cavity (the ~5 kJ/mol destabilization associated with Ile→Val mutations at these positions suggests that only a partial collapse is possible in response to Val mutations). Conversely, the substantial destabilization of the Cys83→Ile mutation indicates that the main-chain region 79–80 is unable to accommodate the increase in the side-chain buried area without causing strain; thus, this main-chain region appears to possess greater rigidity than its symmetry-related regions. The shorter apparent length, and the presence of Pro79, may contribute to the apparent rigidity of this main-chain region. Initial efforts to address this question, first by the introduction of a Pro79→Gly point mutation, and then by combining this with the insertion of an additional residue (Gly) after position 78, failed to stabilize a Cys83→Ile mutation (Table 2). The conformation and flexibility of these main-chain regions is therefore likely determined by more complex interactions involving the side chains within these regions. Ongoing studies are investigating this possibility.
FGF-1 contains three free Cys residues at positions 16, 83, and 117 (not related by the threefold structural symmetry). These free Cys residues are each located at solventinaccessible positions. Functional studies of Cys→Ser mutations at positions 16 and 83 have shown an increase in the protein half-life (in serum-free medium and in the absence of heparin) from 0.26 to 13 h (Ortega et al. 1991). Subsequent thermodynamic studies of these Ser mutants indicated that the increase in functional half-life was not due to an increase in stability, and in fact, individual Ser mutations at these positions destabilized the structure by a significant 8–10 kJ/mol (Culajay et al. 2000). The increase in functional half-life was attributed to the elimination of thiol-mediated chemistry that promotes irreversible denaturation, and buried free Cys residues were postulated to be important regulators of functional half-life (Ortega et al. 1991; Culajay et al. 2000). Thus, the asymmetry of the tertiary structure associated with the packing environment of position 83 results in a stability-based structural requirement for a Cys residue at this position, which subsequently regulates functional half-life. This result is similar to a previously reported heparin-binding functionality of residue positions 120–122 in FGF-1 (Brych et al. 2004). These residues appear as a structural insertion that disrupts the tertiary structure symmetry of FGF-1, and while their excision stabilizes the structure, the ability to bind heparin is substantially diminished (Brych et al. 2004). Furthermore, an invariant Met residue at position 67 within the core of FGF-1 is essential to stabilize the position 120–122 insertion. Thus, it appears to be the case that several regions of apparent asymmetry within the fundamentally symmetric structure of FGF-1 are associated with the accommodation of function.
Materials and methods
Mutagenesis and expression
All studies utilized a synthetic gene for the 140 amino acid form of human FGF-1 (Gimenez-Gallego et al. 1986; Linemeyer et al. 1990; Ortega et al. 1991; Blaber et al. 1996) with the addition of an amino-terminal six residue “His-tag” to facilitate purification (Brych et al. 2001). The reference protein for the current set of mutations is the “SYM6ΔΔ” mutant form of FGF-1 (Table 1). This mutant form exhibits a greater tertiary and primary structure symmetry and is more stable (Table 2) than the wild-type protein (Brych et al. 2004). The QuikChange site-directed mutagenesis protocol (Stratagene) was used to introduce individual or combination mutations using mutagenic oligonucleotides of 25–31 bases in length (Biomolecular Analysis Synthesis and Sequencing Laboratory, Florida State University). All FGF-1 mutants were expressed using the pET21a(+) plasmid/BL21(DE3) Escherichia coli host expression system (Invitrogen Corp.). Mutant construction and expression followed previously described procedures (Blaber et al. 1999; Culajay et al. 2000; Brych et al. 2001). All mutants were purified as previously described using nickel-nitrilotriacetic acid (Ni-NTA) chromatography, followed by FPLC purification on a Superdex- 75 column (Brych et al. 2001).
Isothermal equilibrium denaturation
FGF-1 contains a single buried tryptophan residue (Trp107) that exhibits atypically greater fluorescence quenching in the native versus denatured state, and the X-ray structure of FGF-1 indicates that this quenching of Trp107 fluorescence is due to an adjacent proline residue (Pro121) (Blaber et al. 1996, 1999). The differential fluorescence between the native and denatured states has been used to quantify the unfolding of the FGF-1 protein, and has been shown to be in excellent agreement with unfolding as monitored by CD spectroscopy (Blaber et al. 1999; Brych et al. 2003). The deletion of Pro121 in the SYM6ΔΔ mutant form of FGF-1 (utilized as the background protein for the present mutational study) diminishes the quenching of Trp107 and, subsequently, the differential fluorescence signal between native and denatured states (Brych et al. 2004). Thus, isothermal equilibrium denaturation by GuHCl was quantified using circular dichroism as the spectroscopic probe. CD measurements were collected on an Aviv model 202 circular dichroism spectrometer (Proterion Corp.) equipped with a Peltier-controlled temperature unit maintaining a constant temperature of 298 K, and using a 10-mm path-length cuvette. Protein samples (25 μm) were equilibrated overnight in 50 mM sodium phosphate, 100 mM NaCl, 10 mM ammonium sulfate, 2 mM DTT (pH 7.5) at 298 K in 0.1 M increments of guanidine HCl (GuHCl). Buffer traces were collected, averaged, and subtracted from the sample traces. Data smoothing was performed prior to buffer subtraction using a 5-point fast Fourier transform filter. The denaturation process was monitored by observing the change in CD signal at 227 nm with increasing GuHCl. The general-purpose nonlinear least squares fitting program DataFit (Oakdale Engineering) was used to fit the change in molar ellipticity at 227nmversusGuHCl concentration data to a six-parameter two-state model (Eftink 1994):
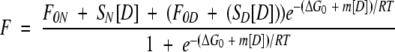 |
(1) |
where [D] is the denaturant concentration, F0N and F0D are the 0 M denaturant molar ellipticity intercepts for the native and denatured states, respectively, and SN and SD are the slopes of the native and denatured state baselines, respectively. ΔG0 and m describe the linear function of the unfolding free energy versus denaturant concentration at 298 K. The effect of a given mutation upon the stability of the protein (ΔΔG) was calculated by taking the difference between the Cm values for wild-type and mutant and multiplying by the average of the m values, as described by Pace and Scholtz (1997):
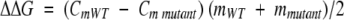 |
(2) |
Folding kinetics measurements
Folding and unfolding kinetic data was collected using previously described methods (Brych et al. 2004). Protein samples (~150μM) were incubated overnight in 3.0 M GuHCl, 50 mM NaPi, 100 mM NaCl, 10 mM (NH4)SO4, 2 mM DTT (pH7.5) at 298 K. Refolding was initiated by a 1:10 dilution of protein solutions into 50 mM sodium phosphate, 100 mM NaCl, 10 mM ammonium sulfate, 2 mM DTT (pH 7.5) containing either 0.1 or 0.05 M increments of GuHCl up to the projected midpoint of denaturation as determined by isothermal equilibrium denaturation experiments. All data was collected using an Aviv model 202 CD spectrometer equipped with a Peltier-controlled temperature unit maintaining a constant temperature of 298 K and an Aviv model SF305 stopped flow system. Data collection times for each protein were designed so as to quantify the CD signal over five half-lives, or>96% of the total expected amplitude.
Unfolding kinetics measurements
Protein samples (~300 μM) were dialyzed against 50 mM sodium phosphate, 100 mM NaCl, 10 mM ammonium sulfate, 2m MDTT (pH 7.5) overnight at 298 K. Unfolding was initiated by 1:10 dilution into 50 mM sodium phosphate, 100 mM NaCl, 10 mM ammonium sulfate, 2 mM DTT (pH 7.5) with final GuHCl concentration in the range from 1.5 to 5.5 M in 0.5 M increments. The unfolding process was quantified by following the change in CD signal at 227 nm using an Aviv model 202 circular dichroism spectrometer equipped with an Aviv model SF305 stopped flow system and a Peltier-controlled temperature unit, maintaining a constant temperature of 298 K. Data collection times for each protein were designed so as to quantify the CD signal over three to four half-lives, or>93% of the total expected amplitude.
Folding and unfolding kinetic analysis
Both folding and unfolding kinetic data were collected in triplicate at each GuHCl buffer condition. In all cases, data from at least three separate experiments were averaged. The kinetic rates and amplitudes versus denaturant concentration were calculated from the time dependent change in tryptophan fluorescence using a single exponential model:
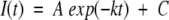 |
(3) |
where I(t) is the intensity of fluorescent signal at time t, A is the corresponding amplitude, k is the observed rate constant for the reaction, and C is a constant that is the asymptote of the CD signal. Folding and unfolding rate constant data were fit to a global function describing the contribution of both rate constants to the observed kinetics as a function of denaturant (“chevron” plot) as described by Fersht (1999):
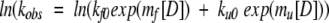 |
(4) |
where kf0 and ku0 are the folding and unfolding rate constants, respectively, extrapolated to 0 M denaturant; mf and mu are the slopes of the linear functions relating ln(kf) and ln(ku), respectively, to denaturant concentration; and [D] is the denaturant concentration.
Mitogenic assay
The mitogenic activity of various mutants was evaluated by a tissue-culture based assay. NIH 3T3 fibroblasts were initially plated in Dulbecco’s modified Eagle’s medium (DMEM) (American Type Culture Collection) supplemented with 10% (v/v) newborn calf serum (NCS) (Sigma), 100 units of penicillin, 100 mg of streptomycin, 0.25 mg of Fungizone, and 0.01 mg/mL of gentamicin (Gibco) (“serum-rich” medium) in T75 tissue culture flasks (Fisher). The cultures were incubated at 37°C with 5% CO2 supplementation. At 80% cell confluence, the cells were washed with 5-mL cold TBS (0.14 M NaCl, 5.1 mM KCl, 0.7 mM Na2HPO4, 24.8 mM Trizma base [pH 7.4]) and subsequently treated with 5 mL of a 0.025% trypsin solution (Invitrogen Corp.). The trypsinized cells were subsequently seeded in T25 tissue culture flasks (Fisher) at a cell density of 3.0×104 cells/ cm2 (representing 20% confluence). Cell synchronization was initiated by serum starvation in DMEM with 0.5% NCS, 100 units of penicillin, 100 mg of streptomycin, 0.25 mg of Fungizone, and 0.01 mg/mL of gentamicin (“starvation” medium). Cultures were incubated for 48 h at 37°C, the medium was then decanted and replaced with fresh medium supplemented with FGF-1 (0–10 μg/mL), and the cultures were incubated for an additional 48 h. After this incubation, the medium was decanted and the cells were washed with 1 mL of cold TBS (pH 7.4). A total of 1 mL of 0.025% trypsin was then added to release the cells from the flask surface, and 2 mL of serum-rich medium was added to dilute and inhibit the trypsin. The cells were counted using a hemacytometer (Hausser Scientific Partnership). Experiments were performed in quadruplicate and the cell densities were averaged. The relationship between the cell number and log concentration of added growth factor was fit to a sigmoid function.
Acknowledgments
We thank Dr. Claudius Mundoma, Physical Biochemistry Facility, Kasha Laboratory, Institute of Molecular Biophysics, and Ms. Pushparani Dhanarajan, Molecular Cloning Facility, Biological Science, for their helpful comments. This work was supported by grant MCB 0314740 from the National Science Foundation.
Abbreviations
FGF-1, human acidic fibroblast growth factor
GuHCl, guanidinium hydrochloride
CD, circular dichroism
DTT, dithiothreitol.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051494405.
References
- Adamek, D., Popovic, A., Blaber, S., and Blaber, M. 1998. Denaturation studies of haFGF. In Biocalorimetry: Applications of calorimetry in the biological sciences (eds. J.E. Ladbury and B.Z. Chowdhry), pp. 235–241. John Wiley & Sons Ltd., Chichester, UK.
- Blaber, M., DiSalvo, J., and Thomas, K.A. 1996. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry 35 2086–2094. [DOI] [PubMed] [Google Scholar]
- Blaber, S.I., Culajay, J.F., Khurana, A., and Blaber, M. 1999. Reversible thermal denaturation of human FGF-1 induced by low concentrations of guanidine hydrochloride. Biophys. J. 77 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, J.D., Wilke, C.O., Arnold, F.H., and Adami, C. 2004. Stability and the evolvability of function in a model protein. Biophys. J. 86 2758–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brych, S.R., Blaber, S.I., Logan, T.M., and Blaber, M. 2001. Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core region of a β-trefoil. Protein Sci. 10 2587–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brych, S.R., Kim, J., Logan, T.M., and Blaber, M. 2003. Accommodation of a highly symmetric core within a symmetric protein superfold. Protein Sci. 12 2704–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brych, S.R., Dubey, V.K., Bienkiewicz, E., Lee, J., Logan, T.M., and Blaber, M. 2004. Symmetric primary and tertiary structure mutations within a symmetric superfold: A solution, and not a constraint, to achieve a foldable polypeptide. J. Mol. Biol. 344 769–780. [DOI] [PubMed] [Google Scholar]
- Culajay, J.F., Blaber, S.I., Khurana, A., and Blaber, M. 2000. Thermodynamic characterization of mutants of human fibroblast growth factor 1 with an increased physiological half-life. Biochemistry 39 7153–7158. [DOI] [PubMed] [Google Scholar]
- Eftink, M.R. 1994. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 66 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, A.E., Baase, W.A., Zhang, X-J., Heinz, D.W., Blaber, M., Baldwin, E.P., and Matthews, B.W. 1992. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255 178–183. [DOI] [PubMed] [Google Scholar]
- Fersht, A.R. 1999. Kinetics of protein folding. W.H. Freeman and Co., New York.
- Gimenez-Gallego, G., Conn, G., Hatcher, V.B., and Thomas, K.A. 1986. The complete amino acid sequence of human brain-derived acidic fibroblast growth factor. Biochem. Biophys. Res. Comm. 128 611–617. [DOI] [PubMed] [Google Scholar]
- Heidary, D.K. and Jennings, P.A. 2002. Three topologically equivalent core residues affect the transition state ensemble in a protein folding reaction. J. Mol. Biol. 316 789–798. [DOI] [PubMed] [Google Scholar]
- Kim, J., Lee, J., Brych, S.R., Logan, T.M., and Blaber, M. 2004. Sequence swapping does not result in conformation swapping for the β4/β5 and β8/ β9 β-hairpin turns in human acidic fibroblast growth factor. Protein Sci. 14 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, D., Thoma, R., Henn-Sax, M., Sterner, R., and Wilmanns, M. 2000. Structural evidence for evolution of the β/α barrel scaffold by gene duplication and fusion. Science 289 1546–1550. [DOI] [PubMed] [Google Scholar]
- Linemeyer, D.L., Menke, J.G., Kelly, L.J., Disalvo, J., Soderman, D., Schaeffer, M.-T., Ortega, S., Gimenez-Gallego, G., and Thomas, K.A. 1990. Disulfide bonds are neither required, present, nor compatible with full activity of human recombinant acidic fibroblast growth factor. Growth Factors 3 287–298. [DOI] [PubMed] [Google Scholar]
- Mach, H. and Middaugh, C.R. 1994. Probing the affinity of polyanions for acidic fibroblast growth factor by unfolding kinetics. Arch. Biochem. Biophys. 309 36–42. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, D. 2000. The molecular evolutionary history of a winged bean α-chymotrypsin inhibitor and modeling of its mutations through structural analysis. J. Mol. Evol. 50 214–223. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Lesk, A.M., and Chothia, C. 1992. β-Trefoil fold. Patterns of structure and sequence in the kunitz inhibitors interleukins- 1β and 1α and fibroblast growth factors. J. Mol. Biol. 223 531–543. [DOI] [PubMed] [Google Scholar]
- Ortega, S., Schaeffer, M.-T., Soderman, D., DiSalvo, J., Linemeyer, D.L., Gimenez-Gallego, G., and Thomas, K.A. 1991. Conversion of cysteine to serine residues alters the activity, stability, and heparin dependence of acidic fibroblast growth factor. J. Biol. Chem. 266 5842–5846. [PubMed] [Google Scholar]
- Pace, C.N. and Scholtz, J.M. 1997. Measuring the conformational stability of a protein. In Protein structure: A practical approach (ed. T.E. Creighton), pp. 299–321. Oxford University Press, Oxford, UK.
- Pellegrini, L., Burke, D.F., von Delft, F., Mulloy, B., and Blundell, T.L. 2000. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407 1029–1034. [DOI] [PubMed] [Google Scholar]
- Plotnikov, A.N., Hubbard, S.R., Schlessinger, J., and Mohammadi, M. 2000. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101 413–424. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. and Russell, R.B. 2000. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all β-trefoil proteins. J. Mol. Biol. 302 1041–1047. [DOI] [PubMed] [Google Scholar]
- Richardson, J. and Richardson, D.C. 1989. The de novo design of protein structures. Trends Biochem. Sci. 14 304–309. [DOI] [PubMed] [Google Scholar]
- Romero, A., Lucena-Pineda, A., and Gimenez-Gallego, G. 1996. X-ray structure of native full-length human fibroblast-growth factor at 0.25- nm resolution. Eur. J. Biochem. 241 453–461. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Buckle, A.M., and Fersht, A.R. 1994. Stability and function: Two constraints in the evolution of barstar and other proteins. Structure 2 945–951. [DOI] [PubMed] [Google Scholar]
- Shoichet, B.K., Baase, W.A., Kuroki, R., and Matthews, B.W. 1995. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. 92 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strewler, G.J. 2001. FGF23, hypophosphatemia, and rickets: Has phosphatonin been found? Proc. Natl. Acad. Sci. 98 5945–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Minasov, G., and Shoichet, B.K. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320 85–95. [DOI] [PubMed] [Google Scholar]
- Wolynes, P.G. 1996. Symmetry and the energy landscapes of biomolecules. Proc. Natl. Acad. Sci. 93 14249–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]