Abstract
A novel potent trypsin inhibitor was purified and characterized from frog Bombina maxima skin. A full-length cDNA encoding the protein was obtained from a cDNA library constructed from the skin. Sequence analysis established that the protein actually comprises three conserved albumindomains. B.maximaserum albumin was subsequently purified, and its coding cDNA was further obtained by PCR-based cloning from the frog liver. Only two amino acid variations were found in the albumin sequences from the skin and the serum. However, the skin protein is distinct from the serum protein by binding of a haem b (0.95 mol/mol protein). Different from bovine serum albumin, B. maxima albumin potently inhibited trypsin. It bound tightly with trypsin in a 1:1 molar ratio. The equilibrium dissociation constants (KD) obtained for the skin and the serum proteins were 1.92 × 10−9 M and 1.55 × 10−9 M, respectively. B. maxima albumin formed a noncovalent complex with trypsin through an exposed loop formed by a disulfide bond (Cys53–Cys62), which comprises the scissile bond Arg58(P1)–His59(P1′). No inhibitory effects on thrombin, chymotrypsin, elastase, and subtilisin were observed under the assay conditions. Immunohistochemical study showed that B. maxima albumin is widely distributed around the membranes of epithelial layer cells and within the stratum spongiosum of dermis in the skin, suggesting that it plays important roles in skin physiological functions, such as water economy, metabolite exchange, and osmoregulation.
Keywords: albumin, skin, trypsin, haem, amphibian, frog
Amphibian skin is a morphologically, biochemically, and physiologically complex organ that fulfills a wide range of functions necessary for amphibian survival. A series of mutually compatible functions take place in the skin, including respiration, water regulation, anti-predator, antimicrobial defense, temperature control, and reproduction (Duellman and Trueb 1986). Although numerous studies have focused on the bioactive components in amphibian skin “secretions,” from which lots of peptides with diverse biological activities have been isolated and characterized, the physiological and functional complexity and the relative biochemical mechanisms of amphibian skin are incompletely understood (Clarke 1997).
Albumin serves as a transport and depot protein for numerous endogenous and exogenous compounds in the circulation system, including metals, fatty acids, amino acids, metabolites, and many drug compounds. Thus, the most important physiological role of albumin is to bring such solutes in the bloodstream to their target organs, as well as to maintain the pH and osmotic pressure of plasma (Dugaiczyk et al. 1982; Peters 1996; Kragh-Hansen et al. 2002). One of the well-studied albumins is human serum albumin (HSA). It is a single-chain protein synthesized and secreted from liver cells. Composed of 585 amino acids with a molecular weight of 66,500, HSA consists of three structurally similar globular domains (labeled I–III); each of which contains two subdomains (A and B) that share common structural elements (Dugaiczyk et al. 1982; He and Carter 1992; Curry et al. 1998). Under severe hemolysis, serum albumin can also become a significant transporter of heme, principally as hemin (FeIII protoporhyrin- IX [Cl]) that binds to a single site within a hydrophobic cavity in subdomain IB (Wardell et al. 2002).
Serine protease inhibitors are widely found in animals, plants, and microorganisms. The primary structures, with the number of amino acids ranging from 29 to ~400, and the structural properties of these inhibitors differ significantly. Based on sequence, topological, and functional similarities, the class of serine protease inhibitors can be grouped into at least 16 different families (Bode and Huber 1992, 2000). They are of broad interest because they play key roles in the modulation of protease physiological functions. They also participate in the innate immune response (Imler and Hoffmann 2000) and in carcinogenesis (Clawson 1996). In particular, several protease inhibitors have been identified and isolated from amphibian skin secretions, such as BSTI and BMTI from the skin secretions of Bombina species (Mignogna et al. 1996; Lai et al. 2002). They are 60 amino acids long and can be grouped into a novel family of serine protease inhibitors based on their unique feature of primary and three-dimensional structures (Rosengren et al. 2001).
The Chinese red belly frog (Bombina maxima) is an endemic amphibian in the mountainous regions of South West China, living in very harsh environments (Fei 1999). Our recent studies showed that its skin secretions contain many bioactive peptides (Lee et al. 2005a,b; Zhang et al. 2005). In this report, we present characterization and molecular cloning of B. maxima albumin from the skin (BmA-skin) and the serum (BmA-serum). Interestingly, the frog albumin was found to be a novel potent trypsin inhibitor, and the major inhibitory mechanism was characterized. Apparently, B. maxima albumin represents the first member of a novel class of trypsin inhibitors.
Results
Purification of BmA-skin
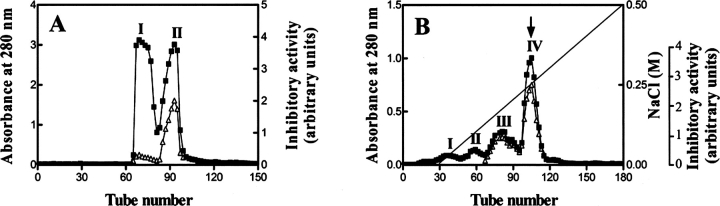
Initiated with 2 g of lyophilized B. maxima skin homogenate (containing 620 mg proteins), DEAE Sephadex A-50 column (pH 7.8) resulted in the separation of five protein peaks. Trypsin inhibitory activity was found between peaks I and II and in peak V (Fig. 1A ▶). BMTI has been purified previously from NaCl-free eluted fractions between peaks I and II (Lai et al. 2002). The peak V of DEAE Sephadex A-50 column was collected and then applied to a Sephadex G-75 gel filtration column equilibrated with 50 mM Tris-HCl (pH 7.8), containing 5 mM EDTA and 0.1 M NaCl. This purification step resulted in the separation of two protein peaks, in which trypsin inhibitory activity was found in peak II. The peak II of Sephadex G-75 was loaded finally on a DEAE Sephadex A-50 column equilibrated with 50 mM Tris-HCl (pH 8.8), yielding only one protein peak that associated again with trypsin inhibitory activity. Finally, 24 mg product was obtained.
Figure 1.
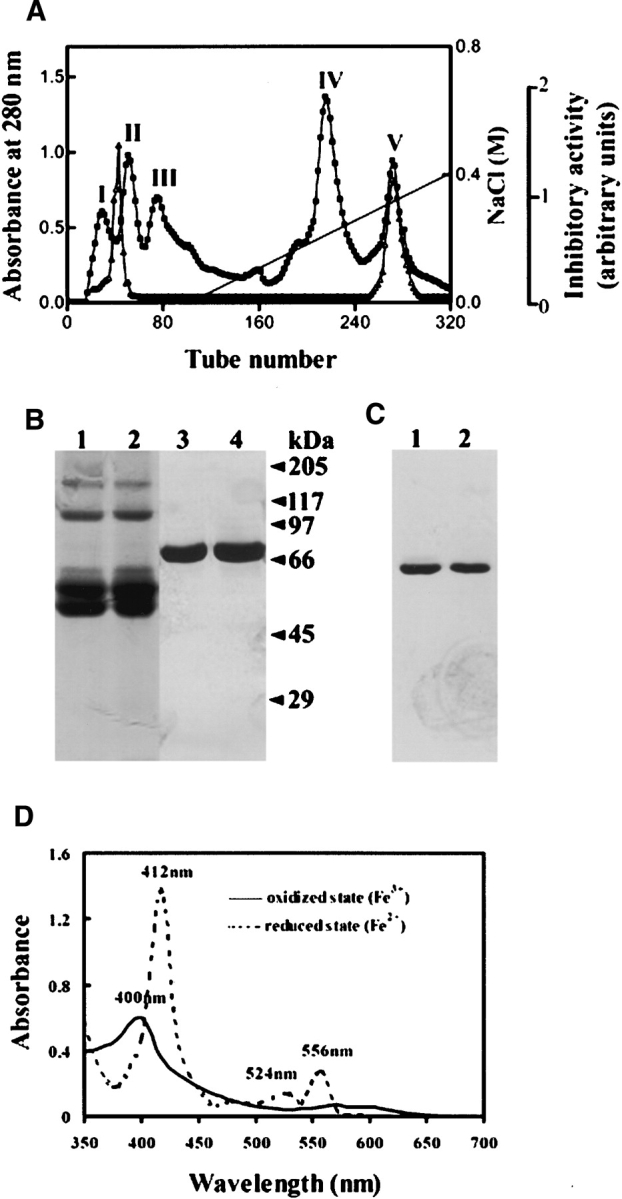
(A) Ion exchange chromatography of B. maxima skin homogenate on a DEAE Sephadex A-50 (Pharmacia) column (2.6 × 50 cm) equilibrated with 50 mM Tris-HCl buffer (pH 7.8), containing 5 mM EDTA. The elution was performed at a flow rate of 20 mL/h with a linear NaCl gradient, collecting fractions of 5 mL per tube. The protein concentration was estimated from the absorbance at 280 nm (▪). Trypsin inhibitory activity (▵) was determined as described in Materials and Methods. (B) SDS-PAGE (10% acrylamide) analysis of purified B. maxima albumin: BmA-skin (lanes 1,3), BmA-serum (lanes 2,4), non-reducing conditions (lanes 1,2), and reducing conditions (lanes 3,4). (C) PAGE in native conditions (pH 8.5) (10% acrylamide): BmA-skin (lane 1) and BmA-serum (lane 2). In both B and C, proteins were silver-stained. (D) Absorption spectra of BmA-skin in oxidized and reduced states.
Molecular characterization of BmA-skin
Purified BmA-skin migrated as a single protein band both in native PAGE and in SDS-PAGE under reducing conditions with silver staining, showing an apparent molecular weight of 67,000 (Fig. 1B,C ▶). However, in SDS-PAGE under nonreducing conditions, the protein migrated as several protein bands, such as two major bands of 50,000 and 55,000 and a minor band of 60,000, which possess the same N-terminal sequence. This might reflect conformational heterogeneity of the protein induced by SDS without breakage of disulfide bonds. Furthermore, after sequence analysis, minor protein bands with apparent molecular weights of 110,000 and 160,000 in SDS-PAGE under nonreducing conditions were proven to be the dimeric and trimeric forms of the protein (Fig. 1B ▶). PAGE, combined with Schiff’s reagent staining (Zacharius et al. 1969), showed that BmA-skin is not a glycoprotein.
BmA-skin is characterized as having an absorption spectrum with a maximum at 400 nm, and a shoulder at 570 nm in the resting state and in the presence of alkaline pyridine (Fig. 1D ▶). In the presence of a reducing reagent dithionite, the absorption maxima at 412 nm (Soret band), 524 nm (β-band), and 556 nm (α-band) appeared, revealing the presence of a haem b factor (Berry and Trumpower 1987). The calculated content of haem b bound in BmA-skin is 0.95 mol/mol protein. The absorption spectrum of the purified protein was the same as that in the presence of an oxidizing reagent K3Fe(CN)6, suggesting that BmA-skin was purified in a completely oxidized form.
Molecular cloning and sequence analysis of BmA-skin
The purified protein was subjected to amino acid sequencing by Edman degradation with an ABI model 476A protein sequencer. The N-terminal 34 amino acids of the protein were determined. In addition, the sequences of nine internal peptides (accounting to 113 amino acid residues), acquired by trypsin digestion (the protein was first denatured by dithiothreitol and S-carboxyamidomethylated) and subsequent HPLC C18 column separation, were also obtained. The partial cDNA sequence obtained by a PCR amplification with primers P1 and P6R greatly facilitated the subsequent molecular cloning of BmA-skin. A cDNA library constructed from B. maxima skin was screened at high stringency by an efficient and rapid PCR-based procedure (Zhang et al. 1995). A positive clone was identified and isolated. Both strands of the clone were sequenced. The cDNA structure of BmA-skin was found to contain a coding region of 1824 nucleotides. The encoded amino acid sequence corresponds to a precursor protein of 607 residues. The determined N-terminal and internal amino acid sequences were found exactly in the deduced protein sequence, thereby unequivocally confirming the identity of the isolated cDNA clone (GenBank AY885649). The N-terminal residue Asp is preceded by a 22-amino-acid signal peptide. A BLASTA search in the data banks showed that the purified protein is an albumin-like protein that exhibits a 47% sequence identity with 68 kDa Xenopus laevis serum albumin (Moskaitis et al. 1989), 39% with HSA (Dugaiczyk et al. 1982), and 38% with bovine serum albumin (BSA) (Hilger et al. 2001).
Purification and molecular cloning of BmA-serum
Gel filtration of B. maxima serum on a Sephadex G-100 column resulted in separation of two protein peaks (Fig. 2A ▶); trypsin inhibitory activity was found mainly in peak II. The peak II of Sephadex G-100 column was further loaded on a Q-Sepharose ion exchange column, resulting in the separation of four protein peaks (Fig. 2B ▶). The trypsin inhibitory activity was found in peak III and IV. Peak IV is BmA-serum, as indicated by an arrow. In both native PAGE and SDS-PAGE, the purified protein showed the same migration patterns as those of BmA-skin (Fig. 1B,C ▶). The N-terminal 31 amino acids of BmA-serum determined are the same as those of BmA-skin. However, BmA-serum is characterized of a much lower content of haem b (0.05 mol/mol protein).
Figure 2.
(A) Gel filtration of B. maxima serum. Four microliters of the frog serum was applied to a Sephadex G-100 (superfine, Pharmacia) column (2.6 × 100 cm) equilibrated with 50 mM Tris-HCl (pH 7.5) and containing 0.1 M NaCl. Elution was achieved with the same buffer at a flow rate of 9 mL/h, collecting fractions of 3 mL per tube. (B) The peak II of Sephadex G-100 column was lyophilized and dialyzed against 50 mM Tris-HCl (pH 7.5), for 24 h at 4°C. The peak was then loaded on a Q-Sepharose (Pharmacia) ion exchange column (2.6 × 30 cm). The elution was performed at a flow rate of 48 mL/h with a linear NaCl gradient, collecting fractions of 3 mL per tube. The absorbance of the eluant was monitored at 280 nm (▪). Trypsin inhibitory activity (▵) was determined as described in Materials and Methods.
BmA-serum coding cDNA obtained from the frog liver is generally the same as that of BmA-skin from the skin, except two nucleotide mutations in protein coding region that resulted in Gly471 to Asn and Ser569 to Asn replacements, as well as an eight-base insertion in 3′ untranslated region in the cDNA of BmA-serum (GenBank AY885650).
Serine protease inhibitory activity
Both BmA-skin and BmA-serum potently inhibited the amidolytic activity of trypsin. At dosages of B. maxima albumin at ~40 nM, the amidolytic activity of trypsin (30 nM) on BApNA was totally blocked. In contrast, BSA did not influence the amidolytic activity of trypsin (Fig. 3A ▶), even with a concentration used up to 5 μM. Figure 3B ▶ illustrates that the inhibition of B. maxima albumin on trypsin activity is time-stable; no recovery of the amidolytic activity of trypsin was observed within 24 h. However, no inhibition of the hydrolysis of S-2238 by thrombin, BTEE by chymotrypsin, S-4760 by elastase, and C-3022 by subtilisin could be observed even with B. maxima albumin concentrations used up to 5 μM.
Figure 3.
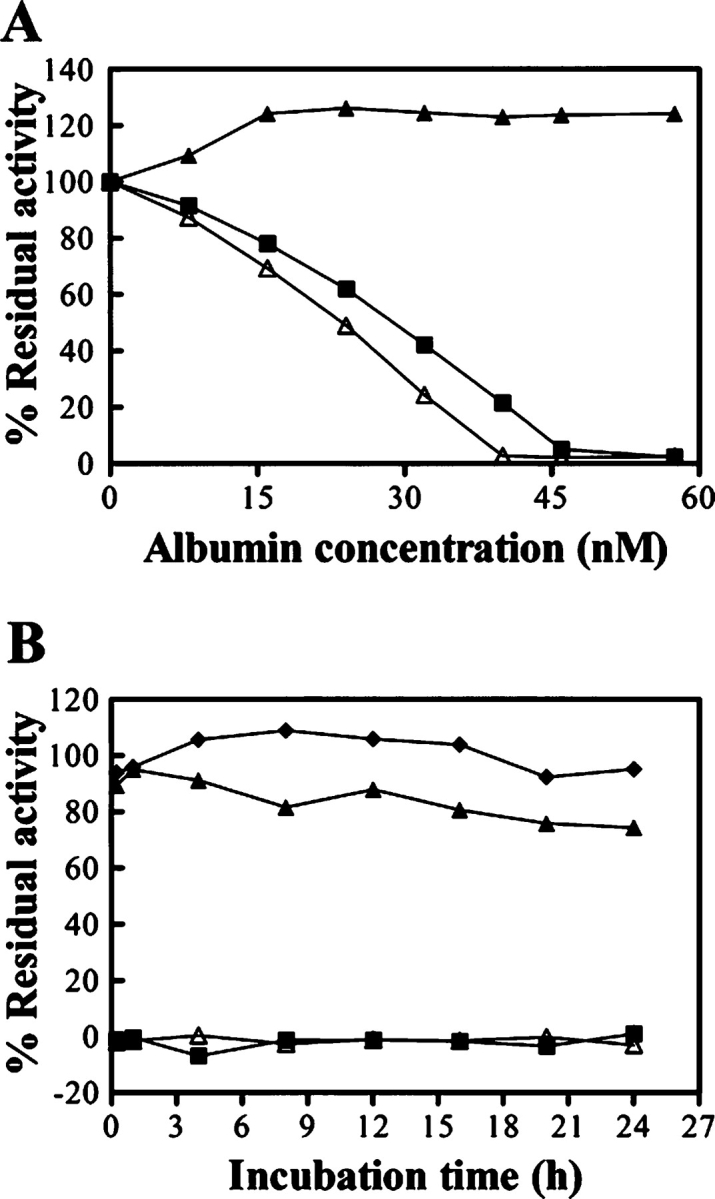
Inhibitory effects of B. maxima albumin on trypsin. (A) Trypsin (30 nM) was incubated with various amounts of BmA-skin (▵), BmA-serum (▪) or BSA (▴) for 15 min at 25°C. The residual amidolytic activity of trypsin was determined. (B) Time stability of the inhibition of B. maxima albumin on trypsin activity. Trypsin (30 nM) was incubated with the buffer (♦), 45 nM of BmA-skin (▵), BmA-serum (▪), or BSA (▴) at 25°C for various times and then the residual amidolytic activity of trypsin was measured.
Albumin–trypsin binding analysis
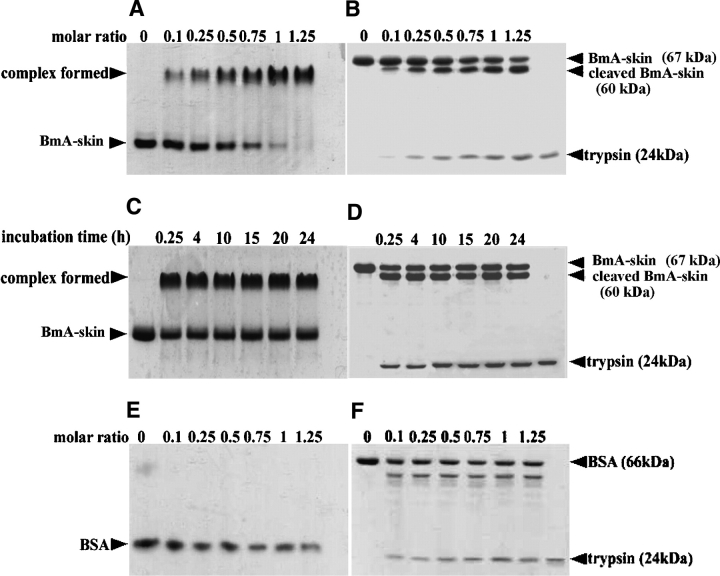
Binding of trypsin to BmA-skin and formation of a stable complex were first observed in native PAGE (Fig. 4A ▶), and Figure 4C ▶ further illustrates that the binding is time-stable. Gel filtration analysis revealed that the apparent molecular weights of BmA-skin and trypsin are 67,000 and 23,000, respectively, while that of the complex formed between them is ~90,000 (data not shown). This result indicated that trypsin binds with BmA-skin in a 1:1 molar ratio. The binding kinetic constants were determined by surface plasmon resonance with BIAcore 3000 (BIAcore AB). The equilibrium dissociation constant (KD) obtained for BmA-skin was 1.92 × 10−9 M. The mechanism by which BmA-skin exerts its trypsin inhibitory activity was further investigated by SDS-PAGE under reducing conditions (Fig. 4B ▶). Dissociation of the complex in SDS-PAGE indicated that the formation of the complex is noncovalent. In addition, cleavage of BmA-skin by trypsin and formation of a new band of 60,000 were observed. Sequence analysis showed that the cleavage occurred at Arg58–His59 bond (HSA numbering system), located in a loop formed by a disulfide bridge Cys53–Cys62 (He and Carter 1992; Curry et al. 1998). Figure 4D ▶ shows that the cleavage of BmA-skin by trypsin was completed at 15 min, and no further change of the proportion between the native and the cleaved protein was observed in 24 h. In contrast, no complex formation was observed between BSA and trypsin in native PAGE under the same conditions (Fig. 4E ▶). With the increase of trypsin dosages, the BSA protein band became faint, and the smaller digestive fragments of BSA could be observed in SDS-PAGE (Fig. 4F ▶). Similar results were obtained when BmA-serum was used in the above experiments, with a KD value of 1.55 × 10−9 M obtained in binding kinetic analysis.
Figure 4.
Analysis of binding complex formed between BmA-skin and trypsin. BmA-skin (A,B) (1.85 μM), or BSA (E,F) (1.85 μM) was incubated with various amounts of trypsin at different molar ratio (1:0.1–1.25) for 15 min at 25°C, or alternatively, BmA-skin (C,D) (2.5 μM) was incubated with trypsin (1.85 μM) at 25°C for various times. The incubated mixtures were analyzed by native-PAGE (A,C,E) and by SDS-PAGE under reducing conditions (B,D,F).
Immunohistochemical study
The distribution of B. maxima albumin in the skin was studied by immunohistochemical method. The integument of the frog consists of an outer layer, the epidermis of ectodermal origin, and an underlying layer, the dermis. The pigment cells, granular glands, and mucous glands imbedded in the dermis were clearly observed (Fig. 5A ▶). With the rabbit anti-BmA-skin serum, immunoreactivity was observed around the membranes of epithelial layer cells and the stratum spongiosum of dermis in the skin (Fig. 5C,D ▶), while no immunoreactivity was detectable in granular and mucous glands. The control rabbit serum did not stain any of these structures (Fig. 5B ▶).
Figure 5.
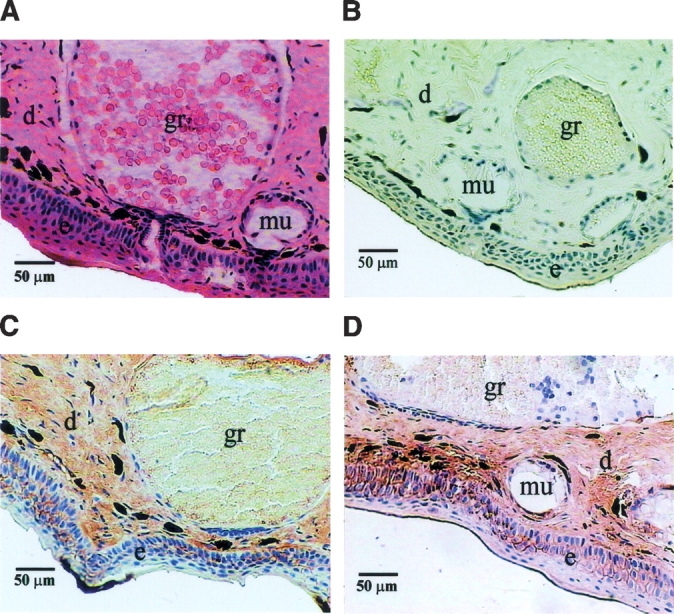
Hematoxylin and eosin (HE) treated and immunoenzymatically stained skin sections of B. maxima observed under a light microscope. (A) Skin section treated with HE. (B) Normal rabbit serum was used as primary antibodies for negative control. (C,D) Positive staining of the antigen (albumin) was recognized with a brown color. e, epidermis; d, dermis; gr, granular glands; mu, mucous glands.
Discussion
In mammals, albumin is normally synthesized by liver cells and secreted into bloodstream (Peters 1996). For the first time, albumin has been characterized and cloned from the skin of amphibian B. maxima. BmA-skin is not secreted into the skin secretions, since Western blotting analysis could not detect its existence in the secretions. Accordingly, the protein was not detectable in granular and mucous glands in immunochemical study (Fig. 5 ▶). Meanwhile, BmA-serum was also purified and cloned from the liver. In PCR cloning of BmA-serum, several pairs of specific primers located in different conserved regions of the protein were used. Eventually, only one albumin protein was cloned from the liver. In addition, BmA-skin and BmA-serum cDNAs differ only in two nucleotides in protein coding regions, resulting in two amino acid mutations. Consequently, BmA-serum shares similar biochemical and immunochemical properties as those of BmA-skin. It is possible that the minor sequence variations found in the skin and the serum proteins are due to allelic variation of a single gene in the frog.
HSA comprises three homologous albumin domains that assemble to form a heart-shaped molecule. Each domain contains five or six internal disulphide bonds (He and Carter 1992; Curry et al. 1998). Similarly, sequence determination established that mature B. maxima albumin is composed of 585 amino acids organized into three internally homologous albumin domains. The number and spacing of cysteine residues in B. maxima albumin are virtually identical to those in mammalian albumins, and by homology, it could be predicted that they are paired in the same pattern.
To our knowledge, there are no reports in the literature concerning albumin proteins acting directly on serine proteases as potent inhibitors. Present studies show that B. maxima albumin possesses a potent and timestable trypsin inhibitory activity. According to different inhibition mechanisms, three different types of serine protease inhibitors can be distinguished: canonical inhibitors, noncanonical inhibitors, and serpins. The canonical inhibitors usually react with cognate enzymes and bind to the enzymes through the exposed and convex binding loop, in which is located the reactive site P1–P1′ peptide bond, according to a common, substrate-like standard mechanism. The reactive site can be selectively hydrolyzed by the enzyme (Bode and Huber 1992, 2000; Laskowski and Qasim 2000). Serpins are larger proteins of ~350 amino acids, occurring in plasma. They are irreversible covalent “suicide” protease inhibitors, interacting with their target protease through a flexible and exposed binding loop in a substrate-like mechanism (Ye and Goldsmith 2001). B. maxima albumin binds noncovalently to trypsin through a short exposed loop, in which is located the reactive site Arg58(P1)–His59(P1′), in a substrate-like mechanism. This is somehow similar to canonical inhibitors. However, it is noteworthy that the smaller size of the loop in B. maxima albumin is itself unusual and not typical of other canonical inhibitors whose loops are much longer (Bode and Huber 2000; Laskowski and Qasim 2000). In addition, while typical canonical serine protease inhibitors are capable of being cleaved by their target enzymes, this cleavage is slow and the dissociation of the complex typically results in regeneration of the native inhibitor. In this line, B. maxima albumin is further distinguished from typical canonical inhibitors by a rapid cleavage at the scissile bond, which accounted for about half of the inhibitor in the equilibrium achieved, and by the tight and stable binding capability of the product formed with the enzyme (Fig. 4C,D ▶).
Based on the inhibitory mechanism that B. maxima albumin adopted to inhibit trypsin, it is interesting to compare the albumin sequences from different sources. At variance with B. maxima albumin, one mutation of Arg58 to Ser58 occurred in HSA and BSA. This may well explain the lack of trypsin inhibitory activity of BSA. In a preliminary study, it was found that purified X. laevis 68-kDa serum albumin also showed significant trypsin inhibitory activity, but the activity was about fourfold lower than that of B. maxima albumin (Y.X. Zhang, W.H. Lee, and Y. Zhang, unpubl.). This might be caused by sequence variations in the region comprising the scissile bond, since the intermolecular interactions of subsites elongated on both sides of the scissile bond are very important in the mechanism of inhibition of serine proteases by protease inhibitors (Bode and Huber 1992). By possessing potent trypsin inhibitory activity, B.maxima albumin could function directly or indirectly as a defensive substance against predators, similar to those of many protease inhibitors in plants serving a defensive function against insect infestation (Laskowski and Qasim 2000), implicating the species’ adaptation to unique environments.
The expression of albumin in skin was also observed in other frog species, such as X. laevis and Bufo andrewsi (Y.X. Zhang, W.H. Lee, and Y. Zhang, unpubl.). It would be interesting to know the biological roles of rich accumulation of albumin in frog skin. Albumin is characterized of playing important roles in the binding and transport of water, cations (such as Ca2+, Na+, and K+), fatty acids, hormones, and metabolites, as well as in maintaining colloidal osmotic pressure (Peters 1996; Kragh-Hansen et al. 2002). Reasonably, wide distribution of such a protein in skin, especially in epidermis, may greatly facilitate the uptake and accumulation of environmental substances needed for frog survival.
There are essentially five residues, Tyr161, Ile142, Tyr138, His146, and Lys190, that are involved in haem binding in HSA (Wardell et al. 2002). In B. maxima albumin, all of these residues are either conserved (Tyr138, His146) or substituted with closely homologous residues, Ile142 to Val, Tyr161 to Phe, and Lys190 to His. Although BmA-skin and BmA-serum possess the same potential haem binding site, an interesting observation at present study is that the binding of a haem b in B. maxima albumin accumulated in the skin, compared with the protein circulated in the serum (0.95 vs. 0.05 mol/mol protein). It has been well documented that cutaneous gas exchange is highly important in all active (as contrasted with resting or dormant) amphibians (Duellman and Trueb 1986). One of possible functions of haem b cofactor bound in BmA-skin might be the participation of the protein in the cutaneous gas exchange of the frog. Even then, there has been no report on the reduction of hemin to heme and the oxygenation of the heme bound to HSA. It has been shown that HSA incorporated with eight molecules of synthetic heme derivative with a covalently bound proximal base is able to reversibly bind and release O2 under physiological conditions (pH 7.3, 37°C) such as hemoglobin (Hb) or myoglobin (Mb) (Tsuchida et al. 1999). It remains to be investigated whether the haem bound in BmA-skin is able to bind oxygen and what its physiological role in frog skin respiration is.
The elucidation of expression and accumulation of albumin in amphibian skin provides useful insights into our understanding of molecular basis of amphibian skin physiological functions, including water economy, metabolite exchange, and osmoregulation. B. maxima albumin also offers a new possibility for the studies related to the structure/function of albumin proteins, including the binding of a haem in the protein and its physiological implications. Being the first member of a novel class of trypsin inhibitors, B. maxima albumin should be a useful tool in the study of protease–inhibitor interactions.
Materials and methods
Materials
Chromogenic substrate H-d-Phe-Pip-Arg-pNA (S-2238) was from Kabi Vitrum. L-Benzoyl-L-arginine-pNA (BApNA), N-Succinyl- Ala-Ala-Ala-pNA (S-4760), N-CBZ-Gly-Gly-Leu-pNA (C-3022), N-Benzoyl-L-tyrosine ethyl ester (BTEE), and 3,3′-diaminobenzidine tetrahydrochloride (DAB) enhance liquid substrate system for immunohistochemistry were from Sigma. Horseradish peroxidase-conjugated goat anti-rabbit IgG polyclonal antibodies were from Santa Cruz Biotechnology. Human thrombin, porcine elastase, bovine α-chymotrypsin (type II), Bacillus licheniformis subtilisin, BSA, and normal goat serum were from Sigma. The protein concentration was determined by a protein assay kit (Bio-Rad) with BSA as a standard. Bovine pancreatic trypsin (type I) was purchased from Sigma. To determine themolar concentration of the active site enzyme, the trypsin was subjected to active site titration with p-nitrophenyl-p′-guanidinobenzoate hydrochloride (Chase and Shaw 1967), using a Lambda Bio 40 UV/VIS spectrometer (Perkin-Elmer). Titration was performed in 0.1 M veronal buffer (pH 8.6), containing 0.02 MCaCl2. Titration of the active site of the trypsin gave a ratio of active site concentration to protein concentration of 0.97 ± 0.02, indicating that the trypsin was fully active.
Preparation of frog skin homogenate
Adult specimens of B. maxima of both sexes (n=6; weight range 30–50 g) were collected in Chuxiong County, Yunnan Province, in southwest China. The frog was anesthetized with ether, the blood was collected by a cardiac puncture, and then the skin was peeled and washed in 50 mM Tris-HCl buffer (pH 7.8), containing 5 mM EDTA and 0.1 M NaCl. The skin was cut into small pieces, and homogenized in the same buffer. The homogenate was centrifuged 5000g for 20 min. The supernatant was collected, lyophilized, and stored in −80°C. The coagulated blood was centrifuged 3000g for 30 min to collect the serum.
Electrophoretic studies
SDS-PAGE and native PAGE were performed as reported (Laemmli 1970). For SDS-PAGE, samples were pretreated in 2.5% SDS alone (nonreducing conditions) or in 2.5% SDS and 5% β-mercaptoethanol (reducing conditions) for 5 min at 100°C. Gels were stained with 0.1% Coomassie brilliant blue R-250 in methanol/acetic acid/water (3/1/6) or silver-stained (Morrissey 1981).
Determination of bound haem b
The assay was performed as described (Berry and Trumpower 1987). Briefly, 0.5 mL of a solution containing 0.2 M NaOH and 40% (v/v) pyridine and K3Fe(CN)6 (0.6 mM) was placed in a 1-mL cuvette. A 0.5 mL aliquot of BmA-skin (15 μM) was added and mixed thoroughly, and the oxidized spectrum was recorded from 350 to 700 nm; 1.5 mg of solid sodium dithionite was added to record the reduced spectrum.
Serine protease inhibition assays
The inhibitory effects of the sample tested on the hydrolysis of synthetic chromogenic substrates by serine proteases were assayed in 50 mM Tris-HCl buffer, containing 1 mM CaCl2, at pH 7.8 (for trypsin, thrombin, chymotrypsin, and elastase) or pH 8.45 (for subtilisin) at 25°C. The protease (final concentrations 30 nM trypsin, 14 nM thrombin, 40 nM chymotrypsin, 463 nM elastase, or 30 nM subtilisin) and different amounts of the inhibitor (final concentrations ranging from 0.01 to 5 μM) were preincubated for 15 min at 25°C. BApNA (final concentration 2 mM) and S-2238 (0.02 mM) were used as a substrate for trypsin and thrombin, respectively; S-4760 (0.2 mM), for elastase; and C-3022 (0.2 mM), for subtilisin. The reaction was initiated by the addition of the substrate. The formation of p-nitroaniline (pNA) was monitored continuously at 410 nm for 2 min. In the case of chymotrypsin, BTEE (0.3 mM) was used as the substrate, and the reaction was monitored at 253 nm. For analysis of time stability of the inhibition on trypsin activity, trypsin (30 nM) was incubated with the inhibitor (45 nM) at 25°C for various times and then the residual amidolytic activity of trypsin was measured as mentioned above.
Binding complex formation
BmA-skin (1.85 μM) or BSA (1.85 μM) was incubated with various amounts of trypsin at different molar ratios (1:0.1 to 1.25) for 15 min at 25°C. Alternatively, BmA-skin (2.5 μM) was incubated with trypsin (1.85 μM) at 25°C for different times. The samples were then analyzed by native PAGE and SDS-PAGE under reducing conditions. For gel filtration analysis of the binding complex, BmA-skin (15 μM) was incubated with trypsin at different molar ratios of 1:0, 0:1, 1:1, and 1:2, for 15 min at 25°C. The sample was loaded on a Sephadex G-100 (Pharmacia) column (2.6 × 50 cm) equilibrated with 50 mM Tris-HCl buffer (pH 7.5) containing 0.1 M NaCl, and eluted at a flow rate of 12 mL/h. Blue dextran was used to estimate the outer volume. The apparent molecular weight was estimated as described (Andrews 1964). The binding kinetic constants were determined by surface plasmon resonance with BIAcore 3000 (BIAcore AB). Trypsin, dissolved in 10 mM NaAc buffer (pH 5.0), was immobilized to a flow cell of a carboxymethyl-dextran CM5 sensor ship with an amine coupling kit according to the method provided by the manufacturer. The binding assay was performed with a constant flow rate of 35 μL/min at 25°C with BIAcore running buffer (10 mM HEPES [pH 7.4], containing 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P2O). BmA-skin (concentration in the range of 25 nM–2 μM) were injected for 3 min, and dissociation data were collected for 5 min. Regeneration after each cycle was performed by using 10 mM glycine-HCl (pH 2.3) for 1 min. The kinetic parameters were obtained by fitting of the sensorgrams to a 1:1 Langmuir binding isotherm with BIAevaluation software version 4.
Cloning of BmA-skin
On the basis of the determined N-terminal and internal peptide sequences of BmA-skin, two oligonucleotide primers, P1 and P6R, were designed for amplification of a cDNA internal fragment by PCR. Primer P1 (5′-GGNGA(AG)GTNTA(CT)A A(AG)AA(AG)GT-3′) is oriented in the sense direction and corresponds to the N-terminal residues 10–16 of the protein. Primer P6R (5′-CCTTCTGCACATGGCATCCT-3′) is oriented in the anti-sense direction and corresponds to the residues 422–428. Total RNA was prepared from the frog skin by a RNA extract kit (Invitrogen). Single-stranded cDNAs were prepared from the mRNAs contained in the total RNA (5 μg) by reverse transcriptase (Invitrogen), using an oligo-d(T)18 primer. A first amplification by PCR with the primers P1 and P6R was carried out by using Taq polymerase. The PCR products were subcloned into a pGEM-T vector (Promega).
From the cDNA fragment sequence obtained from the first PCR amplification, two BmA-skin specific primers, P9 (5′-TG GATACTATAGACTCCGAGG-3′) and P9R (5′- TCAAGTT GATGTTCATATCC-3′) were designed, which are oriented in the sense (P9) and the anti-sense (P9R) directions, corresponding to BmA-skin residues 16–23 and 353– 359, respectively. A directional frog skin cDNA library was constructed with a plasmid cloning kit (SuperScript Plasmid System, GIBCO/BRL) as described previously (Lee et al. 2005a). Primers P9 and P9R were used in a PCR-based method for high-stringency screening of full-length frog albumin clones from the library as described before (Zhang et al. 1995).
Cloning of BmA-serum
5′ and 3′ RACE-RT-PCRs were used to obtain the full-length cDNA sequence encoding BmA-serum from liver with kits of 5′ and 3′ RACE systems for rapid amplification of cDNA ends (Invitrogen). Total RNA was prepared from the frog liver. The reverse transcription was performed by using an adapter primer provided by the manufacturer, which has an additional poly(T)17 tail and contains the sequence of primer P0 (5′- GGCCACGCGTCGACTAGT-3′). A PCR amplification was then carried out by using primers P9 and P0 with Pfu DNA polymerase (Promega). In the case of 5′ RACE-RT-PCR amplification, two BmA-skin specific primers, P11R (5′-TT CTATTAGAATAGGATTTCC-3′) and P10R (5′-TCCATTTC ACAGAATTCTCCC-3′), corresponding to BmA-skin residues 479–485 and 459–465, respectively, were used. The reverse transcription was performed by using primer P11R, and the rest of the steps exactly followed the instructions provided by the manufacturer. The cDNA sequence of BmA-serum was confirmed by sequencing at least five independent PCR-amplified clones. The nucleotide sequence data reported in this article are available from GenBank database with accession numbers AY 885649 (BmA-skin) and AY885650 (BmA-serum).
Immunohistochemical study
Polyclonal rabbit anti-BmA-skin antibodies were raised. Skin tissues of the dorsal regions dissected from adult specimens of B. maxima were fixed in 70% ethanol and cut into 5-μm sections after paraffin embedding. Each section was blocked with 10% normal goat serum in PBS for 1 h at 37°C and then incubated with polyclonal anti-BmA-skin antibodies for 12 h at 4°C. Horseradish peroxidase-conjugated goat anti-rabbit IgG polyclonal antibodies were used as the secondary antibodies. The distribution of the antigen in the tissues was visualized by 3,3′-diaminobenzidine (DAB) substrate solution. The section was then counterstained with hematoxylin. For negative control, rabbit serum without immunization with BmA-skin was used instead of the anti-BmA-skin antibodies.
Electronic supplemental material
Amino acid sequence comparison of BmA-skin with those of X. laevis 68 kDa serum albumin (xP-albumin), HSA, and BSA is given. The HSA numbering system was used. Gaps have been introduced to optimize the sequence homology. Identical residues in all sequences are shown by asterisks. The P1-P1′ site of trypsin inhibition is boxed. The residues, which are important in haem binding in HSA (Wardell et al. 2002), are shaded. Two mutations of Gly471 to Asn and Ser569 to Asn are each caused by one base mutation (G to A), found in the cDNA from the liver, and are bolded and underlined.
Acknowledgments
This work was supported by the grants of Western Light, projects from the Chinese Academy of Sciences (Y.Z., W.H.L.), and grants from National Natural Science Foundation (30170195, 30470380, 30200044) and Yunnan Science and Technology Commission (2003C0066M).
Abbreviations
BmA-skin, B. maxima albumin from skin
BmA-serum, B. maxima albumin from serum
BSA, bovine serum albumin
HSA, human serum albumin
pNA, p-nitroanilide
RACE-RT-PCR, rapid amplification of cDNA ends-reverse transcription PCR
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051551105.
Supplemental material: see www.proteinscience.org
References
- Andrews, P. 1964. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem. J. 91 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, E.A. and Trumpower, B.L. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161 1–15. [DOI] [PubMed] [Google Scholar]
- Bode, W. and Huber, R. 1992. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 204 433–451. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structural basis of the endoproteinase–protein inhibitor interaction. Biochim. Biophys. Acta 1477 241–252. [DOI] [PubMed] [Google Scholar]
- Chase Jr., T. and Shaw, E. 1967. p-Nitrophenyl-p’-guanidinobenzoate HCl: A new active site titrant for trypsin. Biochem. Biophys. Res. Commun. 29 508–514. [DOI] [PubMed] [Google Scholar]
- Clarke, B.T. 1997. The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol. Rev. 72 365–379. [DOI] [PubMed] [Google Scholar]
- Clawson, G.A. 1996. Protease inhibitors and carcinogenesis: A review. Cancer Invest. 14 597–608. [DOI] [PubMed] [Google Scholar]
- Curry, S., Mandelkow, H., Brick, P., and Franks, N. 1998. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 5 827–835. [DOI] [PubMed] [Google Scholar]
- Duellman, W.E. and Trueb, L. 1986. Relationship with the environment. In Biology of amphibians (eds. W.E. Duellman and L. Trueb), pp. 197–228. McGraw-Hill, New York.
- Dugaiczyk, A., Law, S.W., and Dennison, O.E. 1982. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc. Natl. Acad. Sci. 79 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, L. 1999. Atlas of amphibians of China. Henan Science and Technology Press, Zhengzhou, China.
- He, X.M. and Carter, D.C. 1992. Atomic structure and chemistry of human serum albumin. Nature 358 209–215. [DOI] [PubMed] [Google Scholar]
- Hilger, C., Grigioni, F., De Beaufort, C., Michel, G., Freilinger, J., and Hentges, F. 2001. Differential binding of IgG and IgA antibodies to antigenic determinants of bovine serum albumin. Clin. Exp. Immunol. 123 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler, J.L. and Hoffmann, J.A. 2000. Signaling mechanisms in the antimicrobial host defence of Drosophila. Curr. Opin. Microbiol. 3 16–22. [DOI] [PubMed] [Google Scholar]
- Kragh-Hansen, U., Chuang, V.T., and Otagiri, M. 2002. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol. Pharm. Bull. 25 695–704. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lai, R., Liu, H., Lee, W.H., and Zhang, Y. 2002. Identification and cloning of a trypsin inhibitor from skin secretions of Chinese red-belly toad Bombina maxima. Comp. Biochem. Physiol. B 131 47–53. [DOI] [PubMed] [Google Scholar]
- Laskowski, M. and Qasim, M.A. 2000. What can the structures of enzyme-inhibitor complexes tell us about the structures of enzyme substrate complexes? Biochim. Biophys. Acta 477 324–337. [DOI] [PubMed] [Google Scholar]
- Lee, W.H., Li, Y., Lai, R., Li, S., Zhang, Y., and Wang, W. 2005a. Variety of antimicrobial peptides in the Bombina maxima toad and evidence of their rapid diversification. Eur. J. Immunol. 35 1220–1229. [DOI] [PubMed] [Google Scholar]
- Lee, W.H., Liu, S.B., Shen, J.H., Jin, Y., and Zhang, Y. 2005b. Cloning of bradykinin precursor cDNAs from skin of Bombina maxima reveals novel bombinakinin M antagonists and a bradykinin potential peptide. Regul. Pept. 127 207–215. [DOI] [PubMed] [Google Scholar]
- Mignogna, G., Pascarella, S., Wechselberger, C., Hinterleitner, C., Mollay, C., Amiconi, G., Barra, D., and Kreil, G. 1996. BSTI, a trypsin inhibitor from skin secretions of Bombina bombina related to protease inhibitors of nematodes. Protein Sci. 5 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey, J.H. 1981. Silver stain for proteins in polyacrylamide gels: A modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117 307–310. [DOI] [PubMed] [Google Scholar]
- Moskaitis, J.E., Sargent, T.D., Smith Jr., L., Pastori Jr., R.L., and Schoenberg, D.R. 1989. Xenopus laevis serum albumin: Sequence of the complementary deoxyribonucleic acids encoding the 68- and 74-kilodalton peptides and the regulation of albumin gene expression by thyroid hormone during development. Mol. Endocrinol. 3 464–473. [DOI] [PubMed] [Google Scholar]
- Peters Jr., T. 1996. All about albumin: Biochemistry, genetics, and medical application. Academic Press, New York.
- Rosengren, K.J., Daly, N.L., Scanlon, M.J., and Craik, D.J. 2001. Solution structure of BSTI: A new trypsin inhibitor from skin secretions of Bombina bombina. Biochemistry 40 4601–4609. [DOI] [PubMed] [Google Scholar]
- Tsuchida, E., Komatsu, T., Matsukawa, Y., Hamamatsu, K., and Wu, J. 1999. Human serum albumin incorporating tetrakis(o-pivalamido)phenylporphinatoiron( II) derivative as a totally synthetic O2-carrying hemoprotein. Bioconj. Chem. 10 797–802. [DOI] [PubMed] [Google Scholar]
- Wardell, M., Wang, Z., Ho, J.X., Robert, J., Ruker, F., Ruble, J., and Carter, D.C. 2002. The atomic structure of human methemalbumin at 1.9 Å. Biochem. Biophys. Res. Commun. 291 813–819. [DOI] [PubMed] [Google Scholar]
- Ye, S. and Goldsmith, E.J. 2001. Serpins and other covalent protease inhibitors. Curr. Opin. Struct. Biol. 11 740–745. [DOI] [PubMed] [Google Scholar]
- Zacharius, R.M., Zell, T.E., Morrison, J.H., and Woodlock, J.J. 1969. Gly coprotein staining following electrophoresis on acrylamide gels. Anal. Biochem. 30 148–152. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Wisner, A., Xiong, Y.L., and Bon, C. 1995. A novel plasminogen activator from snake venom: Purification, characterization, and molecular cloning. J. Biol. Chem. 270 10246–10255. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y., Wan, S.G., Wei, S.S., Lee, W.H., and Zhang, Y. 2005. Bm-TFF2, a trefoil factor protein with platelet activation activity from frog Bombina maxima skin secretions. Biochem. Biophys. Res. Commun. 330 1027–1033. [DOI] [PubMed] [Google Scholar]