Abstract
Glutathione transferases (GSTs) are a superfamily of enzymes that play a vital functional role in the cellular detoxification process. They catalyze the conjugation of the thiol group of glutathione (GSH) to the electrophilic groups of a wide range of hydrophobic substrates, leading to an easier removal of the latter from the cells. The κ class is the least studied one among various classes within the superfamily. We report here the expression, purification, and crystal structure of human κ class GST (hGSTK), which has been determined by the multiple-isomorphous replacement method and refined to 1.93 Å resolution. The overall structure of hGSTK is similar to the recently reported structure of κ class GST from rat mitochondrion. Each subunit of the dimeric hGSTK contains a thioredoxin (TRX)-like domain and a helical domain. A molecule of glutathione sulfinate, an oxidized product of GSH, is found to bind at the G site of each monomer. One oxygen atom of the sulfino group of GSF forms a hydrogen bond with the hydroxyl group of the catalytic residue Ser16. The TRX-like domain of hGSTK shares 19% sequence identity and structure similarity with human θ class GST, suggesting that the κ class of GST is more closely related to the θ class enzyme within the GST superfamily. The structure of the TRX-like domain of hGSTK is also similar to that of glutathione peroxidase (GPx), implying an evolutionary relationship between GST and GPx.
Keywords: glutathione transferase, crystal structure, active site, glutathione sulfinate, thioredoxin-like domain, glutathione peroxidase
Glutathione transferases (GSTs, EC 2.5.1.18), formerly known as glutathione S-transferases, are a superfamily of enzymes that play a vital role in cellular detoxification process. GSTs catalyze the conjugation of the thiol group of glutathione (GSH), the tripeptide γ-Glu-Cys-Gly, to the electrophilic groups of a wide range of hydrophobic substrates, resulting in greater solubility and easier removal of the hydrophobic substrate from the cells (Mannervik and Danielson 1988; Pickett and Lu 1989; Coles and Ketterer 1990; Armstrong 1991; Tsuchida and Sato 1992; Wilce and Parker 1994; Sheehan et al. 2001). GSTs have been the focus of considerable interest with regard to resistance toward drugs, insecticides, herbicides, and antibiotics. GSTs may also be involved in the intra-cellular storage and transport of a variety of hydrophobic, nonsubstrate compounds (Oakley et al. 1999). GSTs were reported to exhibit glutathione peroxidase (GPx, EC. 1.11.1.9) activity (Hurst et al. 1998; Jowsey et al. 2003), and human κ class GST (hGSTK) was recently identified to be localized in peroxisomes (Morel et al. 2004).
GSTs have been divided into an ever-increasing number of classes based on their amino acid sequence homology in combination with other criteria, such as tertiary structure similarity, substrate specificity, and immunological identity (Mannervik et al. 1992; Sheehan et al. 2001). GSTs generally share greater than 60%sequence identity within a class and less than 30%among distinct classes. Over the past years the three-dimensional structures of soluble GSTs in several classes have been reported (Ji et al. 1992, 1995; Dirr et al. 1994; Cameron et al. 1995; Rossjohn et al. 1998; Board et al. 2000; Polekhina et al. 2001). GSTs function as a dimeric enzyme, and the subunit structure adopts a similar canonical fold consisting of an N-terminal domain, assuming a topology similar to the thioredoxin (TRX) fold and a C-terminal domain comprising several α-helices (Sheehan et al. 2001). The κ class is the least studied one among various classes within the superfamily. Recently, the structure of κ class GST from rat mitochondrion (rGSTK) in complex with GSH was reported, which shows a folding topology different from that of the other GST classes (Ladner et al. 2004).
hGSTK is a homodimer; each monomer consists of 226 amino acids with a molecular mass of 26 kDa. hGSTK shares ~70% sequence identity with both rat and mouse κ class GSTs (Pemble et al. 1996; Morel et al. 2004). We report here the expression, purification, and crystal structure of hGSTK, which has been determined by the multiple-isomorphous replacement (MIR) method and refined to 1.93 Å resolution. The structure of rGSTK at 2.5 Å resolution was reported when the structure refinement of hGSTK was nearly complete. The overall structure of hGSTK is similar to that of rGSTK. At the active site of hGSTK, GSH was found to be in an oxidized state, namely glutathione sulfinate (GSF). Sequence alignment and structure comparison of the TRX-like domain of hGSTK with those of other classes of GSTs indicate that the κ class is more closely related to the θ class than to the other classes within the GST superfamily. The structure comparison of hGSTK with GPx is also discussed.
Results
Overall structure of hGSTK
The crystal structure of hGSTK in complex with GSF, refined to 1.93 Å resolution, yielded a final R-factor of 0.179 and a free R-factor of 0.203, respectively. The final structure model contains 1 homodimer of hGSTK, 2 GSF molecules, and 179 water molecules. Each subunit consists of 218 amino acid residues, and the N-terminal three residues and the C-terminal five residues could not be located due to weak electron density. The side chains of three residues (Met196 in both subunits and His75 in one subunit) exhibit dual conformations. Analysis of the stereochemistry of the protein model using the program PROCHECK (Laskowski et al. 1993) shows that 90.6% and 9.4% of the residues are in the most favored and additional allowed regions of the Ramachandran plot, respectively. The refinement statistics are summarized in Table 1. Figure 1, A and B ▶, shows the overall structure of hGSTK.
Table 1.
Statistics of diffraction data and structure refinement
| Native (form A) | Native (form B) | Hg (OAc)2 Derivative (form B) | K2Pt(CN)4 Derivative (form B) | |
| Diffraction data statistics | ||||
| Space group | C2 | P21212 | P21212 | P21212 |
| Unit cell parameters | ||||
| a (Å) | 98.28 | 224.86 | 224.16 | 224.35 |
| b (Å) | 118.27 | 88.16 | 88.03 | 88.10 |
| c (Å) | 52.59 | 53.95 | 53.87 | 53.88 |
| β (°) | 102.67 | |||
| No. of monomers/asymmetric unit | 2 | 4 | 4 | 4 |
| Resolution (Å) | 1.86 | 2.40a | 2.30 | 2.80 |
| No. of unique reflections | 48,426 | 42,746 | 46,959 | 25,931 |
| Rmerge (%)b | 4.7 (38.3)c | 19.4 (25.5)a | 10.9 (23.8)c | 11.9 (22.6)c |
| Completeness (%) | 99.3 (93.9)c | 99.0 (99.9)a | 96.9 (83.7)c | 95.9 (99.1)c |
| 〈I/σ(I) 〉 | 6.9 (1.3)c | 2.0 (1.9)a | 5.4 (2.1)c | 5.6 (2.9)c |
| ΔFiso (%)d | 14.9 | 9.1 | ||
| ΔFano (%)e | 4.3 | 3.4 | ||
| Refinement statistics | ||||
| No. of amino acid residues | 436 | |||
| No. of GSF | 2 | |||
| No. of solvent molecules | 179 | |||
| R factor (%)f | 17.9 | |||
| Free R factor (%) | 20.3 | |||
| Rms deviation | ||||
| Bond length (Å) | 0.005 | |||
| Bond angles (°) | 1.1 | |||
| Mean temperature factors (Å2) | ||||
| Main-chain atoms | 35 | |||
| Side-chain atoms | 39 | |||
| GSF | 25 | |||
| Solvent | 44 | |||
| Luzzati atomic positional error (Å) | 0.2 | |||
a For native crystal form B, X-ray data were collected to 2.4 Å resolution, but the data in the resolution range higher than 3.0 Å were not used. The numbers in parentheses correspond to the data in the resolution shell 3.02–3.25 Å.
bRmerge = ∑hkl ∑i| Ii (hkl ) − 〈 I(hkl ) 〉|/∑hkl ∑iIi(hkl), where Ii (hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the mean intensity for reflection hkl from multiple measurements.
c The numbers in parentheses correspond to the data in the highest resolution shell (1.86–1.93 Å , 2.30–2.38 Å , and 2.80–2.90 Å for native crystal from A, Hg derivative, and Pt derivative, respectively). For native crystal form A, the data in the shell (1.86–1.93 Å ) were not used for refinement, and 〈I/σ(I) 〉 in the shell (1.93–2.01 Å ) is 1.8.
d Isomorphous difference ratios between the heavy-atom derivatives (Fph) and the native (Fp) were calculated at 3 Å resolution. ΔFiso = 〈|| Fph |−|Fp|| 〉/〈 (|Fph | + |Fp|)/2〉
e Anomalous difference ratios of the heavy-atom derivatives were calculated at 3 Å resolution. ΔFano= 〈||Fph(+)| − | Fph (−)||〉 /〈(|Fph (+) |(+) |Fph (−) |)/2〉.
fR factor= ∑||Fo| − |Fc ||/∑|Fo|.
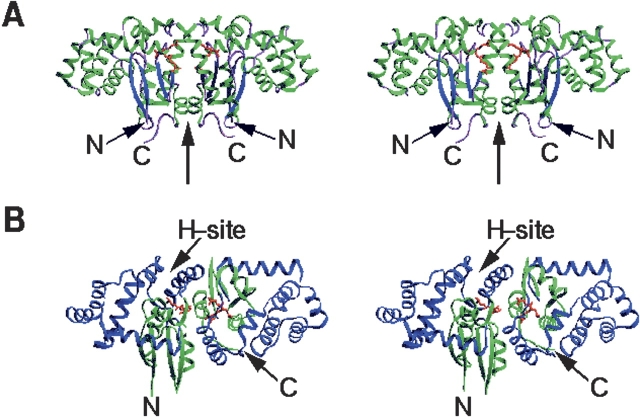
Figure 1.
Overall structure of the dimeric hGSTK. Two GSF molecules are shown as red ball-and-stick models. (A) View perpendicular to the twofold NCS axis (thick arrow), showing the butterfly-like shape of the dimer. The α-helices and β-sheets are shown in green and in blue, respectively. (B) View showing the binding cleft of the H-site. Domains I and II are shown in green and in blue, respectively. These diagrams were prepared using the program SETOR (Evans 1993).
hGSTK shares high sequence homology with rat and mouse GSTK (69.5% and 71.2% sequence identity, respectively, without insertion or deletion) (Morel et al. 2004), and the secondary structure of hGSTK is substantially the same as that of rGSTK (Ladner et al. 2004). The hGSTK monomer comprises two domains (Fig. 1B ▶). Domain I is composed of residues 4–52 and 184–221, with a TRX-like fold (Martin 1995) that is characterized by a βαβ (β1α1β2) motif and a ββα motif (β3β4α10) linked by an α-helix (α2) to form a four-stranded β-sheet surrounded by three α-helices. Domain II is composed of residues 60–178, which folds as seven α-helices (α3–α9). Domain II is inserted between the βαβ and ββα motifs of domain I. The two domains are connected together by two short linkers.
The tertiary structure of hGSTK is also similar to that of rGSTK. The major differences between the two structures are located in the regions of Asn53–Pro60 and Pro84–Lys94. The root-mean-square (RMS) deviation between the two structures is ~0.7 Å for all Cα atoms, and is ~0.5 Å if the aforementioned two regions were omitted in the superposition. In hGSTK residues Asn53–Pro60 form an exposed loop containing a short 310 helix (Pro55–Leu59); the corresponding part in rGSTK forms a β-turn. In hGSTK, the residues Pro84–Lys94 form a β-turn (Pro84–Phe87) and an α-helix (α4, Asp86–Cys93); the corresponding region in rGSTK forms an irregular α-helix. Both regions show low-temperature factors in hGSTK but high temperature factors in rGSTK (Ladner et al. 2004).
Dimer interface
The two subunits of the hGSTK dimer are related by a twofold noncrystallographic symmetry (NCS) axis (Fig. 1A ▶). Superposition of the two subunits yields an RMS deviation of 0.22 Å for all Cα atoms, indicating that the two subunits are in general identical. The dimer interface buries 1386 Å 2 solvent-accessible surface area of each subunit, corresponding to 13%of the surface area of the monomer. The dimer interface is dominated by hydrophobic interactions between residues from domain I of one subunit and domain II of the other, similar to the other classes of GSTs. In addition, a total of 19 salt bridges and hydrogen bonds are formed across the subunit interface, and two NCS-related aspartic acids (Asp201) make stacking interactions with each other at the dimer interface.
Structure of the active site
The physiological substrate GSH is bound at a hydrophilic cleft, designated as the G-site. Similar to the G-sites in other GSTs, the G-site in hGSTK is formed primarily by structural elements of the TRX-like domains from both subunits, including α1, a loop connecting β4 and α10, and the two small linkers connecting the TRX-like domain and the α-helical domain. The G site is located in the dimer interface region and is shielded from solvent by the other subunit.
In the hGSTK structure there is strong electron density at the G-site of each subunit, which shows unambiguously that the bound substrate has an oxidized thiol group (Fig. 2A ▶). In other words, the bound substrate is GSF, the sulfinated GSH. The GSF molecule is well ordered, with a mean B value of 25 Å 2 (Table 1), and is bound in an extended fashion in the G-site. The two GSF molecules of a dimer face each other (Fig. 1A ▶). The amide groups of the γ-glutamyl moieties of the two GSFs are 5.5 Å apart, which is the shortest distance between the two GSFs, and the carboxyl groups of the glycyl moieties point to each other with a distance of about 6–7 Å. This binding mode is similar to those observed in the structures of other GSTs in complexes with GSH or its analogs.
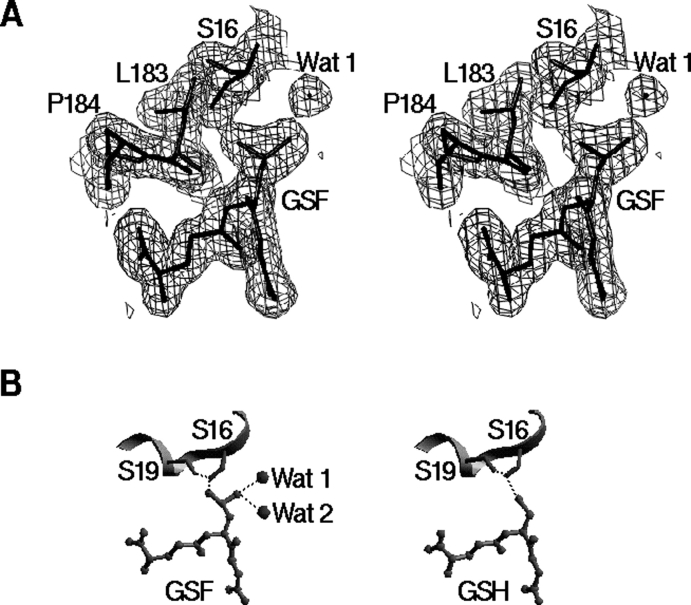
Figure 2.
Structure of the catalytic active G-site. (A) SIGMAA-weighted 2Fo–Fc electron density map (1σ contour level) at the G-site. This diagram was prepared using the program TURBO-FRODO. (B) Hydrogen- bonding interactions of the sulfino group of GSF with Ser16 of hGSTK (left), compared with those of the thiol group of GSH with Ser16 of rGSTK (right). Two water molecules bound to the sulfino group of GSF in hGSTK structure are shown as spheres. Hydrogen bonds are shown as dotted lines. This diagram was prepared using the program SETOR.
The bound GSF makes extensive hydrophilic (both hydrogen bonding and salt bridge) interactions with several residues nearby, which are conserved within the κ class. The γ-glutamyl moiety interacts with both the main chain and side chain of Ser200, the side chains of Asp201 and Arg202* (* denotes the residue of the adjacent subunit). The side chain of the glycyl moiety makes two direct interactions with the side chains of Asn53 and Lys62* and an indirect interaction with the side chain of Lys62 via a water molecule. The main-chain amide and carbonyl groups of the cysteinyl moiety form two hydrogen bonds with the main chain of Leu183. One oxygen atom of the sulfino group of the oxidized cysteinyl moiety is hydrogen bonded with the side chain Oγ atom of Ser16 and the main-chain amide group of Tyr18, and the other oxygen atom forms hydrogen bonds with two water molecules (Fig. 2B ▶). The Ser16 Oγ atom forms an additional hydrogen bond to the Ser19 Oγ atom. The sulfur atom of GSF is 3.7 Å away from the Ser16 Oγ atom, slightly beyond hydrogen-bonding distance.
Near the G-site there is an H-site for binding the hydrophobic substrate (Fig. 1B ▶). In hGSTK, residues from α3, α4, and α6 of domain II form the H-site, and several residues from α1 (along with the β-turn preceding it), α2, and the loop connecting α9 and β3 also make certain contributions to the H-site. Compared with rGSTK (Ladner et al. 2004), most of the residues forming the H-site are highly conserved in hGSTK.
Discussion
Oxidation state of the GSH substrate
In the rGSTK structure the GSH substrate is in a reduced state containing a thiol group, and the sulfur atom of GSH makes a hydrogen bond (2.9 Å ) with the Oγ atom of Ser16 (Fig. 2B ▶) (Ladner et al. 2004). Although the crystals of hGSTK were grown in the presence of GSH, the structure of hGSTK reveals that the bound substrate is GSF, an oxidized product of GSH. In order to accommodate the two additional oxygen atoms of GSF, both the side chain of Ser16 and the sulfur atom of GSF in the hGSTK structure are displaced apart by 0.6–0.7 Å , compared with those in the rGSTK structure. Consequently, the hydrogen bond between the sulfur atom of GSF and the Oγ atom of Ser16 is disrupted (3.7 Å ). Instead, one oxygen atom of the sulfino group of GSF forms two hydrogen bonds with the Ser16 Oγ atom (2.6 Å ) and the main-chain amide group of Tyr18 (3.0 Å ). The other oxygen atom of the sulfino group makes hydrogen bonds (2.9 Å ) with two water molecules (Fig. 2B ▶). The side chain of Ser16 also forms a hydrogen bond with the Oγ atom of Ser19 (2.7 Å ), which is also found in the rGSTK structure. This hydrogen-bonding network appears to stabilize the oxidized sulfino group of GSF.
It was reported that the turnover of the S16A mutant of rGSTK toward its electrophilic substrate 1-chloro- 2,4-dinitrobenzene (CDNB) was about 30-fold less efficient than the wild-type enzyme (Ladner et al. 2004), suggesting that Ser16 is essential for the catalysis. In the structure of rGSTK in complex with GSH, the hydroxyl group of Ser16 makes a hydrogen-bonding interaction with the sulfur atom of GSH, suggesting that it is likely protonated to stabilize the thiolate anion in the catalysis. In both the κ and θ classes of GSTs the catalytic Ser residue is strictly conserved, suggesting that these two classes might share a common catalytic mechanism. In most of the other classes of GSTs, including α, μ, and π classes, the catalytic residue is a tyrosine. In the ω class and bacterial β class of GSTs, a conserved cysteine forms a disulfide with the thiol group of GSH (Nishida et al. 1998; Board et al. 2000).
In the hGSTK structure, the bound substrate is in the oxidized state. The first possibility is that GSH is oxidized in the crystallization solution by oxygen in air, and the oxidized product GSF then binds the enzyme, acting as an inhibitor. The chemical reaction from RS− to RSO2− follows a radical-mediated mechanism involving the RSOO−* radical (Oae and Doi 1991).
Besides GSH-conjugating activity, GSTs can also serve as a peroxidase and is present in peroxisomes where oxygen free radicals, hydroxyl radicals, and hydrogen peroxides can be generated, as previously reported (Jowsey et al. 2003; Morel et al. 2004). The primary biological function of peroxidase enzymes is to oxidize a variety of hydrogen donors at the expense of peroxide or molecular oxygen (Forstrom et al. 1978). hGSTK exhibits low activity (0.173±0.06 and 0.015±0.001 μM · min−1 · mg−1, respectively) of GSH peroxidase towards cumene hydroperoxide and t-butyl hydroperoxide, respectively (Morel et al. 2004). The kinetic study showed a linear dependence of rate with concentration of 1-palmitoyl-2-(13-hydroperoxy-cis-9, trans-11-octadecadienoyl)-L-3-phosphatidylcholine (PLPH-OOH) and a catalytic specificity value Kcat/Km(PLPC-OOH) of 4.2 (μM−1 · S−1) for a recombinant α class GST (Hurst et al. 1998). The structure of hGSTK in complex with GSF suggests the second possibility; i.e., hGSTK catalyzes the oxidation of the substrate GSH by exerting its peroxidase activity, with oxygen in air as an oxidant, and GSF is the product of the enzymatic reaction, which is bound at the active center. However, the detailedmechanism is unclear.
The κ class of GSTs is closely related to the θ class enzyme
Although all classes of mammalian GSTs consist of a TRX-like domain and a helical domain, the secondary structure topology of the κ class GSTs differs substantially from those of the other classes of GSTs. In the other classes the two typical structural motifs (βαβ motif and ββα motif) of the TRX-like domain are linked together by a long loop containing an α-helix (α2), and the C-terminal helical domain consists of a varied number of α-helices (e.g., four α-helices in the π and the μ classes, five in the α class, and six in the θ class). The two domains are connected together by a short linker. However, in the κ class of GSTs the helical domain is inserted between the βαβ and ββα motifs of the TRX-like domain and contains seven α-helices. The dimer of hGSTK adopts a butterfly-like shape with wide wings (Fig. 1A ▶), while in the other classes there is a deep V-shape crevice in the intersubunit interface of the dimer, which does not exist in the κ class enzymes.
It was previously known that the entire sequence of hGSTK showed an absence of sequence homology with any other class of GST, and rGSTK was reported to have limited identity with the θ class at the N terminus (over residues 6–16) (Harris et al. 1991; Pemble et al. 1996; Morel et al. 2004). However, the sequence alignment of the TRX-like domain between hGSTK and various classes of GSTs indicates that the secondary structural elements of this domain can be aligned reasonably well with each other (Fig. 3A ▶). This secondary structure-based alignment reveals three strictly conserved amino acid residues (Pro184, Asp191, and Gly192 in hGSTK) and four highly conserved residues (Leu10, Tyr12, Leu26, and Leu206 in hGSTK) among various classes of GSTs. Pro184 adopts a conserved cis-configuration in all known GST structures (Sheehan et al. 2001). This residue is located in the vicinity of the G site (the shortest distances from Pro184 to both GSF and Ser16 are ~4.5 Å in the hGSTK structure) and right after Leu183, the main chain of which forms a pair of hydrogen bonds with the backbone of the cysteinyl moiety of GSF (Fig. 2A ▶). These hydrogen-bonding interactions are conserved in the structures of all GSTs in complexes with GSH or its analogs. It seems likely that the cis-configuration of Pro184 is required for maintaining Leu183 at a position favorable for binding the GSH substrate.
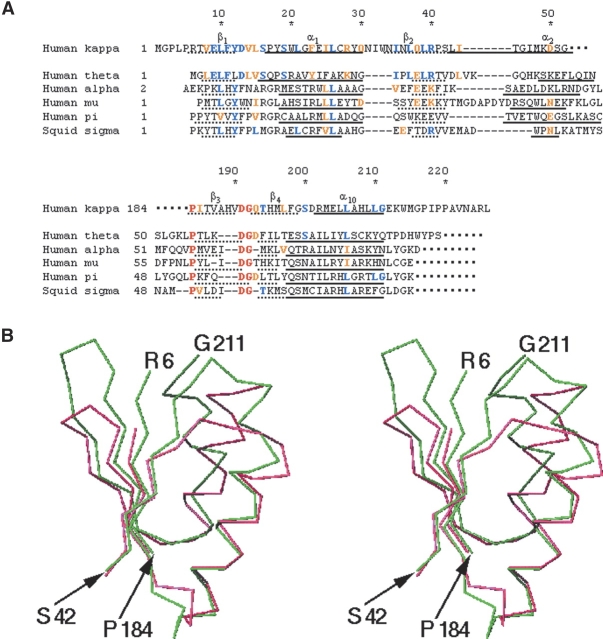
Figure 3.
Comparison of the TRX-like domain of hGSTK with other GSTs. (A) Sequence alignment of the TRX-like domains between hGSTK and the representatives of other classes of GSTs. The GSTs used in comparison are human θ GST (PDB entry code 1LJR), human α GST (1K3L), human μ GST (1HNA), human π GST (5GSS), and squid sigma GST (1GSQ). The residue numbers of hGSTK are shown on top. Residues strictly conserved in all classes are shown in red, and the residues identical to and conserved with those of hGSTK in blue and orange, respectively. (B) The Cα atom superposition of the TRX-like domains of hGSTK (Arg6–Ser42 and Pro184–Gly211 in green) and human θ class GST (Leu3–Val33 and Pro55–Gln79 in pink). This diagram was prepared using the program SETOR.
Moreover, the structure-based alignment of the TRX-like domain indicates that this domain of hGSTK shares 15 identical residues (19%) and additional eight conserved substitutions (10%) with that of human θ class GST (hGSTT), while the identical and conserved residues together are in the range of 15%–19% between hGSTK and the GSTs in any other class (α, μ, π, and σ) (Fig. 3A ▶). Superposition of the TRX-like domains of other classes of GSTs with hGSTK shows that they share structure similarity in this domain except for α2, yielding an RMS deviation of 1.8 Å between κ and θ classes for 56 corresponding Cα atoms (Fig. 3B ▶) and of 2.0–2.1 Å between κ and either of α, μ, π, and σ classes for 55–57 Cα atoms (the calculation is always limited to the βαβ and ββα motifs in this paper). The conformation of the segment Asp13– Trp20 of hGSTK where the catalytic residue Ser16 is located is similar to the corresponding region of the θ class and very different from those of the other classes. In addition, both κ and θ classes of GSTs lack some common features found in other classes of GSTs, such as the pronounced V-shape crevice and a “key-and-lock motif” in three-dimensional structures (Sheehan et al. 2001). Both κ and θ classes have a serine residue as the catalytic active site. The enzymatic activity of the κ classGSTs is limited to conjugation with CDNB and ethacrynic acid (Harris et al. 1991), which is also similar to the θ class GSTs. Based on the structural comparison of rGSTK with members of the canonical GST superfamily, Ladner and coworkers (Ladner et al. 2004) proposed that the protein folds of GSTs diverge from a common thioredoxin/glutaredoxin progenitor via parallel evolutionary pathways with a domain insertion for the former and a domain addition for the latter. Our results further suggest that the κ class is more closely related to the θ class than to the other classes within the GST superfamily.
rGSTK was reported to be more closely related to DsbA, a disulfide bond protein, than to other classes of GSTs (Ladner et al. 2004). The folding topology of DsbA is similar to κ class GSTs, with a TRX-like domain interrupted by a helical domain containing six helices. When the entire sequence of hGSTK is aligned with that of DsbA, α2–α4 of hGSTK corresponds to a 44-residue deletion in DsbA, and the two complete sequences share 10% identity and 6% conservative changes, approximately half in each domain, indicating that the sequence homology between hGSTK and DsbA is higher for the entire sequence and lower for the TRX-like domain, compared with those between hGSTK and any other class ofGSTs. The superposition of the TRXlike domain between hGSTK and DsbA gives an RMS deviation of 2.3 Å for 65 corresponding Cα atoms, larger than those between different classes of GSTs.
Structure comparison of hGSTK with GPx
Although hGSTK shows low activity of a peroxidase, it differs from GPx, which belongs to the selenoprotein family and functions to catalyze the reduction of hydroperoxides using GSH as a reducing substrate (Forstrom et al. 1978). GPx is a tetrameric enzyme, and its subunit structure contains a TRX-like fold and the catalytic site is a selenocysteine (Ren et al. 1997). The two types of enzyme, GST and GPx, share no sequence homology, and are dissimilar in overall structure of the entire sub-unit, but they exhibit similarity in the thioredoxin-like fold structure except for α2, with an RMS deviation of 1.6 Å for 65 Cα atoms when hGSTK is superimposed with bovine erythrocyte GPx (Epp et al. 1983). The relative positions of the catalytic residues in the TRX-like folds in the two enzymes (Ser16 in hGSTK and a selenocysteine in the GPx) are well conserved, which accounts for the recently reported results that incorporation of selenocysteine into GSH-specific binding scaffold using auxotrophic expression system converted the Lucilia cuprina GST to a selenium-containing enzyme that displayed the GPx activity comparable with that of natural GPx, which provided a proof that GST and GPx were evolved from a common “glutathione-binding protein” ancestor (Yu et al. 2005).
Materials and methods
Protein expression and purification
The cDNA corresponding to hGSTK was obtained from the cDNA library of human CD34+ hematopoietic stem/progenitor cells (Zhang et al. 2000). The hGSTK gene was cloned into the NdeI and XhoI restriction sites of the pET- 22b(+) expression plasmid (Novagen) and fused with a hexa-histidine tag at the C terminus. The plasmid was transformed into and expressed in Escherichia coli BL21(DE3) strain (Novagen). The cells transformed with the vector were grown over-night at 310 K in 100 mL of LB medium containing ampicillin (0.1 mg/mL). The culture was added to 2 L of LB medium and incubated until OD600 reached 0.7. After 3 h of expression induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 303 K, the cells were collected by centrifugation at 4000g and suspended in 40 mL of lysis buffer (pH 7.4) containing 50 mMNaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mM EDTA, 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. The cells were lysed on ice by sonication and the cell debris was precipitated by centrifugation at 15,000g.
The hGSTK protein was purified by affinity chromatography using a nickel-nitrilotriacetic acid-agarose column (Qiagen). The lysis extract was loaded on the column and then washed with a washing buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole [pH 7.4]) to elute nonspecific binding proteins. The target protein was eluted with an elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole [pH 7.4]). The fractions containing the hGSTK protein were pooled together and dialyzed extensively against a storage buffer containing 20 mM NaH2PO4 (pH 7.2), 20 mM NaCl, 1 mM EDTA, and 1 mM DTT. Reducing SDS-PAGE analysis of the purified protein showed a single band at 26 kDa. Dynamic light scattering analysis indicated that the protein was a homogeneously dispersed homodimer in both the presence and absence of a substrate (data not shown). The purified protein was further concentrated to about 40 mg/mL in the storage buffer for crystallization experiments. All purification steps were carried out at 277 K.
Crystallization and diffraction data collection
Two forms of hGSTK crystals were grown by the hanging drop vapor diffusion method. Crystals of form A grew at 293 K: A 2 μL protein solution at a concentration of 40 mg/mL (20 mM NaH2PO4 [pH 7.2], 20 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 mM GSH) was mixed with 2 μL reservoir solution containing 10% PEG8000, 10% PEG1000, and 3% glucose. Crystals of form B grew at 277 K: A 2 μL protein solution at a concentration of 10 mg/mL was mixed with 2 μL reservoir solution containing 0.1 M MES (pH 6.0), 10% dioxane, 1.6 M (NH4)2SO4, and 0.01 M trimethylamine.
Native X-ray diffraction data of both crystal forms were collected using a MarCCD detector at Beijing Synchrotron Radiation Facility, to 1.86 Å resolution at 293Kand to 2.4 Å resolution at 100 K for crystals of form A and form B, respectively, and the latter data in the resolution range higher than 3.0 Å were not used because of anisotropy of diffraction. The data were processed using the program AUTOMAR (Klein and Bartels 2000).
To determine the initial phases, two heavy-atom derivatives were prepared by soaking the native crystals of form B in Hg(OAc)2 and K2Pt(CN)4 (0.1 M for 9 h), respectively. The derivative data were collected at 100 K using an in-house Rigaku R-Axis IV++ image-plate detector. Diffraction data processing was performed using the CrystalClear package (Pflugrath 1999). Structure factors of the derivative data were subsequently scaled together with the native data (crystal form B) using the CCP4 program suite (Collaborative Computational Project 1994).
The crystal data and the data collection statistics are summarized in Table 1.
Structure determination and refinement
The initial phases of crystal form B were solved by MIR using the program SOLVE (Terwilliger and Berendzen 1999). The Patterson functions of the two heavy-atomderivatives at 3Å resolution revealed eight Hg atoms and one Pt atom, yielding an overall Z-score of 21.5 and amean figure of merit (FOM) of 0.41. The initial phases were further improved by statistical density modification using the program RESOLVE, yielding an overall FOM of 0.75 (Terwilliger 2002). The RESOLVE program automatically built 673 polyalanine residues out of 904 (226×4) residues in an asymmetric unit and successfully located most of the secondary structural elements. Chain was easily traced, and an initial model was built for one of the four subunits of crystal form B using program O (Jones et al. 1991) and TURBO-FRODO (Roussel and Cambillau 1991). This initial model was then used as the search model to solve the phases of crystal form A by molecular replacement using program AmoRe (Navaza 1994), which produced two outstanding peaks, corresponding to the two subunits in an asymmetric unit of crystal form A.
Crystallographic refinement was performed using the program CNS (Brünger et al. 1998) against the native data of crystal form A, since the crystal in this form diffracts better and contains less molecules in an asymmetric unit than crystal form B. A bulk solvent correction was applied throughout the refinement. Manual model building was carried out using TURBO-FRODO based on SIGMAA-weighted difference Fourier maps (2Fo–Fc and Fo–Fc) and composite omit maps. NCS restraints were imposed during the course of refinement up to 2.2 Å resolution, and were released in the later stage of refinement. In the initial difference Fourier maps there was strong residual electron density at the G-site, which was unambiguously interpreted to be GSF. Water molecules were included in the structure model in the late stage of refinement.
Coordinates
The atomic coordinates of the human κ class glutathione transferase have been deposited with the Protein Data Bank under accession code 1YZX.
Acknowledgments
We are grateful to Drs. Yuhui Dong and Peng Liu of Beijing Synchrotron Radiation Facility, Institute of High Energy Physics, China, for their help with synchrotron data collection. We thank Qiuhua Huang of Shanghai Institute of Hematology, Rui-Jin Hospital, Shanghai Second Medical University, for providing the hGSTK plasmid. We are also grateful to other members of our groups for their technical support and helpful discussion. This work was supported by the National Natural Science Foundation of China Grants 30125011, 30170223, and 30130080 to J.D.; the Ministry of Science and Technology of China Grants 2002BA711A13 to both Z.X and J.D., and 2004AA235091 and 2004CB520801 to J.D.; and the Chinese Academy of Sciences Grant KSCX1-SW-17 to J.D.
Abbreviations
GST, glutathione transferase
hGSTK, human κ class GST
rGSTK, rat mitochondrial κ class GST
GSH, glutathione
GSF, glutathione sulfinate
TRX, thioredoxin
CDNB, 1-chloro-2,4-dinitrobenzene
DTT, dithiothreitol
MIR, multiple-isomorphous replacement
NCS, noncrystallographic symmetry
RMS, root-mean-square
GPx, glutathione peroxidase
PEG, polyethylene glycol.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051463905.
References
- Armstrong, R.N. 1991. Glutathione S-transferases: Reaction mechanism, structure, and function. Chem. Res. Toxicol. 4 131–140. [DOI] [PubMed] [Google Scholar]
- Board, P.G., Coggan, M., Chelvanayagam, G., Easteal, S., Jermiin, L.S., Schulte, G.K., Danley, D.E., Hoth, L.R., Griffor, M.C., Kamath, A.V., et al. 2000. Identification, characterization, and crystal structure of the ω class glutathione transferases. J. Biol. Chem. 275 24798–24806. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse- Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography and NMR systems: A new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Cameron, A.D., Sinning, I., L’Hermite, G., Olin, B., Board, P.G., Mannervik, B., and Jones, T.A. 1995. Structural analysis of human α-class glutathione transferase A1–1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate. Structure 3 717–727. [DOI] [PubMed] [Google Scholar]
- Coles, B. and Ketterer, B. 1990. The role of glutathione and glutathione transferases in chemical carcinogenesis. Crit. Rev. Biochem.Mol. Biol. 25 47–70. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Dirr, H., Reinemer, P., and Huber, R. 1994. Refined crystal structure of porcine class π glutathione S-transferase (pGST P1–1) at 2.1 Å resolution. J. Mol. Biol. 243 72–92. [DOI] [PubMed] [Google Scholar]
- Epp, O., Ladenstein, R., and Wendel, A. 1983. The refined strucutre of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur. J. Biochem. 133 51–69. [DOI] [PubMed] [Google Scholar]
- Evans, S.V.1993. SETOR: Hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11 134–138. [DOI] [PubMed] [Google Scholar]
- Forstrom, J.W., Zakowski, J.J., and Tappel, A.L. 1978. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry 17 2639–2644. [DOI] [PubMed] [Google Scholar]
- Harris, J.M., Meyer, D.J., Coles, B., and Ketterer, B. 1991. A novel glutathione transferase (13–13) isolated from the matrix of rat liver mitochondria having structural similarity to class θ enzymes. Biochem. J. 278 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, R., Bao, Y., Jemth, P., Mannervik, B., and Williamson, G. 1998. Phospholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases. Biochem. J. 332 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X., Zhang, P., Armstrong, R.N., and Gilliland, G.L. 1992. The three-dimensional structure of a glutathione S-transferase from the μ gene class. Structural analysis of the binary complex of isoenzyme 3–3 and glutathione at 2.2-Å resolution. Biochemistry 31 10169–10184. [DOI] [PubMed] [Google Scholar]
- Ji, X., von Rosenvinge, E.C., Johnson, W.W., Tomarev, S.I., Piatigorsky, J., Armstrong, R.N., and Gilliland, G.L. 1995. Three-dimensional structure, catalytic properties, and evolution of a σ class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods. Biochemistry 34 5317–5328. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Jowsey, I.R., Thomson, R.E., Orton, T.C., Elcombe, C.R., and Hayes, J.D. 2003. Biochemical and genetic characterization of a murine class κ glutathione S-transferase. Biochem. J. 373 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, C. and Bartels, K. 2000. Automar, marFLM, marHKL and marXDS: Data reduction software for mar detectors. Acta Crystallogr. A 56 S295. [Google Scholar]
- Ladner, J.E., Parsons, J.F., Rife, C.L., Gilliland, G.L., and Armstrong, R.N. 2004. Parallel evolutionary pathways for glutathione transferases: Structure and mechanism of the mitochondrial class κ enzyme rGSTK1–1. Biochemistry 43 352–361. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Mannervik, B. and Danielson, U.H. 1988. Glutathione transferases— Structure and catalytic activity. CRC Crit. Rev. Biochem. 23 283–337. [DOI] [PubMed] [Google Scholar]
- Mannervik, B., Awasthi, Y.C., Board, P.G., Hayes, J.D., Di Ilio, C., Ketterer, B., Listowsky, I., Morgenstern, R., Muramatsu, M., Pearson, W.R., et al. 1992. Nomenclature for human glutathione transferases. Biochem. J. 282 305–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J.L. 1995. Thioredoxin—A fold for all reasons. Structure 3 245–250. [DOI] [PubMed] [Google Scholar]
- Morel, F., Rauch, C., Petit, E., Piton, A., Theret, N., Coles, B., and Guillouzo, A. 2004. Gene and protein characterization of the human glutathione S-transferase κ and evidence for a peroxisomal localization. J. Biol. Chem. 279 16246–16253. [DOI] [PubMed] [Google Scholar]
- Nishida, M., Harada, S., Noguchi, S., Satow, Y., Inoue, H., and Takahashi, K. 1998. Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: Catalytic roles of Cys10 and His106. J. Mol. Biol. 281 135–147. [DOI] [PubMed] [Google Scholar]
- Oae, S. and Doi, J.T. 1991. Organic sulfur chemistry: Structure and mechanism. CRC Press, Boca Raton, FL.
- Oakley, A.J., Lo Conte, L., Nuccetelli, M., Mazzetti, A.P., and Parker, M.W. 1999. The ligandin (non-substrate) binding site of human π class glutathione transferase is located in the electrophilic binding site (H-site). J. Mol. Biol. 291 913–926. [DOI] [PubMed] [Google Scholar]
- Pemble, S.E., Wardle, A.F., and Taylor, J.B. 1996. Glutathione S-transferase class κ: Characterization by the cloning of rat mitochondrial GST and identification of a human homologue. Biochem. J. 319 749– 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath, J.W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55 1718–1725. [DOI] [PubMed] [Google Scholar]
- Pickett, C.B. and Lu, A.Y. 1989. Glutathione S-transferases: Gene structure, regulation, and biological function. Annu. Rev. Biochem. 58 743– 764. [DOI] [PubMed] [Google Scholar]
- Polekhina, G., Board, P.G., Blackburn, A.C., and Parker, M.W. 2001. Crystal structure of maleylacetoacetate isomerase/glutathione transferase ζ reveals the molecular basis for its remarkable catalytic promiscuity. Biochemistry 40 1567–1576. [DOI] [PubMed] [Google Scholar]
- Ren, B., Huang, W., Akesson, B., and Ladenstein, R. 1997. The crystal structure of seleno-glutathione peroxidase from human plasma at 2.9 Å resolution. J. Mol. Biol. 268 869–885. [DOI] [PubMed] [Google Scholar]
- Rossjohn, J., McKinstry, W.J., Oakley, A.J., Verger, D., Flanagan, J., Chelvanayagam, G., Tan, K.L., Board, P.G., and Parker, M.W. 1998. Human θ class glutathione transferase: The crystal structure reveals a sulfate-binding pocket within a buried active site. Structure 6 309–322. [DOI] [PubMed] [Google Scholar]
- Roussel, A. and Cambillau, C. 1991. TURBO-FRODO, Silicon Graphics geometry partners dictionary. Silicon Graphics, Mountain View, CA.
- Sheehan, D., Meade, G., Foley, V.M., and Dowd, C.A. 2001. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. 2002. Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 58 1937–1940. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, S. and Sato, K. 1992. Glutathione transferases and cancer. Crit. Rev. Biochem. Mol. Biol. 27 337–384. [DOI] [PubMed] [Google Scholar]
- Wilce, M.C. and Parker, M.W. 1994. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta 1205 1–18. [DOI] [PubMed] [Google Scholar]
- Yu, H.-j., Liu, J.-q., Bock, A., Li, J., Luo, G.-m., and Shen, J.-c. 2005. Engineering glutathione transferase to a novel glutathione peroxidase mimic with high catalytic efficiency: Incorporation of selenocysteine into glutathione-binding scaffold using auxotrophic expression system. J. Biol. Chem. 280 11930–11935. [DOI] [PubMed] [Google Scholar]
- Zhang, Q.H., Ye, M., Wu, X.Y., Ren, S.X., Zhao, M., Zhao, C.J., Fu, G., Shen, Y., Fan, H.Y., Lu, G., et al. 2000. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 10 1546–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]