Summary
Snail family genes are conserved among species during evolution and encode transcription factors expressed at different stages of development in different tissues. These genes are involved in a broad spectrum of biological functions; cell differentiation, cell motility, cell cycle regulation, and apoptosis. However, little is known about the target genes involved in these functions. Here we show that mouse Snail family members, Snail (Sna) and Slug (Slugh), are involved in chondrocyte differentiation by controlling the expression of type II collagen (Col2a1) and aggrecan. In situ hybridization analysis of developing mouse limb demonstrated that Snail and Slug mRNAs were highly expressed in hypertrophic chondrocytes. Inversely, the expression of collagen type II mRNA disappeared during hypertrophic differentiation. Snail and Slug mRNA expression was downregulated during differentiation of the mouse chondrogenic cell line ATDC5 and overexpression of exogenous Snail or Slug in ATDC5 cells inhibited expression of collagen type II and aggrecan mRNA. Reporter analysis revealed Snail and Slug suppressed the promoter activity of Col2a1, and the E-boxes in the promoter region were the responsible element. Gel shift assay demonstrated the binding of Snail to the E-box. Since type II collagen and aggrecan are major functional components of extracellular matrix in cartilage, these results suggest an important role for Snail-related transcription repressors during chondrocyte differentiation.
Keywords: Aggrecans; Animals; Cell Differentiation; genetics; Cell Line; Chondrocytes; cytology; metabolism; Collagen Type II; genetics; DNA-Binding Proteins; physiology; E-Box Elements; Embryo, Mammalian; Extracellular Matrix; Extracellular Matrix Proteins; Gene Expression Regulation, Developmental; Lectins, C-Type; Mice; Promoter Regions (Genetics); Proteoglycans; RNA, Messenger; analysis; Repressor Proteins; physiology; Tibia; Transcription Factors; physiology
Keywords: Snail, transcription factor, chondrocyte, collagen type II, aggrecan
Introduction
During skeletal development, condensed mesenchymal cells give rise to chondrocytes and form cartilage primordia, which serve as templates for endochondral bone formation. In cartilage primordia, chondrocytes undergo further differentiation, starting as proliferating chondrocytes, becoming prehypertrophic chondrocytes and ending as hypertrophic chondrocytes. Chondrocytes secrete abundant extracellular matrix (ECM) into the extracellular space and the composition of ECM changes during the sequential differentiation steps. Type II collagen and aggrecan are major components of the cartilage extracellular matrix produced by proliferating and prehypertrophic chondrocytes, and Type X collagen is a major collagen type produced by hypertrophic chondrocytes. The differentiation into to hypertrophic chondrocytes is accompanied with switching of collagen types from type II to type X and the disappearance of aggrecan and link protein (1,2). Dysfunction of these extracellular macromolecules in mutant mice results in striking skeletal malformation, suggesting their important roles in skeletal formation (3–7).
Type II collagen is a homotrimer of the alphal (II) chain (Col2a1). Two elements for the regulation of Col2a1 transcription are known; one is the enhancer located within the first intron that mediates cartilage-specific expression of Col2a1, and the other is in the promoter region that has both silencers and promoter elements (8–12). One of the E-boxes (CAGGTG) in the promoter region is involved in the negative regulation of Col2a1 but the transcription factors which bind to that E-box remain unknown (13).
Drosophila Snail was the first member identified among the Snail family transcription factors and encodes a zinc finger-type transcription factor that is necessary for gastrulation and mesoderm formation (14). Snail family members have been evolutionarily conserved among many species. They share highly conserved C2H2 type zinc finger domains, which bind to the E-box and repress the transcription of target genes (reviewed in 15). Snail-related transcription repressors were shown to play roles in a broad spectrum of biological functions such as cell differentiation, cell adhesion, cell movement, cell cycle regulation and apoptosis (16–26, reviewed in 15,27). Mouse Snail family members include Snail (Sna), Slug (Slugh) and Smuc (28). Expression patterns of mouse Snail and Slug in the early developmental stage suggest involvement of Snail and Slug in chondrogenesis (24, 28–30). Mouse Snail mRNA is expressed in condensed precartilage mesenchyme but not detected in chondrocytes at 14.5 days except in the distal phalanges of the hind limbs, which are the only sites of precartilage in the limbs at this stage (24). In the 3 day old rat, mRNA of rat Slug is detected in condensing mesenchymal cells corresponding to cartilage precursors, and at 5 days, the cartilage primordium itself was negative for the expression (29). In this study we found an inverse correlation between the gene expression of Snail and Slug and type II collagen and aggrecan in vivo and in vitro during chondrocyte differentiation in the growth plate of mouse embryos. For the experiments in vitro, we have used well-characterized chondrogenic cell line, ATDC5, which is derived from embryonal teratocarcinoma and recapitulate the sequential steps of chondrocyte differentiation (29–32). Using ATDC5 cells we found that overexpression of Snail or Slug reduced the expression level of type II collagen mRNA and aggrecan mRNA. Reporter analysis using the Col2a1 promoter revealed that Snail and Slug suppress the activity. We found that E-boxes, which are binding motifs for the Snail family, are involved in the negative regulation of Col2a1 transcription (13). Reporter analysis showed Snail suppressed the transcriptional activity of Col2a1 through the E-boxes. Gel shift assay demonstrated the binding of Snail to the E-box. These results suggest that Snail and Slug regulate chondrocyte differentiation by repressing the genes, the Col2a1 gene and possibly the aggrecan gene, that encode major ECM molecules in cartilage.
Experimental Procedures
Cell Culture
ATDC5 cells were maintained in DMEM/F-12 medium (LifeTechnology) supplemented with 5% FBS and antibiotics in plastic dishes at 37 °C under 5% CO2. For the induction of chondrocyte differentiation in three-dimension culture, the cells were trypsinized and embedded in alginate beads in a differentiation medium consisting of DMEM/F-12, 10% FBS, 10ug/ml human insulin, 10ug/ml human transferrin, 10ng/ml sodium selenite (Roche) and antibiotics. To embed the cells into beads, trypsinized cells were centrifuged for 5 min at 1,500 rpm, washed once with a 150mM NaCl solution and suspended in a alginate solution (1.2% alginate, 150 mM NaCl, 1mM CaCl2, and 20mM HEPES) at a cell density of 1.0 × 106 cells/ml. This mixture was poured into a plastic syringe and dropped through a 22-gauge needle into 40ml of a 50mM CaCl2 solution. The CaCl2 solution was decanted and the beads were washed once with DMEM/F-12 medium. Beads were placed at 37°C in 100mm dishes with 20ml of the differentiation medium. For the cell recovery, the culture medium was discarded and the beads were washed once with PBS and dissociated in 150mM NaCl. The cells were pelleted from the suspension.
Transfection Assay
ATDC5 cells were transiently transfected using FuGENE 6 (Roche). First, 250,000 cells were seeded per 100mm dish. Then after 16h culture, they were transfected with 12μg of plasmids mixture. The medium was renewed 24 hr after the transfection. For Northern blotting, the cells were harvested 48 hr after transfection. Stable transfectants of ATDC5 expressing exogenous Snail or Slug were established following transfection with Lipofectamine 2000 (Life Technologies). At 16 hr after the seeding of 80,000 cells per well onto 24-well plates, cells were transfected with 0.9μg of plasmid. Stable transfectants were selected in 400 mg/ml of G418 for 10 days. pcDNA3 mSna was kindly provided by A. Cano (18) and pcDTet-mtmSna by J. Cross. PCR3 Mslug S was previously described (26). For tetracycline-induced exogenous Snail expression, ATDC5 cells transfected with pcDTet-mtmSna were cultured in beads for 48 hours with 3 ng/ml of doxycyclin in the differentiation medium. At this concentration, doxycyclin was not toxic to cells undergoing chondrogenic differentiation in the three dimension culture or in monolayer culture.
In Situ Hybridization
Templates for 35S -Labeled riboprobes for type II collagen, type X collagen, Snail and Slug mRNAs were used as described previously (18,29,33). Antisense[35S]cRNAs were synthesized using the Gemini Transcription Kit (Promega, Madison, WI) and [35S]UTP(1289 Ci/mmol; Perkin Elmer Life Sciences). In situ hybridization was performed as described previously (33). Slides were dipped into NTB-2 (Eastman Kodak Co.) and stored at 4°C for three to four days to visualize signals. After development, sections were counterstained with hematoxylin and eosin and mounted.
Northern Blot Analysis
Total RNA was extracted from ATDC5 cells using an RNeasy Mini Kit (Qiagen). The 8μg RNA was applied to an agarose gel for electrophoresis, then transferred onto a Hybond N membrane (Amersham). cDNAs were labeled with [32P] dCTP using a Random Primed DNA Labeling Kit (Promega) and used for probes. The membranes were hybridized with the labeled probe at 68°C, washed first at room temperature in 2X SSC and 0.1% sodium dodecyl sulfate (SDS) and then at 65°C in 0.2X SSC and 0.1% SDS, and analyzed with a Cyclone Phosphorimager (Packard). Probes for type II collagen, type X collagen, and the PTH/PTHrP receptor were prepared from plasmids described previously (33). Probes for Snail and Slug were prepared from fragments of the plasmids used for in situ hybridization. Sox9 probes were made from mouse cDNA (1070–1324).
Reporter Assays
The plasmid constructs pCII-312, pCII 977, pCII-312E, and pCII-977E have been described previously (34). Briefly, these constructs contain the 5′-flanking sequence from the rat Col2a1 gene upstream of the CAT coding sequence. pCII-312E and pCII-977 also contains a 1,500-bp region from the first intron that has enhancer activity. For CAT assays, ATDC5 cells were transfected in 100mm dishes using Lipofectamine 2000 (Life Technologies) or FuGENE 6 (Roche) according to the manufacturer’s instructions. The cells were cotransfected with Snail or Slug expression plasmid, 2.5μg of reporter plasmid (pCII-312, pCII-977, pCII-312E, or pCII-977E), 2.5μg of Beta-galactosidase expression plasmid (CMV beta-gal) and pcDNA3.1. For Lipofectamine 2000, the total amount of DNA was kept at 20μg with pcDNA3.1 and cells were exposed to the DNA solution for 2h and then cultured in fresh DMEM/F-12 medium supplemented with 5% FBS. For FuGENE6, the total amount of DNA was kept 12μg with pcDNA3.1 and the DNA solution was added to the culture medium and left for 24 hr. The medium was then renewed. For three-dimension culture, the cells were embedded in alginate beads 24 hours after transfection and cultured in the differentiation medium for 48 hr. For monolayer culturem, the cells were cultured in the maintaining medium for 48 hours after transfection. Nuclear extracts were prepared for the analysis of CAT activity (35). Equivalent amounts of cellular proteins were incubated in the reaction buffer (2.5M Tris-HCl, pH 7.8, 1mM acetyl-CoA (Roche), and 2μCi/ml [14C]chloramphenicol (NEN)) for 2 hr at 37°C. The level of acetylation was examined by TLC followed by quantitation using a Cyclone Phosphorimager (Packard). CAT activity was normalized to beta-galactosidase activity with a Galacto-Light Plus system (Tropix). For luciferase assays, ATDC5 cells were transfected in six-well plates using FuGENE6 (Roche) according to the manufacturer’s instructions. The EBx1 Luc construct was generated by inserting a PCR-amplified 283 bp promoter region into pE1b-luc (previously described by A. Hata) and was verified by sequencing. mutEBx1 Luc was generated by PCR using a mismatch primer. 977-luc reporter was generated by inserting a PCR-amplified 977 bp sequence from pCII-977 into pE1b-luc, and the E-box mutant series was made by introducing mutations in each E-box using a mismatch primer to CATGCG. All plasmids were verified by sequencing. Luciferase assays were carried out with a Luciferase Assay System (Promega) and Berthold luminometer. A beta-galactosidase expression vector was co-transfected for normalization and assayed with a Galacto-Light Plus kit (Tropix). pCR3 Mslug S and antisense Slug plasmid were previously described (26)
Electromobility Shift Assay (EMSA)
The expression plasmid for HA-tagged Snail (17) was transfected into ATDC5 cells and nuclear extracts were prepared 48h after the transfection We used double-stranded oligonucleotide probes, which contained Snail-related transcription factor binding sites (E-box4) plus unique flanking sequences. In addition, we used oligonucleotide competitors with either an intact or mutated E-box4 site in competition EMSA experiments. The sequences of the oligonucleotides used in all binding reactions are as follows.
WT probe, sense strand; 5′-GGCCTTGGCAGGTGTGGGCTCTGG-3′, WT probe, antisense strand; 5′-CCAGAGCCCACACCTGCCAAGG-3′. Competitor probe, sense strand; 5′-CCTTGGCAGGTGTGGGCTCTGG-3′, Competitor probe antisense strand; 5′-CCAGAGCCCACACCTGCCAAGG -3′, MUT probe sense strand; 5′-GGCCTTGGAAAAAATGGGCTCTGG-3′, MUT probe antisense strand; 5′-CCAGAGCCCATTTTTTCCAAGG-3′. The single-stranded sense and antisense oligonucleotides were annealed in equimolar amounts. The double stranded WT probe and MUT probe were subsequently labeled using Klenow DNA polymerase and [32P]dCTP(NEN). The labeled oligonucleotides were separated from the unincorporated nucleotides using a Sephadex G-50 gravity flow column (Amersham Pharmacia Biotech). Aliquots of 10,000 cpm of the probe were incubated with 2μg of nuclear protein for 30 min on ice in a 20-μl reaction mixture containing 20mM HEPES, pH 7.9, 10% glycerol, 150mM KCl, 3mM MgCl2, 10uM ZnCl2, 0.3mg/ml BSA, and 100ng of Poly[dI-dC] (Roche). Subsequently, 1.0μl of the probe (100,000 cpm) was added and incubated for 30 min on ice. For supershift experiments, 200ng of the anti- HA antibody (Y-11, Santa Cruz) or the rabbit IgG was incubated with the nuclear extract for an additional 20 min prior to the addition of the labeled probe. HA-Snail plasmid was kindly provided by AG Herreros. For competition assay, a 50-fold molar excess of the indicated unlabeled double-stranded competitor oligonucleotides was preincubated with the nuclear extracts in the binding reaction prior to the addition of the probe. In all cases, the final binding reaction mixture was loaded onto a 5% nondenaturing acrylamide gel in 0.5X TBE buffer and electrophoresed at 150V. Gels were dried and analyzed using Phosphorimager (Packard)
Western Bloting
ATDC5 cells were transfected with 12μg of DNA in a 10cm dish or 2 μg of DNA in a 6-well plate using Fugene 6 (Roche). At 48h after the transfection, the cells were harvested and lysed in sample buffer and sheared by passing through a 22-gauge needle. Samples were mixed with loading buffer and separated on a SDS-10% polyacrylamide gel, then transferred onto nitrocellulose membranes by electroblotting. Detection was performed using a polyclonal anti-HA antibody (Y-11, Santa Cruz) against HA epitope, and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz). The bands were visualized with the enhancer chemiluminescence protocol (Amersham).
Results
Expression of Snail, Slug, and type II collagen mRNAs during chondrocyte differentiation in the growth plate
We examined the mRNA expression of Snail, Slug, and collagen type II during chondrocyte differentiation by in situ hybridization. In the growth plate, the sequential steps in differentiation of chondrocytes are easily distinguished in sections stained with HE. The chondrocytes are categorized into four types, resting, proliferating, prehypertrophic and hypertrophic chondrocyte. In situ hybridization revealed that both Snail and Slug were highly expressed in the hypertrophic chondrocytes in the limb of day 16 mouse embryos, whereas the expression level was low in resting and proliferating chondrocytes (Fig. 1A, B, D and E). The profile of expression in the hypertrophic chondrocyte differed slightly between Snail and Slug in that Slug expression is confined to a narrower zone of early hypertrophic chondrocytes. Smuc mRNA was expressed not only in hypertrophic chondrocytes but also in skeletal muscles (data not shown). In the spinal cord and mesodermal tissues in the early stages, Snail and Slug riboprobes showed signals, which did not overlap, indicating the specificity of the probes (data not shown). Type II collagen mRNA was expressed in proliferating and prehypertrophic chondrocytes but not in hypertrophic chondrocytes (Fig. 1H–I and 44). The expression of Aggrecan mRNA has been also reported to disappear in hypertrophic chondrocytes (1).
Fig. 1. Snail and Slug expression in the mouse tibia at embryonic stages.
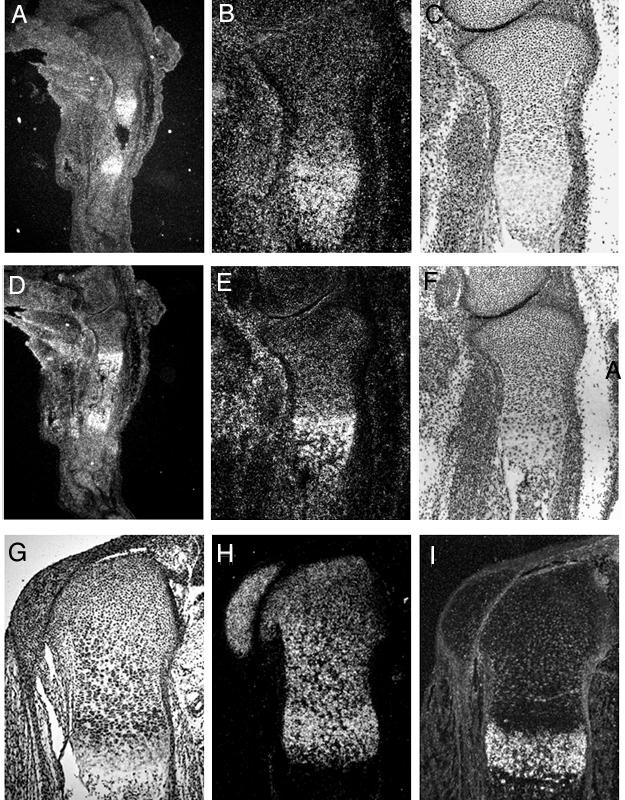
In situ hybridization of longitudinal sections of the tibia in the hindlimb of 16 day mouse embryos with antisense Snail (A, B) and Slug (D, E) 35S-Labeled riboprobes. Panels B and E are higher magnification images of the proximal tibia in panel A and D, respectively. Serial sections were stained with Hematoxylin and Eosin (C, F). Weak signals for both Snail and Slug were detected ubiquitously in soft tissues and strong signals were detected in hypertrophic chondrocytes in dark field views (A, B, D, E). In situ hybridization of longitudinal sections of the proximal humerus in the forelimb of a 17 day-old mouse with antisense Col2a1(H) and Col10a1(I) 35S-labeled riboprobes. Dark field views (H, I) and the corresponding bright field view (G) are shown. Strong signal for type II collagen was observed in resting, proliferating and prehypertrophic chondrocytes but not in hypertrophic chondrocytes (H). Strong signal for typeX collagen (I) expression was seen in hypertrophic chondrocytes. Skin was removed during tissue preparation.
The expresson levels of Snail/Slug and type II collagen are inversely changed during chondrocyte differentiation in ATDC5 cells
To explore the function of Snail/Slug transcription repressors during chondrocyte differentiation, we used the mouse chondrogenic cell line ATDC5. This cell line has been well characterized and shown to be a model for studying gene expression and morphological changes during the normal differentiation of precursor mesenchymal cells into terminally differentiated chondrocytes. (31,32,36,37). With insulin supplementation, the ATDC cells form discrete condensations and express type II collagen in confluent monolayer culture. The condensed cells further differentiate to form cartilage nodules and to express type X collagen; a marker for fully differentiated chondrocytes.
In three-dimension culture systems, once dedifferentiated primary chondrocytes cultured in monolayers recover the chondrocyte phenotype. Therefore three-dimension culture is thought to mimic the situation in vivo better than monolayer culture. We used an alginate beads culture system (38–40), in which the ATDC5 cells are embedded in alginate beads and cultured with insulin. After two days in the three-dimension culture, ATDC5 cells started to express type II collagen mRNA which is a hallmark of chondrocyte differentiation (Fig. 2A). In monolayer cultures of ATDC5 cells, chondrocytes form few condensations and only a minor expression of type II collagen mRNA was detected four days after confluence (32,37). Therefore, this three-dimension culture system using ATDC5 cells is a useful in vitro system with which to study chondrocyte differentiation.
Fig. 2. Overexpression of Snail and Slug reduced collagen typeII expression in ATDC5 cells.
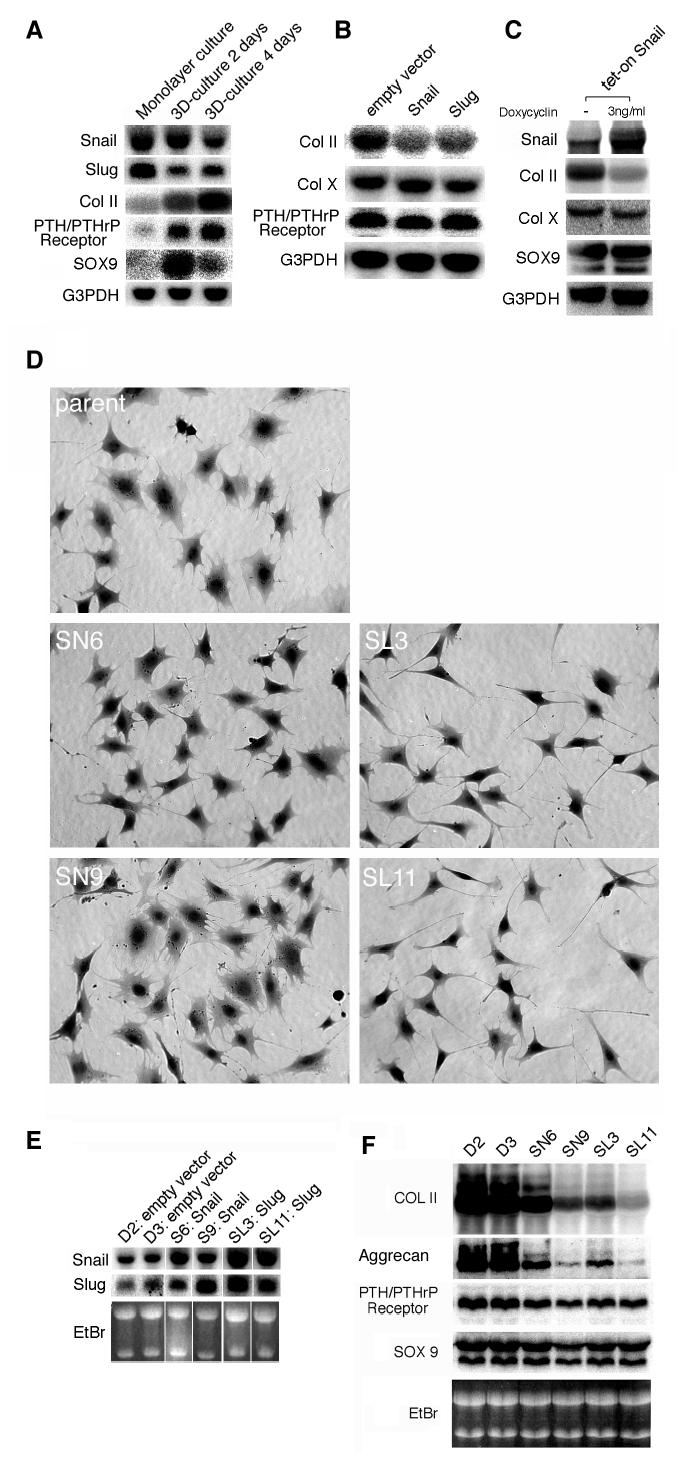
A, B, and C, Expression of Snail, Slug, chondrocyte differentiation markers (type II and type X collagen, PTH/PTHrP receptor and Sox9) and G3PDH was analyzed by Northern blotting.
A. Undifferentiated monolayer ATDC5 cells expressed high levels of Snail and Slug mRNAs, but the mRNA for chondrocyte markers such as type II collagen, PTH/PTHrP receptor and Sox9 was not expressed. When ATDC5 cells were induced to differentiate in a three-dimension culture, they started to express type II collagen mRNA in 2 days. Whereas the level of type II collagen mRNA increased during the 4 days of culture, the levels of Snail and Slug decreased. B and C. Overexpression of Snail or Slug on transient transfection suppressed the expression of type II collagen in ATDC5 cells. (B) Snail or Slug expression plasmid was transfected into monolayer-cultured ATDC5 cells. At 24 hours after transfection the cells were transferred into three-dimension culture system and left to differentiate for 48 hours before being subjected to Northern blot. Although the mRNA of chondrocyte markers PTH/PTHrP receptor and type X collagen, both chondrocyte marekers were detected, the expression level of collagen type II mRNA was decreased compared to the control transfected with empty vector. (C) Tet-activator-regulated Snail expression plasmid was tested in the same setting as described in B. To induce expression of Snail, doxycycline was added to the culture medium at a concentration of 3ng/ml after transfection. Although collagen type X and Sox9 were detected independently of doxycycline treatment, type II collagen transcripts were only detected at low levels in doxycycline-treated ATDC5 cells. D. Morphology and expression of chondrocyte markers in Snail or Slug-transfected ATDC5 cells. Morphology of stable transfectants after antibiotic selection and cloning. The cells were stained with trypan blue after fixation. Compared to the parent and Snail transfectants, the Slug transfectants had a distinctive morphology with long and thin cytoplasmic extensions. E. Northern blot analysis showed a 2–3 fold increase in expression of Snail mRNA and Slug mRNA in Snail-transformants (SN6, SN9) and Slug-transformants (SL3, SL11), respectively, as quantified by phosphorimaging. Note that the expression level of both Snail and Slug mRNA was increased in both transformants. F. The level of type II collagen and aggrecan mRNA was decreased in Snail or Slug - transfected ATDC5 cells. Northern blot analysis of differentiation markers in transformants after 2 days of three-dimension culture. Although PTH/PTHrP receptor and Sox9 mRNA remained at the same level in all clones, the levels of collagen type II mRNA and aggrecan mRNA were decreased in Snail and Slug transformants. EtBr: Agarose gels were stained with ethidium bromide and the photo was taken under an UV illuminator.
We examined the expression of Snail family transcription repressor mRNAs by Northern hybridization during the differentiation of ATDC5 cells. In subconfluent monolayer cultures, mRNAs of chondrocyte differentiation markers; type II collagen, PTH/PTHrP receptor and Sox9 were undetectable (36,37,41). Two days after initiating the three-dimension culture, chondrocyte differentiation markers were detected (Fig. 2A). After four days of culture, the level of type II collagen and PTH/PTHrP receptor mRNA were enforced, whereas the levels of Snail and Slug mRNA were reduced to 60% and 50%, respectively when quantified with a phosphorimager. These results show inverse correlation of expression levels between Snail/Slug mRNA and type II collagen mRNA during chondrocyte differentiation in vitro. Smuc mRNA was not detected in ATDC5 cells by Northern bloting.
Overexpression of Snail/Slug mRNA by transient transfection reduced the collagen II mRNA expression in ATDC5 cells
As Snail/Slug are transcriptional repressors, the inverse relation between the expression of Snail/Slug and that of type II collagen led us to hypothesize that Snail and Slug are involved in regulating Col2a1 gene. To address this hypothesis, we first tested whether overexpression of Snail and Slug affect chondrocyte differentiation. Subconfluent ATDC5 cells were transfected with the Snail or Slug expression vector. After 24 hr, transfected cells were embedded into alginate beads and cultured for two more days. The cells were harvested and the expression of chondrocyte markers was analyzed by Northern blot. The results revealed that, although the PTH/PTHrP receptor was expressed, the expression level of collagen type II mRNA was reduced in cells overexpressing Snail or Slug (Fig. 2B). We also looked for this reduction in type II collagen mRNA using a tetracycline-inducible Snail expression plasmid (Fig. 2C). Interestingly, mRNA of Sox9, which is a positive regulator of Col2a1 gene, was expressed normally despite that level of type II collagen mRNA was reduced. In our system, the level of Sox9 mRNA increased upon differentiation and the timing coincided well with the onset of type II collagen mRNA expression (Fig. 2A). These results suggested that when overexpressed Snail and Slug can overcome the positive regulatory effect of Sox9 and can regulate chondrocyte differentiation by counteracting Sox9.
Next we analyzed ATDC5 cells stably transfected with Snail or Slug expression plasmids. After selection for drug resistance, ten clones were picked and expanded. The expression level of Snail or Slug mRNA in each clone cultured in monolayers was tested by Northern blotting. All the clones showed higher level of expression of Snail or Slug mRNA than the mock transfectant (data not shown). For further analysis, we chose clones expressing two to three times more Snail or Slug mRNA than parental cells (Fig. 2E, quantified by phosphorimaging). In the stable transformants, the expression of both Snail and Slug was increased in the undifferentiated state, indicating some cross-regulatory pathway between the two genes.
In the undifferentiated state, the morphology of Slug transformants was distinct from that of the untransfected or Snail-transfected clones. The Slug transfected cells displayed longer cellular protrusions and a thiner cell body (Fig. 2D). Stable clones were induced to differentiate in three-dimension culture for two days and the expression of the chondrocyte markers was analyzed by Northern Blotting (Fig. 2F). In transformants, the expression levels of collagen type II mRNA were dramatically reduced in spite of the expression of other markers; PTH/PTHrP receptor and Sox9. This finding was consistent with that of the transient transfection, both results showing that overexpression of Snail and Slug prevents the expression of collagen type II mRNA in differentiating ATDC5 cells, but not the expression of PTH/PTHrP receptor and Sox9 mRNAs. In addition to type II collagen, aggrecan mRNA was also reduced which was not demonstrated by the previous transient transfection experiment (Fig. 2F and data not shown).
Transcription of Col2a1 was repressed by Snail/Slug
To address the mechanism of the decrease in type II collagen mRNA after overexpression of Snail or Slug, we carried out a reporter assay testing their effects on the regulatory regions of Col2a1. The Col2a1 gene has a tissue-specific enhancer in the first intron and a silencer in the promoter region. First, we searched for a putative binding site for Snail family transcription repressors, E-box (reviewed in 17) in the 1 kb sequence of the promoter region of the human, rat and mouse Col2a1 gene (GenBank;X58709, M10613, M651611). Three E-boxes were found in the human gene, and four E-boxes in the rat and mouse genes (Fig. 4A and 4B). Rat E-box2 and 3 (CAGGTG) were conserved among the species both in sequence and in relative location (Fig. 3A and 3B). Then we took advantage of well-characterized reporter constructs containing promoter and enhancer regions of rat col2a1 (34). pCII-977E and pCII-312E are CAT reporter constructs containing −977 to +110 and −312 to +110 of the genomic sequence respectively and a 1500-bp first intron sequence from the rat Col2a1 gene (Fig. 3C). pCII-977E has four E-boxes and pCII-312E has one. It has been reported that the 977-bp genomic fragment contains not only promoter activity but also silencing activity, which is observed in chicken embryonic fibroblast and HeLa cells (38).
Fig. 4. Analysis of E-boxes in the promoter region.

A. The 977-bp region of the promoter of the rat Col2a1 gene was subcloned into pGL2-basic vector with or without a mutation in each E-box (977-luc, mut E-box 1 to 4). ATDC5 cells were cotransfected with reporter plasmid and pSV-beta gal. The mutation in E-box 2 and 4 increased the transcription activity while that in E-box 3 slightly increased the activity compared to the activity of 977-luc. pGL2 basic was transfected as a control (lane 6). B. The E-box located at −167 to −162 and its flanking sequence in the rat collagen 2a1 gene is conserved among the human, rat and mouse genes. A 283bp (−173 to +110) of rat genomic fragment containing the E-box was subcloned into the luciferase reporter (EBx1 Luc). The same fragment with mutations in the E-box was also subcloned into the luciferase reporter (mutEBx1 Luc) and used as a control. C. Mutation in the E-box restored the reporter activity in undifferentiated ATDC5 cells. ATDC5 cells were cotransfected with reporter plasmid and pSV-beta gal. D. Swapping the repressive domain of Snail with the activation domain of VP16 increased the reporter activity through the E-box. A chimeric molecule was made by combining the DNA binding domain of Snail with the activation domain of VP16. This molecule is expected to bind the DNA target of Snail and function as an activator. Line graph; Representative change in reporter is shown. The reporter activity of EBx1-luc was increased in a dose-dependent manner by VP16-Snail. For the reporter with a mutated E-box, no dose-dependent change was observed. The dose of VP16-Snail expression plasmid was 0μg (lane 1), 0.1μg (lane 2), 0.2μg (lane 3), 0.5μg (lane 4), and 1.0μg (lane 5). The total amount of plasmids was adjusted to 2μg with empty pcDNA3.1 plasmid. The experiment was triplicated.
Fig. 3. Conservation of E-boxes in the promoter of Col2a1 among human, rat and mouse and repressive effects of Snail/Slug on transcription.
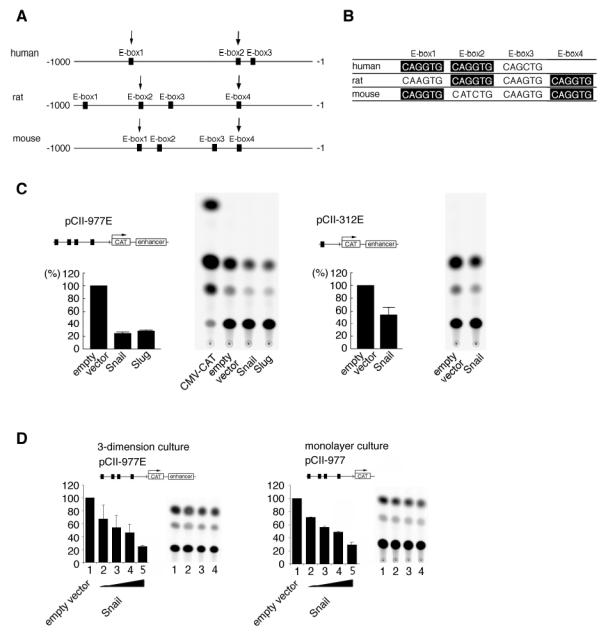
A. Three E-boxes were located in the promoter region of the collagen type2a1 gene in human and four E-boxes in rat and mouse. B. A comparison shows that all species have two conserved E-boxes (CAGGTG, arrow in the figure and highlightened in the table) and the relative positions in the promoter region are similar. C. Snail and Slug suppressed the transcriptional activity of the col2a1 gene promoter and/or enhancer. CAT reporter constructs are described above the graph. The percent activity relative to mock transfection was calculated. The suppressive activity was stronger with pCII-977E, which has a longer promoter region than pCII-312E. CAT constructs, pCII-977E and pCII-312 contain the collagen type2al gene fragment (−977 to +110) and (−312 to +110) respectively. Both reporter constructs had a chondrocyte-specific enhancer. Snail or Slug expression plasmid was cotransfected with CAT constructs and pSV-beta-gal as standard. CMV-CAT was a positive control. The experiment was done in triplicate. D. Snail suppressed the Col2a1 promoter activity both in differentiated and in undifferentiated ATDC5 cells in a dose-dependent manner. ATDC5 cells were cotransfected with the Snail expression plasmid, a CAT construct (pCII-977E) and pSV-beta gal. At 24 hr after transfection, the cells were put into a three-dimension culture system for 2 days to differentiate before harvesting. The activity was suppressed by the Snail expression plasmid in a dose dependent-manner (lane 1–5). ATDC5 cells were cotransfected with the Snail expression plasmid, the CAT construct without an enhancer (pCII-977) and pSV-beta gal and cultured in a monolayer for 2 days. The activity was suppressed by the Snail expression plasmid in a dose-dependent manner (lane 1–5).
The dose of Snail plasmid is 0μg (lane 1), 0.5μg (lane 2), 1.0μg (lane 3), 2.5μg (lane 4) and 12μg (or 20 lane 5). The transfection experiment was triplicated.
We transiently transfected ATDC5 cells with the reporter constructs, and Snail or Slug expression plasmid. CAT activity was repressed when Snail or Slug was co-transfected, but not when the empty vector was co-transfected. (Fig. 3C). These results suggest that Snail and Slug suppress the transcription of Col2a1 through the promoter and/or the enhancer. The promoter region is more likely to include the responsive elements, because pCII-977E had weaker activity than pCII-312E (Fig. 3C), indicating that the length of the promoter affected the suppressive effect of Snail/Slug. pCII-977E has three more E-boxes (E-box 1–3 in Fig. 3A ) than pCII-312E (E-box 4 in Fig. 3A) in the promoter region, therefore, we speculated that these E-boxes may have the additive suppressive effect.
The enhancer sequence was dispensable for suppressive activity of Snail/Slug
Next, we tested the possibility that Snail/Slug can counteract the positive regulation mediated by the enhancer in the reporter constructs. The enhancer contains the binding sites for Sox9 (42), which is a chondrocyte-specific positive regulator of the Col2a1 gene in chondrocytes, and the enhancer also contains E-boxes. We compared the activities of reporter constructs with or without the enhancer: pCII-977E had the enhancer and pCII-977 lacked it. The activities of pCII-977 and pCII-977E were suppressed by Snail and Slug almost equally in both undifferentiated and differentiated ATDC5 cells (Fig. 3D and data not shown). These results suggest that Snail and Slug suppress the activity through the promoter and not through the enhancer. Because of the significant similarity among the Snail family transcription repressors in the DNA binding domains, they are thought to regulate the same target genes (20). The identity of the last four zinc fingers between Snail and Slug is 82%. In this study, overexpression of Snail and Slug (also Smuc, data not shown) gave similar results using the reporter. Therefore, we used only the effect of Snail in further analysis.
Suppressive effect of Snail was mediated by E-box
To test whether each E-box is involved in the regulation, a mutation was introduced into each of the E-boxes in the rat 977-bp promoter sequence and the sequence was subcloned into luciferase vectors. Reporter assay revealed that a mutation in E-box1, 2 and 4 increased the activity whereas a mutation in E-box 3 had no effect on transcription (Fig. 4). The sequence and the location of rat E-box 3 were not conserved well among the three species (Fig. 3A and 3B). These data suggested that rat E-box 3 was not involved in the regulation of Col2a1. The strong desuppressive effect of disrupton of E-box 2 and 4 indicated the importance of these two conserved sites. It has been reported that mutation in E-box 4 desuppressed the activity of the reporter construct containing −307 to +110 of Col2a1 in chicken limb mesenchyme cells and transregulatory factors which bind to this site remained unknown (13). Therefore to study the mechanism, we focused on E-box 4 located at −167 to −162.
To test whether Snail is a candidate transcription factor, we made two luciferase constructs, EBx1-Luc and mutEBx1-Luc, both containing the −173 to +110 stretch of the promoter sequence of rat Col2a1 and mutEBx1-Luc having a mutation in the E-box 4 (CAGGTG to CATGCG, Fig. 4B) that abrogates the binding of Snail and Slug. We compared the activity of the two constructs and found the mutation in the E-box abolished the represser activity in ATDC5 cells (Fig. 4C).
Next we examined the effect of the chimeric protein VP16-Snail, in which the N-terminal half of Snail was removed and the DNA binding domain of Snail was fused with the transcription activator domain of VP16. This chimeric protein can work as a competitor for endogenous Snail-related repressors and can counteract their suppressive activity (23). Mutation in the E-box abolished this effect of VP16 (Fig. 4D). These results suggested that VP16-Snail competed for binding to the E-box with endogenous factors in a dose-dependent manner and desuppressed the activity through the E-box in ATDC5 cells.
Major suppression of Col2a1 transcription was mediated by the Snail family
To further define the negative regulator of the E-box, we examined the effect of antisense Slug mRNA expression, which has been reported to inhibit the functions of endogenous Slug (26). It was reported that injection of antisense Slug mRNA into Xenopus embryo reduced not only XSlug but also Xsnail expression. This is thought to be due to the high homology between XSnail and Xslug (43,44). Therefore, we checked the effect of antisense Slug on Snail protein expression by Western bloting. HA-tagged Snail protein expression was reduced when the Snail expression plasmid was co-transfected with the antisense Slug expression plasmid (Fig. 5A) in a dose-dependent manner. This result suggested that the gene product of mouse Snail can be reduced by the introduction of Slug antisense mRNA. This result was similar to the phenomenon observed in the experiment on Xenopus embryos. Next we examined if mouse Snail protein can reverse the desuppressive effect of antisense Slug mRNA. Fig. 5B shows that antisense Slug mRNA had a desuppressive effect on the 977-luc reporter (Fig. 5B, lane2) and cotransfection with HA-Snail reversed the suppressive effect in dose dependent manner (Fig. 5B, lane 3 to 5), indicating the change in the amount of Snail family protein affected the expression of the Col2a1 gene. We also tested the effect of antisense Slug mRNA on EBx1 and mutEBx1 reporters. Antisense mRNA treatment resulted in increased activity of the EBxl reporter, but had no effect on the mutEBx1 reporter (data not shown). These results suggest that the suppression of Col2a1 transcription was mainly dependent on the effect of the Snail family through E-boxes.
Fig. 5. Antisense Slug mRNA treatment restored the gene expression of Col2a1.
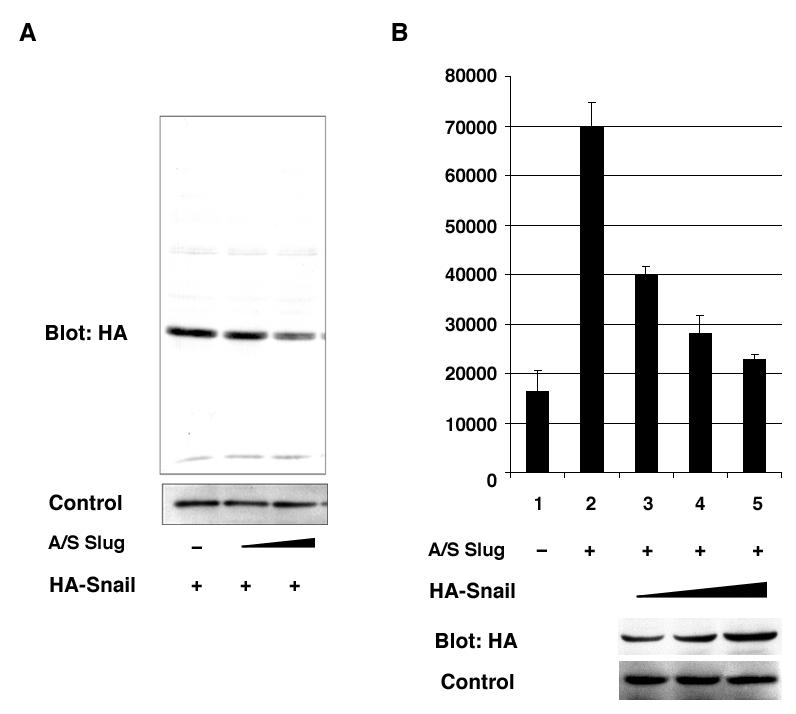
A. The effect of antisense Slug mRNA was tested using HA-tagged Snail expression vector. Undifferentiated ATDC5 cells were transfected with 0.25 μg of HA-Snail expression vector together with antisense Slug mRNA vector. The dose of antisense Slug mRNA expression vector was 0 μg (lane 1), 0.125 μg (lane 2), and 0.25 μg (lane 3). The total amount of plasmids was adjusted to 2μg with pcDNA3.1 empty plasmid. The experiment was triplicated and a representative result is shown. B. Cotransfection of the reporter and antisense-Slug vectors with HA-tagged Snail expression vector recovered the suppressive effect in a dose-dependent manner. Undifferentiated ATDC5 cells were transfected with 977-luc vector, antisense Slug vector, and HA-tagged Snail vector. Then, 0.25 μg of antisense Slug mRNA vector was transfected. The dose of HA-Snail vector was 0 μg (lane 1 and 2), 0.05 μg (lane 3), 0.1 μg (lane 4), and 0.2 μg (lane 5). At the same time, the amount of HA-Snail Hg (lane 3), 0.1 μg (lane 4), and 0.2 μg (lane 5). At the same time, the amount of HA-Snail protein was checked by Western blotting. Protein was extracted 2 days after transfection from the cells in duplicated wells in the plate for luciferase assay.
Snail binds to the E-box in the Col2a1 promoter
We tested whether Snail protein interacts directly with the E-box using electrophoretic mobility-shift assays. The 24-bp oligonucleotides corresponding to the −174 to −151 sequence containing the E-box 4 formed two retarded bands when incubated with the nuclear extract prepared from ATDC5 cells transfected with HA-tagged Snail expression plasmid (Fig. 6). A supershifted band was observed when anti-HA antibody was added to the reaction mixture but not when the control antibody was added. The shifted band disappeared only when cold competitor was added to the reaction and not when mutated oligonucleotide was added. Thus the lower band (Fig. 6, arrow) contained HA-tagged Snail in the complex whereas the upper band (Fig. 6, dot) was non-specific. These results suggest that Snail protein binds to the E-box of Col2a1 in a sequence-specific manner.
Fig. 6. Electromobility shift assays revealed specific binding of Snail to a conserved E-box.
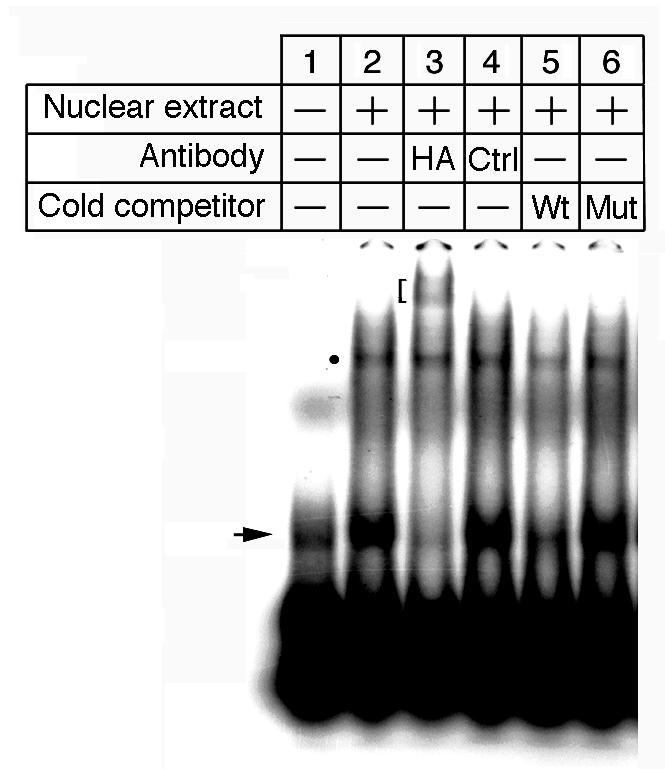
HA-tagged Snail was expressed in ATDC5 cells and the nuclear extract was incubated with double-stranded radiolabeled 24-mer oligonucleotide probes containing E-box4 (lane 2–6).
Shifted bands (arrow and dotted) were detected when the probes were incubated with the nuclear extract (lane 2) but not without the nuclear extract (lane 1). For supershift analysis, anti-HA antibody (lane. 3) or non-specific IgG (lane 4) was added. A supershifted band (blanket) was detected only when the probes were incubated with anti-HA antibody. For competition analysis, a 50-fold molar excess of unlabeled wild type probe (lane 5) or 50-fold molar excess of unlabeled mutated probe was added (lane 6). The shifted band disappeared when the probes were incubated with wild type probe (lane 5), but not when incubated with mutated probes. The non-specific band is marked with a dot.
Discussion
Snail-related transcription factors are repressors of the Col2a1 gene in chondrocyte differentiation
In this study, we demonstrated that Snail family transcription repressors function as negative regulators of Col2a1 transcription. The expression pattern in the growth plate of mouse embryo and ATDC5 chondrocytes showed that the expression of Snail/Slug mRNA inversely correlated with that of type II collagen mRNA. In situ hybridization showed that Snail and Slug were strongly expressed in hypertrophic chondrocytes and weakly expressed in resting, proliferating and prehypertrophic chondrocytes. On the other hand, type II collagen was highly expressed in proliferating and prehypertrophic chondrocytes but not in hypertrophic chondrocytes. During hypertrophic differentiation, the extracellular matrix self-organizes prior to the formation of bone. A switch from collagen type II to collagen type X occurs, and both aggrecan and link protein disappear (1,2). Studies in stable transformants revealed that overexpression of Snail and Slug prevented the expression of major chondrocyte extracellular matrix, collagen type II and aggrecan when transfected cells were induced to differentiate.
Sox9 is one of the major activators of the Col2a1 gene and its specific binding site locates in the enhancer in the first intron (4,8,10,41). Northern blot analysis showed that, although Sox9 mRNA was expressed, type II collagen mRNA was only weakly expressed in Snail or Slug-transfected ATDC5 cells when they were induced to differentiate in a three-dimension culture. Reporter analysis showed the suppressive activity of Snail both in undifferentiated and in differentiated ATDC5 cells. These results suggested a dominant suppressive effect of Snail over Sox9 in differentiating ATDC5 cells and also showed that the enhancer sequence (8) was unnecessary for the suppressive activity of Snail.
We studied the mechanism involved by focusing on the E-box in the promoter region of the Col2a1 gene, which is highly conserved among species. The E-box has been reported to negatively regulate Col2a1 gene transcription (13). In addition, it is a binding site of Snail-related repressors. We found that (1) a chimeric Snail protein which is a fusion of the VP16-activation domain and Snail DNA binding domain transactivated the reporter activity of the reporter construct containing the promoter region via the E-box. (2) antisense Slug mRNA expression desuppressed the reporter activity of the construct with the E-box, and (3) HA-tagged Snail bound to the E-box. These results suggest that Snail family transcription factors can function as repressors of the Col2a1 gene by binding to the E-box of the regulatory sequence. It will be of special interest to investigate whether members of the Snail family also regulate the expression of the aggrecan gene or not. This gene is evolutionary well conserved, but the regulatory regions have not been described yet.
Snail-related transcription factors modulate ECM
We found that Snail and Slug can modulate the expression of collagen type II and aggrecan during chondrocyte differentiation. The transition of these components of ECM is an essential step for chondrocyte differentiation. Other than chondrocytes, Snail/Slug is expressed in premigratory neural crests and plays important roles during their migration. Aggrecan is present along their migratory pathway and guides their migration (46). Collagen type II is transiently present at the interface between epithelial cells and mesenchymal cells during the development of neurocranium (46). These expression patterns suggested that Col2a1 and Aggrecan are regulated by Snail/Slug in neural crest cells. In addition, Snail is expressed at the invasive front of cancer cell lines and primary tumors induced in mice (11). Therefore, we propose that regulating the ECM is another fundamental role of Snail family. Such a mechanism in addition to the cell-cell adhesion and cytoskeleton modulations induced by Snail factors (reviewed in 41) could be involved in cancer development and metastasis..
Acknowledgments
We thank A. Cano, A.G. de Hereros, and J. C. Cross for expression plasmids and Y. Yamada for reporter constructs. We thank G. V. Segre and M. Ikeya for helpful suggestions and H. Valles for fine technical help. We thank all the members of the endocrine unit
References
- 1.Chen Q, Johnson DM, Haudenschild DR, Goetinck PF. Dev Biol. 1995;172:293–306. doi: 10.1006/dbio.1995.0024. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka K, Matsumoto Y, Nakatani Iwamoto FY, Yamada Y. Mol Cell Biol. 2000;20:4428–4435. doi: 10.1128/mcb.20.12.4428-4435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Garofalo S, Vuorio E, Metsaranta M, Rosati R, Toman D, Vaughan J, de Crombrugghe B. Proc Natl Acad Sci USA. 1991;88:9648–9652. doi: 10.1073/pnas.88.21.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacenko O, LuValle PA, Olsen BR. Nature. 1993;365:56–61. doi: 10.1038/365056a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee B, Vissing H, Ramirez F, Rogers D, Rimoin D. Science. 1989;244:978–980. doi: 10.1126/science.2543071. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe H, Nakata K, Kimata K, Nakanishi I, Yamada Y. Proc Natl Acad Sci USA. 1997;94:6943–6947. doi: 10.1073/pnas.94.13.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe H, Yamada Y. Nat Genet. 1999;21:225–229. doi: 10.1038/6016. [DOI] [PubMed] [Google Scholar]
- 8.Horton H, Miyashita T, Kohno K, Hassel JR, Yamada Y. Proc Natl Acad USA. 1987;84:8864–8868. doi: 10.1073/pnas.84.24.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krebshbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y. J Biol Chem. 1996;271:4298–4303. doi: 10.1074/jbc.271.8.4298. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre V, Zhou G, Mukhopadhyay K, Smith CN, Zhang Z, Eberspaecher H, Zhou X, Sinha S, Maity SN, Crombrugghe de B. Mol Cell Biol. 1996;16:4512–23. doi: 10.1128/mcb.16.8.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung KKH, Ng KJ, Ho KKY, Tam PPL, Cheah KSE. J Cell Biol. 1998;141:1291–1300. doi: 10.1083/jcb.141.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savagner P. Bio Essays. 2001;23:1–12. [Google Scholar]
- 13.Murray D, Precht P, Balakir R, Horton WE., Jr J Biol Chem. 2000;5:3610–3618. doi: 10.1074/jbc.275.5.3610. [DOI] [PubMed] [Google Scholar]
- 14.Boulay JL, Dennefeld C, Alberga A. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- 15.Hemavathy K, Ashraf SI, Ip YT. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 16.Ashraf SI, Hu X, Roote J, Ip YT. EMBO. 1999;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia Anatonio de Hereros. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 18.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barro MG, Portillo FM, Nieto A. Nat Cell Biol. 2000;2:1–8. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 19.Fuse N, Hirose S, Hayash S. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- 20.Fuse F, Hirose S, Hayashi S. Development. 1996;122:1059–1067. doi: 10.1242/dev.122.4.1059. [DOI] [PubMed] [Google Scholar]
- 21.Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, Mao M, Inaba T, Look AT. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 22.LaBonne C, Bronner-Fraser M. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama H, Scott IC, Cross JC. Dev Biol. 1998;199:150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- 24.Nieto MA, Bennett MF, Sargent MG, Wilkinson DG. Development. 1992;116:227–237. doi: 10.1242/dev.116.1.227. [DOI] [PubMed] [Google Scholar]
- 25.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 26.Savagner P, Yamada KM, Thiery JP. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arias AM. Cell. 2001;105:425–431. doi: 10.1016/s0092-8674(01)00365-8. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka H, Murayama T, Yokode M, Mori S, Sano H, Ozaki H, Yokota Y, Nishikawa SI, Kita T. Nuc Acid Res. 2000;28:626–633. doi: 10.1093/nar/28.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savagner P, Karavanova I, Perantoni A, Thiery JP, Yamada KM. Dev Dyn. 1998;213:182–187. doi: 10.1002/(SICI)1097-0177(199810)213:2<182::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 30.Smith DE, del Amo FF, Gridley T. Deveopment. 1992;116:1033–1039. doi: 10.1242/dev.116.4.1033. [DOI] [PubMed] [Google Scholar]
- 31.Atsumi T, Miwa Y, Ikawa Y. Cell Differ Dev. 1990;30:109–116. doi: 10.1016/0922-3371(90)90079-c. [DOI] [PubMed] [Google Scholar]
- 32.Newman B, Gigout LI, Sundre L, Grant ME, Wallis GA. J Cell Biol. 2001;154:659–666. doi: 10.1083/jcb.200106040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K, Lanseke B, Karaplis AC, Deeds JD, Kohno H, Nissenson RA, Kronenberg HM, Segre GV. Endocrinology. 1996;137:5109–5118. doi: 10.1210/endo.137.11.8895385. [DOI] [PubMed] [Google Scholar]
- 34.Savagner P, Miyashita T, Yamada Y. J Biol Chem. 1990;265:6669–66. [PubMed] [Google Scholar]
- 35.Andrews NC, Faller DV. Nuc Acid Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukunami C, Shigeno C, Atsumi T, Ishizeki K, Suzuki F, Hiraki Y. J Biol Chem. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukunami C, Ishizeki K, Atsumi T, Ohta Y, Suzuki F, Hiraki Y. J Bone Mine Res. 1997;12:1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 38.Bonaventure J, Kadhom N, Cohen-Solal L, Ng KH, Bourguignon KH, Lasselin C, Freisinger P. Exp Cell Res. 1994;212:97–104. doi: 10.1006/excr.1994.1123. [DOI] [PubMed] [Google Scholar]
- 39.Lemare F, Streimberg N, le Griel C, Demignot S, Adolphe M. J Cell Physiol. 1998;176:303–313. doi: 10.1002/(SICI)1097-4652(199808)176:2<303::AID-JCP8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Petit B, Masuda K, D’Soua AL, Otten L, Pietryla D, Hartmann DJ, Morris NP, Uebelhart D, Schmid TM, Thonar EJMA. Exp Cell Res. 1996;225:151–161. doi: 10.1006/excr.1996.0166. [DOI] [PubMed] [Google Scholar]
- 41.Bi W, Deng M, Zhang Z, Behringer RR, de Crombrugghe B. Nat Genet. 1999;22 doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 42.*****vre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. Mol Cell Biol. 1997;17:1336–1346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carl TF, Dufton C, Hanken J, Klymkowsky MW. Development. 1999;213:101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- 44.Mayor R, Guerrero N, Young RM, Gomez-Skarmeta JL, Cuellar C. Mech Dev. 2000;97:47–56. doi: 10.1016/s0925-4773(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 45.Bell DM, Leung KKH, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tarn PPL, Cheah KSE. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 46.Perissinotto D, Iacopetti P, Bellina I, Doliana R, Colombatti A, Pettway Z, Bronner-Fraser M, Shinomura T, Kimata K, Morgelin M, Lofberg J, Perris R, Shinomura T, Kimata K, Morgelin M, Lofberg J, Perris R. Development. 2000;127:2823–2842. doi: 10.1242/dev.127.13.2823. [DOI] [PubMed] [Google Scholar]
- 47.Thorogood P, Bee J, von der Mark K. Dev Biol. 1986;116:497–509. doi: 10.1016/0012-1606(86)90150-8. [DOI] [PubMed] [Google Scholar]


