SUMMARY
Ligand-specific negative regulation of cytokine-induced signaling relies on down regulation of the cytokine receptors. Down regulation of the IFNAR1 sub-unit of the Type I interferon (IFN) receptor proceeds via lysosomal receptor proteolysis, which is triggered by ubiquitination that depends on IFNAR1 serine phosphorylation. While IFN-inducible phosphorylation, ubiquitination and degradation requires the catalytic activity of the Tyk2 Janus kinase, here we found the ligand- and Tyk2-independent pathway that promotes IFNAR1 phosphorylation, ubiquitination, and degradation when IFNAR1 is expressed at high levels. A major cellular kinase activity that is responsible for IFNAR1 phosphorylation in vitro does not depend on either ligand or Tyk2 activity. Inhibition of ligand-independent IFNAR1 degradation suppresses cell proliferation. We discuss the signaling events that might lead to ubiquitination and degradation of IFNAR1 via ligand-dependent and independent pathways and their potential physiologic significance.
Keywords: cytokine, interferon, receptor, ubiquitination, degradation
INTRODUCTION
Ligand-stimulated ubiquitination and degradation of signaling receptors plays an important role in limiting the amplitude and duration of respective intracellular pathways. Cytokines of Type I interferon family (IFN, including IFNα and IFNβ) exert their anti-proliferative and anti-viral effects by engaging a cognate receptor on the cell surface followed by activation of Jak-Stat signaling proteins and expression of IFN-stimulated genes. The receptor consists of two subunits IFNAR1 and IFNAR2 (reviewed in [1]). IFNAR1 plays a key role in all aspects of IFNα-mediated effects in vitro and in vivo [2-4], and anti-proliferative activity of Type I IFN variants directly correlates with affinity of their binding to IFNAR1 [5].
Down regulation and degradation of IFNAR1 in response IFNα treatment is a pivotal mechanism limiting the extent of cellular responses to IFNα [6, 7]. Turnover of IFNAR1 requires its ubiquitination by the SCFβ-Trcp/HOS E3 ubiquitin ligase [8], which recognizes the conserved phosphorylated 534DSGNYS destruction motif [9]. Previously we reported that phosphorylation of IFNAR1 on Ser535 within this motif (essential for recruitment of β-Trcp) is increased upon stimulation of cells with IFNα [10], and catalytic activation of Tyk2 is required for such an increase [11]. Here we describe ligand- and Tyk2-independent pathway that regulates phosphorylation of IFNAR1 on Ser535 as well as IFNAR1 ubiquitination and degradation.
MATERIALS AND METHODS
Materials
Recombinant Human IFNα (Roferon) was from Hoffmann La-Roche). Recombinant pan-species specific IFNα, ATP, puromycin, methylamine HCl, N-ethylmaleimide were from Sigma; Jak Inhibitor I, (JI,) and AG490 were from Calbiochem and Toronto Research Chemicals, Inc., respectively. Cell media and sera, as well as G418 were from Invitrogen.
DNA constructs
Mammalian vectors for expression of human and mouse pCDNA3-IFNAR1-Flag (wild type and S535A mutants) as well as vectors for bacterial expression of GST-IFNAR1S535A were reported previously [9]. A bacterial expression vector for GST-IFNAR1WT protein [12, 13] was a generous gift of Dr. John J. Krolewski (University of California, Irvine, CA). The pRC-Tyk2 expression vector was described elsewhere [14].
Tissue culture and transfections
293T human embryo kidney cells (kindly provided by Z. Ronai, The Burnham Institute, San Diego, CA) and mouse embryo fibroblasts (MEFs) generated from Tyk2 knockout mice [15], were grown in DMEM in the presence of 10% fetal bovine serum (FBS) and antibiotics at 37°C and 5% CO2. Tyk2-null human fibrosarcoma 11,1-derived clones expressing either wild type (WT) or kinase deficient (KR) Tyk2 [11, 16] were grown in the same medium with addition of G418 (400μg/ml). Transfections were performed using Lipofectamine Plus (Invitrogen) for 293T cells and MEFs, or FuGene (Roche) for 11,1 derivatives at 24−48h before treatment and harvesting. Stable mass cultures of cells expressing IFNAR1 proteins were obtained by co-transfecting IFNAR1 constructs with pBABE-puro vector followed by selection in medium containing puromycin (1μg/ml).
Antibodies and immunotechniques
Antibodies specific for Flag (M2, Sigma), GST (Santa Cruz), phospho-Stat1 and Stat1 (Cell Signaling Technologies) were purchased. Antibodies against Tyk2 [14], as well as antibodies recognizing total IFNAR1 (EA12 and GB8) or IFNAR1 phosphorylated on Ser535 (in human receptor) or Ser526 (in murine receptor) [10, 17] were described previously. Antibody against ubiquitin (FK2 mAb) was from Biomol. Secondary antibodies conjugated to horseradish peroxidase were from Chemicon purchased. Immunoprecipitation and immunoblotting procedures are described elsewhere [18]. Densitometry data were obtained and analyzed using Scion Image Software (version Beta 4.0.2) and the digital images were prepared using Adobe Photoshop 7.0 Software.
In vitro IFNAR1 kinase assay
Recombinant GST-IFNAR1 was produced in bacteria and purified using glutathione Sepharose (GE Healthcare). An in vitro kinase activity assay (phosphorylation of Ser535) was carried out at 30°C for 30 min in a 20μl volume reaction mixture containing 10μg of cell lysate, 1μg of GST-IFNAR1, 2.5mM ATP, 25mM Tris-HCl pH 7.4, 10mM MgCl2, and 2mM NaF. The samples were analyzed by SDS-PAGE and immunoblotted with anti-phospho-IFNAR1 (pS535) and IFNAR1 antibodies.
Ubiquitination and degradation assays
For in vivo ubiquitination assays, cells were harvested and lysed in a buffer containing 150mM NaCl, 50mM Tris-HCl pH 7.6, 50mM NaF, 1% NP40, 0.5mM EDTA, 1 mM orthovanadate, 10 mM N-ethylmaleimide, and protease inhibitors cocktail (Sigma). Endogenous or transiently expressed IFNAR1 was immunopurified using either EA12 or M2 antibody and analyzed for conjugated ubiquitin using FK2 antibody. For the degradation assays, the cells were treated with cycloheximide (50μg/ml, Sigma) with or without IFNα for the indicated periods of time and the levels of IFNAR1 analyzed by immunoprecipitation followed by immunoblotting with the indicated antibodies.
Cell proliferation
293T and KR stable cultures were seeded into 96-well plates (4×103 trypan blue-negative cells per well) in complete medium that contained puromycin, and were washed with fresh medium every 24h thereafter to remove potentially secreted and autocrine acting cytokines. Cell proliferation was assessed after two days of incubation using a colorimetric WST-1 Cell Proliferation kit (Roche) as described previously [19].
RESULTS AND DISCUSSION
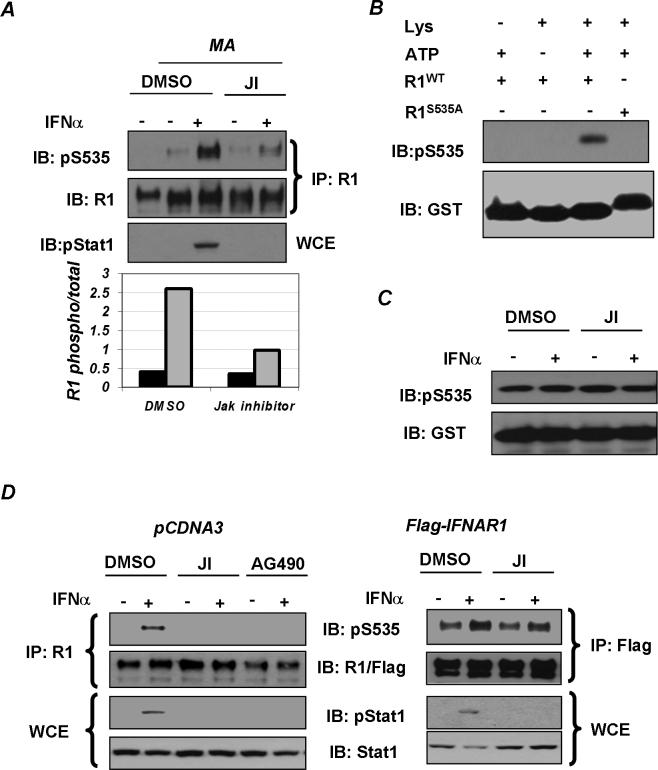
Phosphorylation of IFNAR1 on Ser535 is essential for the recruitment of the βTrcp-containing E3 ubiquitin ligase and for subsequent IFNAR1 ubiquitination and degradation that limits the magnitude and duration of IFNα signaling [9]. This phosphorylation has been previously demonstrated to be induced by treatment of cells with the ligand [10]. Intriguingly, in cells treated with an inhibitor of the lysosomal pathway, methylamine hydrochloride (MA), we detected a modest but reproducible ligand-independent basal phosphorylation of endogenous IFNAR1 on Ser535 in addition to IFNα-stimulated phosphorylation. While pre-treatment of cells with the Jak inhibitor I (JI, Calbiochem) dramatically decreased the level of ligand-induced Ser535 phosphorylation of endogenous IFNAR1, a major fraction of basal phosphorylation of IFNAR1 (∼80−85%) was insensitive to Jak inhibitor (Figure 1A). These results indicate that, besides ligand-induced phosphorylation, IFNAR1 also undergoes basal phosphorylation that does not require Jak activity and can occur on the endogenous IFNAR1, when it accumulates to high levels.
Figure 1. Basal phosphorylation of IFNAR1 in cells and in vitro.
A. Phosphorylation of endogenous IFNAR1 in 293T cells pre-treated (or not – in the left lane) with inhibitor of the lysosomal pathway methylamine (MA, 10mM, 3h) followed by treatment with Jak inhibitor (JI, Calbiochem, 0.5μM, 1hr) and by IFNα (6000IU/ml, 30 min) as indicated was analyzed by IFNAR1 immunoprecipitation (using EA12 antibody) followed by immunoblotting with a phospho-Ser535-specific antibody and with GB8 antibody against total IFNAR1. Tyrosine phosphorylation of Stat1 in whole cell extract (WCE) was analyzed by immunoblotting. Ratio of signals of Ser535 phosphorylated IFNAR1/total IFNAR1 in cells treated (grey bars) or not (black bars) with IFNα was calculated and depicted in the graph. B. In vitro kinase assay with components (added as indicated) including ATP, lysates (“Lys”) from 293T cells treated with IFNα (6000 IU/ml, 30 min) as a source of kinase, and GSTIFNAR1 proteins (wild type or S535A mutant) as substrates. Note that S535A migrates slower as it contains additional amino acids after the GST sequence. This reaction was analyzed by immunoblotting with a phospho-Ser535-specific antibody (upper panel) and with anti-GST antibody (lower panel). C. In vitro kinase assay with lysates from 293T cells treated with JI and IFNα as indicated was analyzed as in panel B. D. 293T cells were transfected with an empty vector (left panels) or Flag-IFNAR1 (right panels), pre-treated for 30min with either Jak Inhibitor I (“JI”, 0.5μM) or AG490 (50μM) and treated with or without IFNα for a further 30min (as indicated). Endogenous (left panels) and Flag-tagged (right panels) IFNAR1 proteins were immunoprecipitated with either EA12 or M2 antibodies and analyzed by immunoblotting with the indicated antibodies. Tyrosine phosphorylation of Stat1 and total Stat1 levels were analyzed by direct immunoblotting on whole cell extracts (WCE) using indicated antibodies.
We established an in vitro kinase assay to detect phosphorylation of GST-IFNAR1 protein on Ser535 using the lysates from IFNα-treated 293T cells (Figure 1B). Remarkably, lysates from untreated cells were equally effective in this assay, and pretreatment of cells with JI did not affect this activity (Figure 1C). This result suggests that cells contain a basal kinase activity that is not regulated by IFNα and does not rely on Jak activity; this activity might be responsible for basal phosphorylation of IFNAR1 in cells.
Previously we demonstrated that exogenously expressed IFNAR1 is synthesized, processed and subcellularly distributed, as well as internalized, sorted and degraded via a lysosomal pathway identically to endogenous IFNAR1 [9-11, 16, 20]. Thus, we further investigated the role of Jak in phosphorylation of IFNAR1 when the latter is expressed at high levels using transfection of Flag-tagged IFNAR1 in 293T cells. As expected, pre-treatment of cells with Jak inhibitors (JI or AG490) inhibited activation of Stat1 in cells transfected either with an empty vector or with Flag-IFNAR1 (Figure 1D). Furthermore, Jak inhibitors efficiently decreased IFNα-induced phosphorylation of endogenous IFNAR1 on Ser535. However, basal phosphorylation of transfected Flag-IFNAR1 was not affected by the Jak inhibitor (Figure 1D, right panel). This data indicates that the activity of a constitutively active kinase(s) that is responsible for phosphorylating highly expressed IFNAR1 is not regulated by Janus kinases.
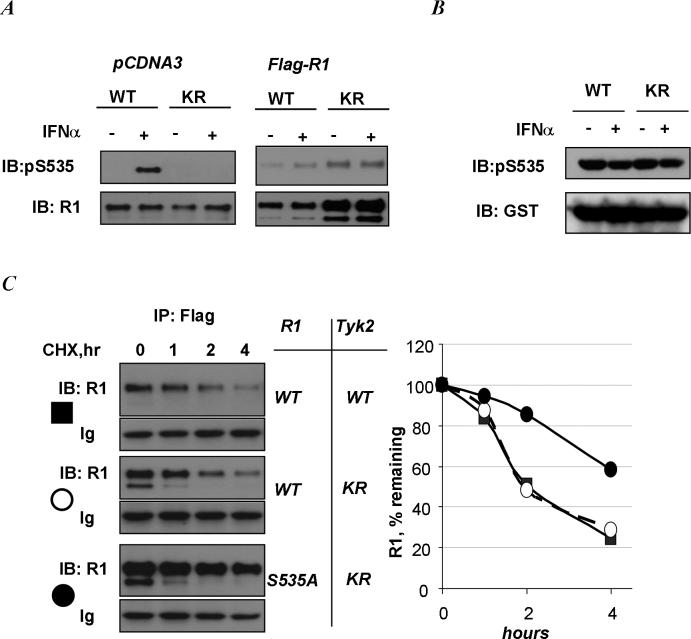
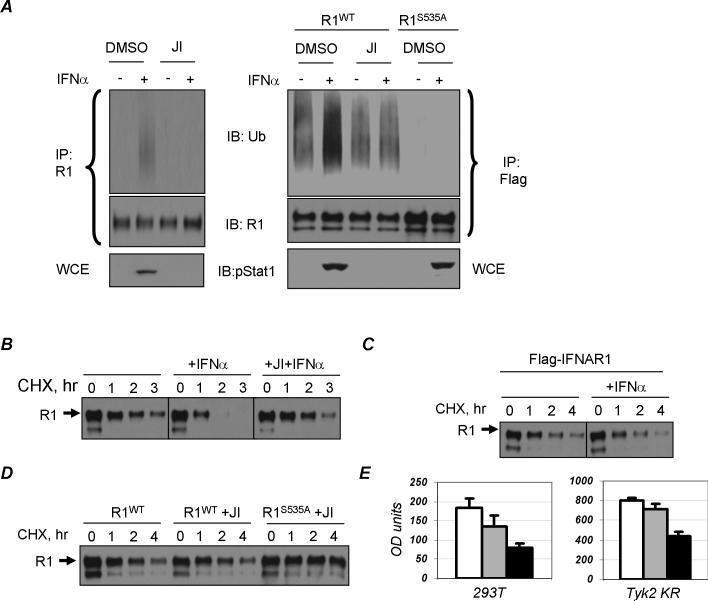
This conclusion was further corroborated using human Tyk2-null 11,1 cell-derived clones reconstituted with either wild type (WT) of kinase-deficient (KR) Tyk2 [16]. While ligand-induced phosphorylation of endogenous IFNAR1 on Ser535 was not observed in KR cells, exogenous Flag-tagged human IFNAR1 exhibited basal phosphorylation that was not inhibited in these cells (Figure 2A). Similar results were obtained in mouse embryo fibroblasts from Tyk2 knockout mice (unpublished data). Conversely, an efficient basal Ser535 kinase activity was detected in the lysates from KR cells (Figure 2B). Furthermore, analysis of IFNAR1 degradation using cycloheximide (CHX) chase in cells incubated without ligand (to prevent activation of a ligand-dependent pathway) demonstrated that the rate of exogenous IFNAR1 proteolysis is comparable in KR and WT cells (Figure 2C). Importantly, mutant IFNAR1S535A is degraded less rapidly than the wild type protein in KR cells that express catalytically inactive Tyk2. These results indicate that ligand- and Tyk2 activity-independent Ser535 phosphorylation of IFNAR1 plays an important role in regulating IFNAR1 stability in the absence of the ligand. Ligand-stumulated ubiquitination of endogenous IFNAR1 in 293T cells was decreased upon treatment with JI (Figure 3A). Overexpressed wild type Flag-IFNAR1 (but not ubiquitination-deficient Flag-IFNAR1S535A mutant) exhibited a basal level of ubiquitination that was not affected by treatment of cells with the Jak inhibitor (Figure 3A, right panel). While adding IFNα to 293T cells treated with CHX robustly stimulated the turnover of endogenous IFNAR1, pre-treatment of cells with JI prevented this stimulation (Figure 3B). On the other hand, the rate of degradation of transfected Flag-IFNAR1 only modestly increased in the presence of IFNα (Figure 3C). Furthermore, in untreated cells, treatment with JI did not inhibit exogenous wild type IFNAR1 degradation rate, which was noticeably higher than that of the IFNAR1S535A mutant (Figure 3D). Taken together, these results point to the existence of a ligand- and Jak-independent pathway; this pathway mediates ubiquitination and degradation of IFNAR1 but yet functions in a manner that is dependent on IFNAR1 phosphorylation within its destruction motif.
Figure 2. Ligand- and Jak-independent IFNAR1 kinase activity and phosphorylation of exogenously expressed IFNAR1.
A. Phosphorylation of endogenous IFNAR1 (left panel, transfected with an empty pCDNA3 vector) or exogenous Flag-IFNAR1 in Tyk2 WT or KR cells and treated with IFNα as indicated was analyzed by IFNAR1 immunoprecipitation (using EA12 or M2 antibody) followed by immunoblotting with a phospho-Ser535-specific antibody and with GB8 antibody (or M2 antibody) against total IFNAR1. B. In vitro kinase assay with lysates from Tyk2-null 11,1 cells reconstituted with either wild type (“WT”) or kinase-dead (“KR”) Tyk2, analyzed as in Figure 1C. C. Degradation of Flag-IFNAR1WT expressed in Tyk2 WT (closed squares) or KR (open circles) cells as indicated was measured by cycloheximide chase (CHX, 50μg/ml) in the absence of IFNα followed by immunoprecipitation with Flag antibody and immunoblotting analysis with IFNAR1 (GB8) antibody. Levels of heavy immunoglobulin chain (“Ig”) serve as a loading control. Degradation of IFNAR1S535A mutant in KR cells (closed circles) is also shown. This graph depicts the level of IFNAR1 remaining at each time point after cycloheximide treatment (in % relative to time point 0) quantitated from this experiment.
Figure 3. Ligand-independent ubiquitination and degradation of IFNAR1.
A. 293T cells were transfected either with an empty vector (left panel) or Flag-IFNAR1 constructs (right panels, wild type R1WT or mutant R1S535A), pre-treated for 30min with JI (0.5μM) and treated with or without IFNα for a further 30min (as indicated). Endogenous (left panels) and Flag-tagged (right panels) IFNAR1 proteins were immunoprecipitated with either EA12 or M2 antibodies, respectively. Ubiquitination (upper panel) and total levels of IFNAR1 were analyzed by immunoblotting with indicated antibodies. Tyrosine phosphorylation of Stat1 was analyzed by direct immunoblotting on whole cell extracts (WCE, lower panel) antibodies. B. 293T cells were transfected with an empty vector, pre-treated with JI and treated with cycloheximide (CHX, 50μg/ml) and IFNα as indicated. Endogenous IFNAR1 was immunoprecipitated using EA12 antibody and analyzed by immunoblotting using GB8 antibody. C. 293T cells were transfected with Flag-IFNAR1 and treated with CHX and IFNα as indicated. Exogenous IFNAR1 was immunoprecipitated using M2 (anti-Flag) antibody and analyzed by immunoblotting using GB8 antibody (against IFNAR1). D. 293T cells were transfected with either wild type (R1WT) Flag-IFNAR1 or IFNAR1S535A mutant (R1S535A), pre-treated with Jak inhibitor (as indicated) and treated with cycloheximide. Levels of IFNAR1 were analyzed as in panel C. E. Growth of 293T or Tyk2-KR cells stably transfected with an empty vector (white bar), or with wild type IFNAR1 (gray bar) or with IFNAR1S535A (black bar). Cells were plated at 4000 per well in 96-well plates without IFN and washed with fresh medium after plating and every 24hr thereafter. Growth of cells at 48hr after plating was measured using WST-1 assay (Roche) and displayed as absorbance units (minus the background). Average results of three independent experiments (each in 5−10 parallel replicates) are plotted.
Considering that forced expression of IFNAR1 inhibited proliferation of K562 cells even in the absence of the ligand [21, 22], it is plausible that an increase in IFNAR1 levels might activate a ligand-independent pathway leading to proteolysis of IFNAR1 and to alleviating its effect on cell growth. To test this hypothesis, we compared the growth inhibitory effect of expression of wild type and mutant IFNAR1S535A in the absence of ligand and in the Tyk2 KR background. If, under these conditions, the ligand/Tyk2-independent pathway is activated and helps the cells to cope with high levels of IFNAR1, the mutant receptor should elicit a stronger growth inhibition than the wild type receptor. Indeed, the growth inhibitory effect of the IFNAR1S535A mutant expression was significantly higher than that of wild type in both 293T and Tyk2 KR cells cultured without Type I IFNs (Figure 3E). These data indicate that phosphorylation of IFNAR1 on Ser535 via the alternative pathway (that does not involve ligand and Tyk2 activity) alleviates the anti-proliferative effect of IFNAR1, most likely via phosphorylation-dependent ubiquitination and degradation of IFNAR1.
We have previously demonstrated the role of the catalytic activity of Tyk2 in ligand-induced phosphorylation, ubiquitination, and degradation of IFNAR1 [11]. Here we describe the identification of an alternative pathway for ubiquitination and degradation of this receptor chain. Data from genetic, biochemical, and pharmacologic analyses demonstrate the presence of a constitutive, Jak-independent IFNAR1 kinase activity in cells and show that phosphorylation, ubiquitination, and degradation of IFNAR1 could be triggered by high levels of IFNAR1 in an IFN/Jak-independent manner. Additional stimuli that trigger the IFN/Jak-independent pathway may exist and are yet to be identified.
Future studies are also required to identify kinases that phosphorylate IFNAR1 in ligand/Tyk2-dependent and independent manners. The end result of both of these activities would be phosphorylation of critical serines to enable the recruitment of β-Trcp. A similar complex regulation is proposed for another β-Trcp substrate, the inhibitor of NF-κB (IκB). Phosphorylation of serines within the destruction motif of IκB required for β-Trcp binding and ubiquitination is mediated not only by the cytokine-inducible IκB kinases (reviewed in [23]), but also by mitogen-stimulated S6/p90Rsk1 kinase [24], and by constitutively active casein kinase 2 [25].
While the physiologic function of the alternative ligand-independent pathway largely remains to be determined, hypothetically, this second pathway may provide the means to limit the level of IFNAR1 - and, therefore, protect cells from anti-proliferative effects of high level of IFNAR1 expression [22]. Given that cells expressing IFNAR1 mutants that lack the negative regulatory domain exhibit increased signaling [7, 26], and a recent report showing that the anti-proliferative potency of Type I IFN variants is largely determined by their affinity to IFNAR1 [5], it is plausible that more than one control mechanism may exist to down regulate IFNAR1 and, hence, allow cells to cope with otherwise potentially deleterious effects of ligand-independent signaling and of future exposure to the ligands. Such or similar Jak-independent mechanisms might be potentially exploited by tumor cells or by infectious agents to evade Type I IFN control, and accordingly, could be targeted to increase the efficiency of IFN used as a therapeutic.
Acknowledgements
We thank Dr. John J. Krolewski for providing reagents. This work was supported by Public Health Service grant CA 092900 from the National Cancer Institute (to S. Y. F.) and by the grant 3158 of the Association pour la Recherche sur le Cancer (to S.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 2.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci U S A. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Constantinescu SN, Croze E, Wang C, Murti A, Basu L, Mullersman JE, Pfeffer LM. Role of interferon alpha/beta receptor chain 1 in the structure and transmembrane signaling of the interferon alpha/beta receptor complex. Proc Natl Acad Sci U S A. 1994;91:9602–9606. doi: 10.1073/pnas.91.20.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 5.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, Uze G, Schreiber G. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constantinescu SN, Croze E, Wang C, Murti A, Basu L, Mullersman JE, Pfeffer LM. Role of interferon alpha/beta receptor chain 1 in the structure and transmembrane signaling of the interferon alpha/beta receptor complex. Proc Natl Acad Sci U S A. 1994;91:9602–9606. doi: 10.1073/pnas.91.20.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu L, Yang CH, Murti A, Garcia JV, Croze E, Constantinescu SN, Mullersman JE, Pfeffer LM. The antiviral action of interferon is potentiated by removal of the conserved IRTAM domain of the IFNAR1 chain of the interferon alpha/beta receptor: effects on JAK-STAT activation and receptor down-regulation. Virology. 1998;242:14–21. doi: 10.1006/viro.1997.9002. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 9.Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. Embo J. 2003;22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 11.Marijanovic Z, Ragimbeau J, Kumar KG, Fuchs SY, Pellegrini S. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem J. 2006;397:31–38. doi: 10.1042/BJ20060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H, Krishnan K, Lim JT, Contillo LG, Krolewski JJ. Molecular characterization of an alpha interferon receptor 1 subunit (IFNaR1) domain required for TYK2 binding and signal transduction. Mol Cell Biol. 1996;16:2074–2082. doi: 10.1128/mcb.16.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Krishnan K, Greenlund AC, Gupta S, Lim JT, Schreiber RD, Schindler CW, Krolewski JJ. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. Embo J. 1996;15:1064–1074. [PMC free article] [PubMed] [Google Scholar]
- 14.Ragimbeau J, Dondi E, Vasserot A, Romero P, Uze G, Pellegrini S. The receptor interaction region of Tyk2 contains a motif required for its nuclear localization. J Biol Chem. 2001;276:30812–30818. doi: 10.1074/jbc.M103559200. [DOI] [PubMed] [Google Scholar]
- 15.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 16.Gauzzi MC, Velazquez L, McKendry R, Mogensen KE, Fellous M, Pellegrini S. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 17.Goldman LA, Zafari M, Cutrone EC, Dang A, Brickelmeier M, Runkel L, Benjamin CD, Ling LE, Langer JA. Characterization of antihuman IFNAR-1 monoclonal antibodies: epitope localization and functional analysis. J Interferon Cytokine Res. 1999;19:15–26. doi: 10.1089/107999099314379. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs SY, Chen A, Xiong Y, Pan ZQ, Ronai Z. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IkappaB and beta-catenin. Oncogene. 1999;18:2039–2046. doi: 10.1038/sj.onc.1202760. [DOI] [PubMed] [Google Scholar]
- 19.Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904–1908. doi: 10.1158/0008-5472.CAN-04-2597. [DOI] [PubMed] [Google Scholar]
- 20.Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. Embo J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colamonici OR, Porterfield B, Domanski P, Constantinescu S, Pfeffer LM. Complementation of the interferon alpha response in resistant cells by expression of the cloned subunit of the interferon alpha receptor. A central role of this subunit in interferon alpha signaling. J Biol Chem. 1994;269:9598–9602. [PubMed] [Google Scholar]
- 22.Colamonici OR, Porterfield B, Domanski P, Handa RK, Flex S, Samuel CE, Pine R, Diaz MO. Ligand-independent anti-oncogenic activity of the alpha subunit of the type I interferon receptor. J Biol Chem. 1994;269:27275–27279. [PubMed] [Google Scholar]
- 23.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 24.Schouten GJ, Vertegaal AC, Whiteside ST, Israel A, Toebes M, Dorsman JC, van der Eb AJ, Zantema A. IkappaB alpha is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. Embo J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilker R, Freuler F, Pulfer R, Di Padova F, Eder J. All three IkappaB isoforms and most Rel family members are stably associated with the IkappaB kinase 1/2 complex. Eur J Biochem. 1999;259:253–261. doi: 10.1046/j.1432-1327.1999.00028.x. [DOI] [PubMed] [Google Scholar]
- 26.Gibbs VC, Takahashi M, Aguet M, Chuntharapai A. A negative regulatory region in the intracellular domain of the human interferon-alpha receptor. J Biol Chem. 1996;271:28710–28716. doi: 10.1074/jbc.271.45.28710. [DOI] [PubMed] [Google Scholar]