Abstract
Acute-phase sera from >5 % of cases of haemorrhagic fever with renal syndrome occurring annually in Korea have been found to exhibit a fourfold or higher antibody titre to Puumala virus (PUUV) than to Hantaan virus (HTNV) by double-sandwich IgM ELISA, suggesting the existence of a PUUV-related hantavirus. Based on the phylogenetic relationships among arvicolid rodents, the royal vole (Myodes regulus) was targeted as a likely reservoir host of hantavirus. Using RT-PCR, a genetically distinct hantavirus, designated Muju virus (MUJV), was detected in lung tissue of royal voles, captured in widely separated geographical regions in Korea during 1996–2007. Pairwise analysis of the full-length S (1857 nt) and M (3634 nt) segments of MUJV indicated approximately 77 % sequence similarity with PUUV. At the amino acid level, MUJV differed from PUUV by 5.5–6.9 % (nucleocapsid) and 10.0–11.6 % (Gn and Gc envelope glycoproteins). Interstrain variation of MUJV sequences from royal voles captured in different regions suggested geographic-specific clustering. Neutralizing antibody titres against PUUV were two- to sixfold higher than to HTNV in sera of MUJV-infected Myodes regulus. Although virus isolation attempts were unsuccessful, the collective data indicate that MUJV is a distinct hantavirus species.
INTRODUCTION
An outbreak of haemorrhagic fever with renal syndrome (HFRS) occurring among United Nations troops during the Korean War first brought this disease entity into the vernacular of Western medicine (Gajdusek, 1956). Since that time, several hundred HFRS cases (with mortality rates of 4–8 %) have been reported annually among military personnel and civilians in Korea. The majority of these clinically severe HFRS cases in Korea are caused by Hantaan virus (HTNV), originally isolated from lung tissue of the striped field mouse (Apodemus agrarius) captured in Songnaeri, Gyeonggi province, Korea (Lee et al., 1978), and Seoul virus (SEOV), harboured by the Norway rat (Rattus norvegicus) (Lee et al., 1982). Soochong virus (SOOV), recently isolated from the Korean field mouse (Apodemus peninsulae), also accounts for cases of HFRS (Baek et al., 2006). By contrast, the bank vole (Myodes glareolus, formerly Clethrionomys glareolus), which serves as the reservoir host of Puumala virus (PUUV), the cause of a milder form of HFRS (known as nephropathia epidemica) throughout Scandinavia and Europe (Brummer-Korvenkontio et al., 1980; Yanagihara et al., 1984), is absent in Korea. Nevertheless, approximately 7 % of HFRS cases in Korea exhibit fourfold or higher antibody titres to PUUV than to HTNV by double-sandwich IgM capture ELISA. Based on these ‘atypical’ ELISA reactivities, the existence of one or more antigenically distinct, arvicolid rodent-borne hantaviruses has long been suspected in the Korean Peninsula.
By analysing the phylogenetic relationships between Myodes glareolus and other arvicolid rodents, the royal vole (Myodes regulus, formerly Eothenomys regulus) appeared to be a likely candidate as a reservoir of a PUUV-related hantavirus in Korea. Using RT-PCR, a genetically distinct arvicolid rodent-borne hantavirus, designated Muju virus (MUJV), was detected in the tissues of Myodes regulus captured in widely separated geographical regions in Korea during 1996–2007. The discovery of MUJV adds to the growing list of hantaviruses in Korea and may account for HFRS cases that cannot be attributed to HTNV, SOOV or SEOV infection.
METHODS
Rodent trapping and serology
Live trapping of royal voles was conducted in 13 counties in six provinces during 1996–2007 (Table 1; Fig. 1). Lung and spleen tissues of royal voles, collected with separate sterile instruments, were frozen at −70 °C until used for gene amplification and virus isolation.
Table 1. Prevalence of hantavirus infection in Myodes regulus captured in Korea, 1996–2007.
Vole sera, diluted 1 : 32 in PBS, were examined for IgG antibodies against PUUV by an indirect immunofluorescent antibody technique.
| Capture site
|
Year | No. captured | No. seropositive | Prevalence (%) | |
|---|---|---|---|---|---|
| Province | County | ||||
| Jeollabuk | Muju | 1996 | 8 | 2 | 25 |
| 1997 | 5 | 0 | 0 | ||
| 1998 | 1 | 0 | 0 | ||
| 1999 | 44 | 7 | 16 | ||
| 2000 | 23 | 3 | 13 | ||
| 2001 | 11 | 0 | 0 | ||
| 2002 | 6 | 1 | 17 | ||
| 2004 | 10 | 2 | 20 | ||
| Gangwon | Cheolwon | 2002 | 7 | 0 | 0 |
| Hongcheon | 1995 | 100 | 12 | 12 | |
| 1997 | 13 | 0 | 0 | ||
| 1998 | 8 | 1 | 13 | ||
| 1999 | 25 | 4 | 16 | ||
| 2000 | 4 | 0 | 0 | ||
| 2001 | 1 | 0 | 0 | ||
| 2002 | 11 | 1 | 9 | ||
| Inje | 1997 | 16 | 3 | 19 | |
| 1999 | 7 | 1 | 14 | ||
| 2004 | 6 | 1 | 17 | ||
| Pyungchang | 2001 | 33 | 0 | 0 | |
| 2002 | 30 | 1 | 3 | ||
| Whacheon | 1998 | 7 | 1 | 14 | |
| Gyeonggi | Gapyung | 1998 | 3 | 0 | 0 |
| Paju | 2005 | 1 | 0 | 0 | |
| 2007 | 4 | 0 | 0 | ||
| Pocheon | 1997 | 2 | 0 | 0 | |
| 1998 | 1 | 0 | 0 | ||
| 2001 | 1 | 0 | 0 | ||
| 2002 | 3 | 0 | 0 | ||
| 2003 | 4 | 0 | 0 | ||
| 2004 | 12 | 0 | 0 | ||
| 2005 | 6 | 0 | 0 | ||
| Yeonchon | 2005 | 2 | 0 | 0 | |
| Chungcheongnam | Yesan | 1997 | 7 | 0 | 0 |
| Gyeongsangbuk | Munkyung | 1998 | 13 | 0 | 0 |
| Jeollanam | Yeongkwang | 2003 | 4 | 0 | 0 |
Fig. 1.
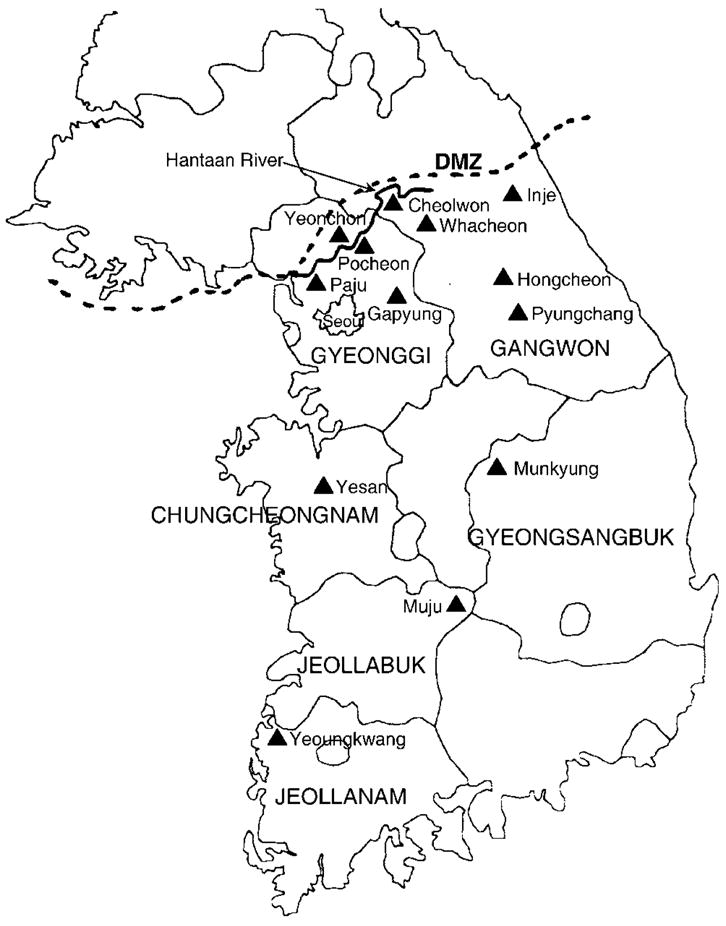
Map of Korea, showing the six provinces (Chungcheongnam, Gangwon, Gyeonggi, Gyeongsangbuk, Jeollabuk and Jeollanam) and 13 counties (▲), where Myodes regulus voles were trapped during 1996–2007. Mt Deogyu (1614 m) and Suseongdae (1240 m), in Muju county, Jeollabuk province, located about 190 km south and 70 km east of Seoul, were two principal capture sites. The nine trapping sites in Gangwon province comprised Damte valley in Cheolwon county; Mt Gyebang (1577 m) in Hongcheon county; Mt Jeombong (1424 m), Mt Gachilbong (1241 m), Mt Hanseg (1104 m), Garibong (1519 m) and Hangyeryung (1307 m) in Inje county; Unduryeong (1089 m) in Pyungchang county; and Mt Whaag (1468 m) in Whacheon county. Other capture sites were Mt Myungseong (923 m), Mt Unbong (868 m), Gapyung, Pocheon and Yeonchon counties and Paju city in Gyeonggi province; Mt Gaya in Yesan county, Chungcheongnam province; Mt Joryeong (1017 m) in Munkyung county, Gyeongsangbuk province; and Yeongkwang county, Jeollanam province.
Royal vole sera, diluted 1 : 32, were examined for IgG antibodies against PUUV by an indirect immunofluorescent antibody (IFA) technique (Lee et al., 1985). In addition, sera from IFA-seropositive voles were tested for neutralizing antibodies against PUUV, HTNV, SEOV and Prospect Hill virus (PHV) by a plaque-reduction neutralization test (PRNT) (Baek et al., 2006; Lee et al., 1985). Briefly, serial twofold dilutions of sera from royal voles captured in Jeollabuk and Gangwon provinces were incubated with 50–100 p.f.u. PUUV, PHV, HTNV or SEOV at 4 °C overnight. Thereafter, virus/ serum mixtures were inoculated onto confluent monolayers of Vero E6 cells (ATCC CRL-1586) grown in 96-well flat-bottomed tissue culture plates, adsorbed at 37 °C for 1 h and then overlaid with Dulbecco’s modified Eagle’s medium (DMEM) containing 0.7 % methylcellulose (Sigma). After incubation for 7 (HTNV and SEOV) or 14 (PUUV and PHV) days, monolayers were fixed with 2 % paraformaldehyde and plaques were counted after immunostaining with rat antiserum against the respective hantavirus and horseradish peroxidase-conjugated goat anti-rat IgG and diaminobenzidine (0.7 mg ml−1). PRNT titres were expressed as the reciprocal of the highest serum dilution giving an 80 % reduction.
Virus isolation
Subconfluent monolayers of Vero E6 cells were inoculated with 5 % suspensions of Myodes regulus lung and spleen tissue in which MUJV sequences were detected by RT-PCR. Inocula were adsorbed by centrifugation at 670 g for 2 h at 25 °C. Subsequently, cells were maintained in DMEM supplemented with 5 % heat-inactivated fetal bovine serum and subcultured at 10- to 14-day intervals, at which time cells were examined for hantavirus antigens by IFA test, using convalescent-phase sera from HFRS patients and rat and mouse antisera specific for PUUV. Vero E6 cells at each passage were also examined for MUJV sequences by RT-PCR. In further experiments, suckling Mongolian gerbils (Meriones unguiculatus) were inoculated by the intraperitoneal route with tissue homogenates (Yanagihara et al., 1985) and their tissues analysed for MUJV sequences by RT-PCR.
RT-PCR amplification of hantavirus
Total RNA, extracted from 20–50 mg of each lung tissue of PUUV-seropositive Myodes regulus using RNAzol (Gibco-BRL), was reverse transcribed using a Superscript II RNase H− reverse transcriptase kit (Gibco-BRL). Hantavirus sequences were then amplified using newly designed and previously described (S+20, M+1) oligonucleotide primers (see Supplementary Table S1, available in JGV Online) (Song et al., 2004; Baek et al., 2006). Gene amplification reactions were performed in 50 μl reaction mixtures containing 200 mM dNTPs, 0.5 U Supertherm polymerase (PureTech Co.), 1 μg cDNA and 10 pM each primer. Initial denaturation at 98 °C for 5 min was followed by touchdown cycling with denaturation at 98 °C for 1 min, annealing from 48 to 38 °C for 1 min and elongation at 72 °C for 1 min 30 s, followed by 20 cycles of denaturation at 98 °C for 1 min, annealing at 42 °C for 1 min and elongation at 72 °C for 1 min 30 s in a Mastercycler ep gradient S (Eppendorf AG). PCR products were size fractionated by electrophoresis on 1–1.5 % agarose gels containing ethidium bromide at 0.5 mg ml−1 and purified using a Wizard PCR Preps DNA Purification System (Promega). DNA sequencing was performed in both directions using a dye-termination cycle-sequencing ready reaction kit (Applied Biosystems) on an automated sequencer (model 377, Perkin Elmer).
PCR amplification of mitochondrial DNA
Total DNA was extracted from fresh vole liver tissue using a QIAamp tissue kit (Qiagen). To study the phylogenetic relationship of Myodes regulus from various geographical regions in Korea, the cytochrome b region of mtDNA was amplified by PCR using previously described universal primers that amplify a 482 bp product: +L14115 (5′-CGAAGCTTG-ATATGAAAAACCATCGTTG-3′); and −L14532 (5′-GCAGCCCCT-CAGAATGATATTTGTCCAC-3′) (Smith & Patton, 1991). PCR was performed in 50 μl reaction mixtures containing 200 μM dNTP and 1.25 U rTaq polymerase (Takara). The initial denaturation step was at 95 °C for 4 min, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and elongation at 72 °C for 1 min in a PTC-200 DNA Engine Peltier thermal cycler (MJ Research). Amplicons were cloned using the pSTBlue-1 vector (Novagen) and sequenced as described above.
Phylogenetic analysis
MUJV sequences from 13 royal voles were aligned and compared with previously published sequences of PUUV strains (Horling et al., 1995; Lundkvist et al., 1998; Plyusnin et al., 1994b; Reip et al., 1995; Sironen et al., 2001; Xiao et al., 1993) and other arvicolid rodent-borne hantaviruses, including PUUV-related viruses (Tobetsu) from Myodes rufocanus in Hokkaido, Japan (Kariwa et al., 1995), Khabarovsk virus (KHAV) from Microtus fortis (Horling et al., 1996), and Tula virus (TULV) from Microtus arvalis (Plyusnin et al., 1994a; Song et al., 2004) and Pitymys subterraneus (Song et al., 2002). Alignment of the full-length S and M genomic segments was carried out using CLUSTAL W (Lasergene program version 5; DNASTAR). For phylogenetic analysis, the neighbour-joining and maximum-parsimony methods (PAUP version 4.0; Sinauer Associates) were used (Swofford, 2003). Topologies were evaluated by bootstrap analysis of 1000 iterations.
RESULTS
Serology and virus isolation
During trapping expeditions between 1996 and 2007, 439 royal voles were captured in mountainous regions in Muju county, Jeollabuk province; Yeongkwang county, Jeollanam province; Hongcheon, Inje, Whacheon, Pyungchang and Cheolwon counties, Gangwon province; Pocheon, Yeonchon and Gapyung counties and Paju city, Gyeonggi province; Yesan county, Chungcheongnam province; and Munkyung county, Gyeongsangbuk province. Sera from 9.1 % (40/439) exhibited IgG antibodies to PUUV, as determined by IFA (Table 1). The IFA seropositivity rates were 13.8 % (15/108) and 9.3 % (25/268) in Myodes regulus captured in Jeollabuk and Gangwon provinces, respectively. None of the 63 Myodes regulus captured in the other four provinces had serological evidence of hantavirus infection.
Multiple attempts to isolate MUJV in Vero E6 cell cultures and in Mongolian gerbils were unsuccessful. However, in tests of sera from anti-PUUV IFA-positive Myodes regulus, captured in Jeollabuk and Gangwon provinces, for neutralizing antibodies against PUUV, PHV, HTNV and SEOV, PRNT titres against PUUV were consistently two-to sixfold higher (reciprocal titres of 320–1280), supporting the existence of a PUUV-like hantavirus.
Sequence analysis of MUJV
Genetically distinct hantaviral sequences, amplified by RT-PCR from lung tissue of 13 PUUV-seropositive Myodes regulus, were designated MUJV. The full-length S segment, a partial 208 nt region of the S segment, a full-length M segment and a 241 nt region of the Gc glycoprotein-encoding M segment were sequenced from four, eight, one and seven MUJV strains, respectively. The GenBank accession numbers for the S and M genomic sequences are provided in Table 2.
Table 2. Muju virus sequences from Myodes regulus captured in Jeollabuk and Gangwon provinces, Korea.
Elevations: Mt Deogyu (1614 m), Suseongdae (1240 m), Mt Gyebang (1577 m), Mt Gachilbong (1241 m) and Mt Jeombong (1424 m).
| Capture site
|
MUJV strain | GenBank accession no.
|
||||
|---|---|---|---|---|---|---|
| Province | County | Mountain | Date | S segment | M segment | |
| Jeollabuk | Muju | Deogyu | 31/10/96 | Muju 96-1 | DQ138133* | DQ138132 |
| 31/10/96 | Muju 96-5 | ND | DQ138134 | |||
| 31/05/99 | Muju 99-27 | DQ138140* | DQ138139 | |||
| 31/05/99 | Muju 99-28 | DQ138142* | DQ138141 | |||
| 14/05/00 | Muju 00-18 | DQ138128* | DQ138127 | |||
| 15/06/04 | Muju 04-4 | DQ138129 | EF198313* | |||
| Suseongdae | 01/03/99 | Muju 99-7 | DQ138138 | DQ138137 | ||
| 15/06/04 | Muju 04-10 | DQ138130 | ND | |||
| Gangwon | Hongcheon | Gyebang | 14/11/99 | Muju 99-63 | DQ138143 | ND |
| 14/11/99 | Muju 99-68 | DQ138144 | ND | |||
| 14/11/99 | Muju 99-72 | DQ138135 | ND | |||
| Inje | Gachilbong | 07/08/97 | Muju 97-32 | DQ138136 | DQ138125 | |
| Jeombong | 19/06/04 | Muju 04-11 | DQ138131 | ND | ||
Full-length S or M segment sequences.
ND, Not determined.
The full-length S genomic segment of MUJV strains 96-1, 99-27, 99-28 and 00-18 was 1857 nt, with a predicted nucleocapsid protein of 433 aa starting at nt 43 and a 511 nt 3′ non-coding region (NCR). The 18 and 17 nt of the 5′ and 3′ ends of the S and M segments were determined empirically. Sequence analysis of the entire S genomic segment showed that MUJV differed from PUUV strains by 23.1–23.7 % and 5.5–6.9 % at the nucleotide and amino acid levels, respectively (Table 3). A hypothetical second open reading frame was identified, as for PUUV, at nt 83–355 and encoding 90 aa. Further analysis of a 208 nt region of the S genomic segment showed that MUJV differed by 18 and 26 % at the nucleotide level, respectively, from the PUUV-related TOB and CRF110 strains, amplified from Myodes rufocanus captured in Hokkaido, Japan, and Far East Russia. The interstrain variation among MUJV strains from Jeollabuk (seven strains) and Gangwon (five strains) provinces based on a 208 nt region of the S genomic segment was 16.8–19.2 % and 1.4–2.9 % at the nucleotide and amino acid levels, respectively. In the hypervariable region of the nucleocapsid protein, between aa 244 and 269, MUJV diverged by 3–5 aa from PUUV strains. However, the functional significance of these substitutions is unknown.
Table 3.
Nucleotide and amino acid sequence similarities (%) of the full-length S segment of MUJV strain 96-1 and the full-length M segment of MUJV strain 04-4 and other arvicolid rodent borne-hantaviruses
| Virus strain | S segment
|
M segment
|
||
|---|---|---|---|---|
| 1857 nt | 433 aa | 3634 nt | 1142 aa | |
| PUUV Sotkamo | 76.9 | 94.2 | 77.5 | 88.9 |
| PUUV Cg1820 | 76.7 | 93.1 | 77.1 | 88.4 |
| PUUV Kami8Cr95* | 76.3 | 94.5 | 73.5† | 90.0† |
| TULV T53 | 67.6 | 80.5 | 72.6 | 79.5 |
| KHAV Mf43 | 71.6 | 86.4 | 73.8 | 83.4 |
| IVV Mc47‡ | 71.4 | 80.6 | 75.4§ | 78.6§ |
| PHV PH-1 | 70.5 | 79.5 | 69.8 | 76.0 |
Also known as Hokkaido virus.
Nucleotide and amino acid sequence homologies are based on a 979 nt region of the Gc glycoprotein-encoding M segment.
I VV, Isla Vista virus.
Nucleotide and amino acid sequence homologies are based on a 337 nt region of the Gc glycoprotein-encoding M segment.
The entire M genomic segment of MUJV strain 04-4 was 3634 nt, with a predicted glycoprotein of 1142 aa starting at nt 40, and a 165 nt 3′ NCR. Pairwise analysis of the entire M segment of MUJV indicated 73.5–77.5 % sequence similarity to PUUV strains. At the amino acid level, MUJV differed from PUUV by 10.0–11.6 %. Sequence analysis of a 241 nt region spanning the Gc glycoprotein-encoding M segment revealed that the interstrain variation among MUJV strains from Jeollabuk (seven strains) and Gangwon (one strain) provinces was 19.5–20.0 % and 7.9–9.3 % at the nucleotide and amino acid levels, respectively. These values were higher than those calculated for PUUV strains from Finland and Russia (Plyusnin et al., 1995). The genetic distance of MUJV and PUUV strains was 19.9–25.7 % at the nucleotide level and 8.8–11.3 % at the amino acid level.
Phylogenetic analyses of MUJV
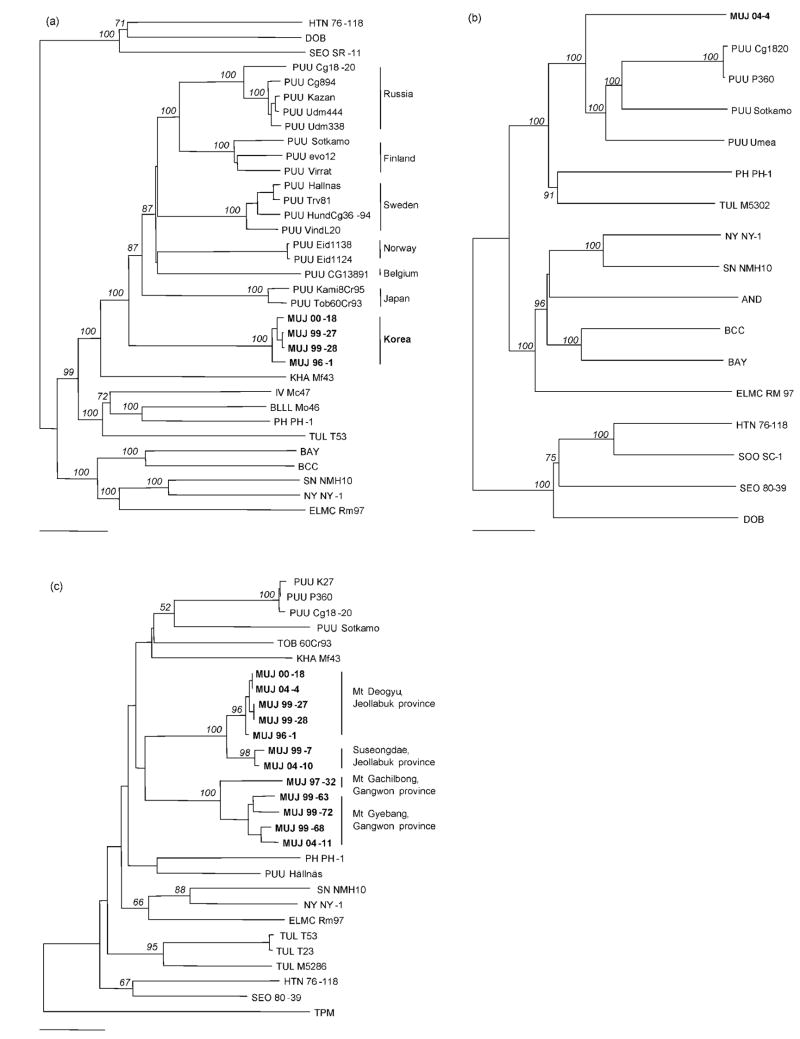
A neighbour-joining phylogenetic tree based on the entire S segment showed that MUJV was genetically distinct from PUUV strains (Fig. 2a). Similar topologies were found in phylogenetic trees based on the entire M segment of MUJV (Fig. 2b). Phylogenetic trees based on a 208 nt region of the S segment (Fig. 2c) and a 241 nt region of the Gc glycoprotein-encoding M segment (data not shown) indicated clustering of MUJV strains by geographical origin. Within the MUJV lineage, strains from Jeollabuk and Gangwon provinces were phylogenetically distinguishable (Fig. 2c). Moreover, sublineages of MUJV were found in voles captured at Mt Deogyu and Suseongdae (Jeollabuk province) and at Mt Gyebang and Mt Gachilbong (Gangwon province).
Fig. 2.
Neighbour-joining phylogenetic trees based on the full-length S segment (a), the full-length M segment (b) and a 208 nt region (nt 1032–1239) of the S segment (c) of MUJV strains and other arvicolid rodent-borne hantaviruses, including Puumala (PUU), Tula (TUL), Khabarovsk (KHA), Isla Vista (IV), Bloodland Lake (BLLL) and Prospect Hill (PH) viruses. The phylogenetic positions of hantaviruses harboured by murid rodents, including Hantaan (HTN), Seoul (SEO), Soochong (SOO) and Dobrava (DOB) viruses, and those carried by neotomine and sigmodontine rodents, including Sin Nombre (SN), New York (NY), El Moro Canyon (ELMC), Black Creek Canal (BCC), Bayou (BAY) and Andes (AND) viruses, are also shown. Thottapalayam (TPM) virus is a prototype shrew-borne hantavirus. The numbers at each node are percentage bootstrap probabilities, as determined for 1000 iterations using PAUP version 4.0. The GenBank accession numbers for the MUJV sequences are provided in Table 2. Bars, 0.05 substitutions per site.
mtDNA sequence analysis
The identities of all Myodes regulus voles in which MUJV sequences were detected, as well as five Myodes regulus voles in which MUJV RT-PCR was negative, were verified by mtDNA sequence analysis. Phylogenetic analysis, based on a 426 nt cytochrome b region of mtDNA, showed that Myodes regulus in Korea clustered together and were evolutionarily distinct from Myodes glareolus, Myodes rufocanus, P. subterraneus and Microtus arvalis (Fig. 3). Myodes regulus shared the same ancestral node as other Myodes voles but formed a different node from Microtus and Pitymys voles.
Fig. 3.
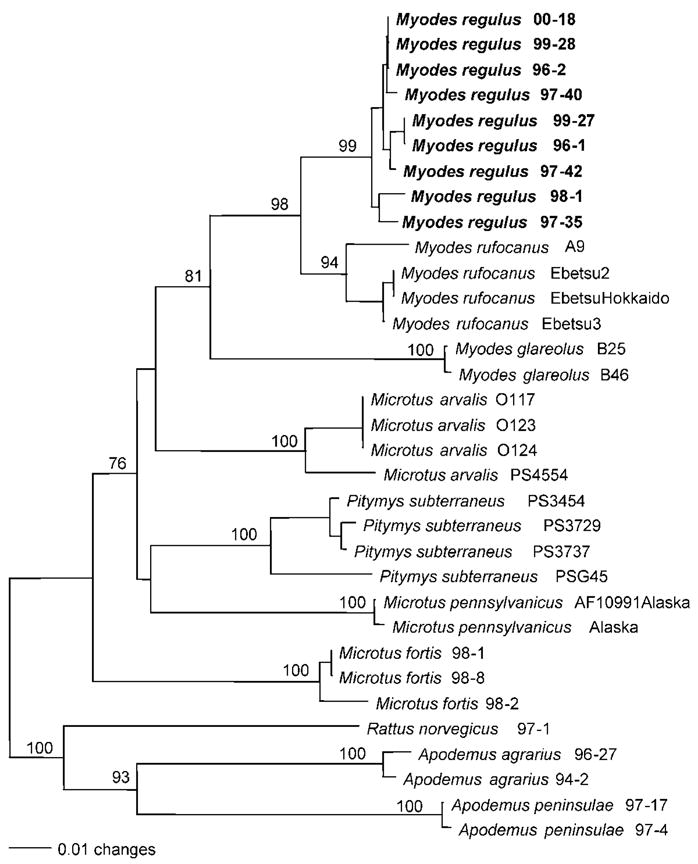
Phylogenetic tree based on the 426 nt cytochrome b region of mitochondrial DNA sequences of arvicolid rodents, showing the phylogenetic position of Myodes regulus in relation to Myodes rufocanus, Myodes glareolus, P. subterraneus, Microtus arvalis, Microtus fortis and Microtus pennsylvanicus. The phylogenetic positions of murid rodents (Rattus norvegicus, Apodemus agrarius and Apodemus peninsulae) are included. Myodes mtDNA sequences were derived from royal voles in which MUJV sequences had been detected. The numbers at each node are percentage bootstrap probabilities, as determined for 1000 iterations using PAUP version 4.0. The GenBank accession numbers for the nine Myodes regulus mtDNA sequences are DQ138116–DQ138124.
DISCUSSION
Phylogenetic analyses of full-length viral genomic sequences indicate that hantaviruses segregate into clades that parallel the evolution of their murid, arvicolid, neotomine and sigmodontine rodent reservoir hosts (Plyusnin et al., 1996; Vapalahti et al., 2003). Thus, by constructing phylogenetic trees based on mitochondrial or nuclear gene DNA sequences of rodents in defined subfamilies, the robust rodent–hantavirus association should allow targeted discovery of new hantaviruses. In applying this predictive paradigm to Apodemus mice, a new hantavirus from the Korean field mouse (A. peninsulae) was recently isolated (Baek et al., 2006). By extending this approach to arvicolid rodents, the royal vole (Myodes regulus) was hypothesized as harbouring a hantavirus because of its close phylogenetic proximity to bank voles (Myodes glareolus) and grey-sided voles (Myodes rufocanus), which are known reservoirs. Thus, the search for a PUUV-like hantavirus in Korea was focused on Myodes regulus as a means of ascertaining the hantavirus involved in HFRS cases occurring annually in Korea (Lee, 1982; Sachar et al., 2003), which show a higher antibody titre to PUUV than to HTNV by double-sandwich IgM ELISA.
Earlier efforts to detect hantaviruses that might account for this seroreactivity in Korea were unsuccessful. Such efforts focused largely on the reed vole (Microtus fortis) (Lee et al., 1978). In this report, royal voles from widely separated regions in Korea were found to harbour a genetically distinct hantavirus (designated MUJV). Phylogenetically intermediate between Microtus and other Myodes voles (Nowak, 1999), Myodes regulus, a member of the subfamily Arvicolinae, inhabits mountainous regions at elevations above 500 m in Korea and north-eastern China.
A. agrarius, the natural reservoir of the prototype HTNV, is the predominant species of field mouse in Korea (Baek et al., 2002; Song et al., 2000). A. peninsulae, which usually inhabits mountainous areas, is the second most common field mouse species in Korea and harbours SOOV, a genetically distinct, HFRS-causing hantavirus (Baek et al., 2006). In Scandinavia, Myodes glareolus is the primary natural reservoir of PUUV, which has also been found in Myodes rufocanus (Brummer-Korvenkontio et al., 1980; Niklasson et al., 1995; Yanagihara et al., 1984). A PUUV-related hantavirus, called Hokkaido virus, has been identified in Myodes rufocanus in Hokkaido, Japan (Kariwa et al., 1995). That Myodes regulus would harbour a hantavirus is not unexpected based on its close phylogenetic relationship with other known arvicolid rodent reservoir species. Also, that MUJV strains would share a common ancestry with PUUV and yet be evolutionarily distinct from PUUV strains is congruent with the co-evolution of these hantaviruses and their arvicolid rodent reservoir hosts.
Geographic-specific clustering has been recognized for arvicolid rodent-borne hantaviruses, including PUUV and TULV (Plyusnin et al., 1994a, b, 1995; Song et al., 2002, 2004). Recently, sequence and phylogenetic analyses of the partial M genomic segment of TULV, isolated from the European common vole (Microtus arvalis) in Poland, revealed that the genotypic segregation of Microtus-borne strains of TULV was dependent on their geographical origin (Song et al., 2004). In this study, phylogenetic trees based on the partial S and M segment sequences similarly showed geographic-specific clustering of MUJV strains from royal voles captured in Suseongdae, located 18 km north-north-west of Mt Deogyu in Jeollabuk province, as well as from royal voles captured at Mt Gachilbong, located 45 km east of Mt Gyebang, in Gangwon province.
Traditionally, the cross-PRNT has been the accepted standard for the serological classification of hantaviruses (Lee et al., 1985). The inability to isolate MUJV, despite intensive attempts over many years, is not unusual for this group of viruses, which are notoriously difficult to isolate in cell culture. In the absence of a MUJV isolate, however, sera from anti-PUUV IFA-positive Myodes regulus exhibited PRNT titres against PUUV that were two- to sixfold higher than against HTNV, SEOV or PHV. Moreover, when hantavirus isolates are unavailable, taxonomic relationships have been gleaned by using phylogenetic approaches. For example, phylogenetic analysis of partial M segment sequences has correlated well with cross-neutralization data and therefore may be a useful adjunct to classifying hantaviruses (Xiao et al., 1994). Based on the genetic divergence from PUUV and other PUUV-like hantaviruses from northern Japan, as well as the phylogenetic analyses of full-length S and M segment sequences and the PRNT data, MUJV is likely to be a new hantavirus species. However, future studies are warranted to ascertain whether MUJV causes human infection and disease.
Acknowledgments
This work was supported in part by grants from MOST (KOSEF), Korea (no. R21-2005-000-10017-0) and from the National Center for Research Resources, National Institutes of Health (P20RR018727 and G12RR003061), as well as grants from the US Department of Defense, Global Emerging Infections Surveillance and Response System (GEIS), Silver Spring, MD, and the Armed Forces Medical Intelligence Center (AFMIC), Ft Detrick, MD.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the S and M genomic sequences of Muju virus determined in this study are DQ138125, DQ138127–DQ138144 and EF198313.
A supplementary table showing oligonucleotide primers for amplification of the S and M segments of MUJV is available with the online version of this paper.
References
- Baek LJ, Song JW, Park KS, Kho EY, Ryu SH, Yanagihara R, Song KJ. Seroepizootiology of hantavirus infection in indigenous rodents in Korea, during 1995–2000. J Microbiol Biotechnol. 2002;12:53–58. [Google Scholar]
- Baek LJ, Kariwa H, Lokugamage K, Yoshimatsu K, Arikawa J, Takashima I, Chung SY, Lee EJ, Moon SS, et al. Soochong virus: a genetically distinct hantavirus isolated from Apodemus peninsulae in Korea. J Med Virol. 2006;78:290–297. doi: 10.1002/jmv.20538. [DOI] [PubMed] [Google Scholar]
- Brummer-Korvenkontio M, Vaheri A, Hovi T, von Bonsdorff CH, Vuorimies J, Manni T, Penttinen K, Oker-Blom N, Lähdevirta J. Nephropathia epidemica: detection of antigen in bank voles and serologic diagnosis of human infection. J Infect Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. [DOI] [PubMed] [Google Scholar]
- Gajdusek DC. Hemorrhagic fevers in Asia: a problem in medical ecology. Geog Rev. 1956;46:20–42. [Google Scholar]
- Horling J, Lundkvist A, Persson K, Mullaart M, Dzagurova T, Dekonenko A, Tkachenko E, Niklasson B. Detection and subsequent sequencing of Puumala virus from human specimens by PCR. J Clin Microbiol. 1995;33:277–282. doi: 10.1128/jcm.33.2.277-282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling J, Chizhikov V, Lundkvist A, Jonsson M, Ivanov L, Dekonenko A, Niklasson B, Dzagurova T, Peters CJ, et al. Khabarovsk virus: a phylogenetically and serologically distinct hantavirus isolated from Microtus fortis trapped in far-east Russia. J Gen Virol. 1996;77:687–694. doi: 10.1099/0022-1317-77-4-687. [DOI] [PubMed] [Google Scholar]
- Kariwa H, Yoshizumi S, Arikawa J, Yoshimatsu K, Takahashi K, Takashima I, Hashimoto N. Evidence for the existence of Puumula-related virus among Clethrionomys rufocanus in Hokkaido, Japan. Am J Trop Med Hyg. 1995;53:222–227. doi: 10.4269/ajtmh.1995.53.222. [DOI] [PubMed] [Google Scholar]
- Lee HW. Korean hemorrhagic fever. Prog Med Virol. 1982;28:96–113. [PubMed] [Google Scholar]
- Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis. 1978;137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- Lee HW, Baek LJ, Johnson KM. Isolation of Hantaan virus, the etiologic agent of Korean hemorrhagic fever from wild urban rats. J Infect Dis. 1982;146:638–644. doi: 10.1093/infdis/146.5.638. [DOI] [PubMed] [Google Scholar]
- Lee PW, Gibbs CJ, Jr, Gajdusek DC, Yanagihara R. Serotypic classification of hantaviruses by indirect immunofluorescent antibody and plaque-reduction neutralization tests. J Clin Microbiol. 1985;22:940–944. doi: 10.1128/jcm.22.6.940-944.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist A, Wiger D, Horling J, Sjolander KB, Plyusnin A, Mehl R, Vaheri A, Plyusnin A. Isolation and characterization of Puumala hantavirus from Norway: evidence for a distinct phylogenetic sublineage. J Gen Virol. 1998;79:2603–2614. doi: 10.1099/0022-1317-79-11-2603. [DOI] [PubMed] [Google Scholar]
- Niklasson B, Hornfeldt B, Lundkvist A, Bjorsten S, LeDuc J. Temporal dynamics of Puumala virus antibody prevalence in voles and of nephropathia epidemica incidence in humans. Am J Trop Med Hyg. 1995;53:134–140. doi: 10.4269/ajtmh.1995.53.134. [DOI] [PubMed] [Google Scholar]
- Nowak RM. Walker’s Mammals of the World. Baltimore: Johns Hopkins University; 1999. [Google Scholar]
- Plyusnin A, Vapalahti O, Lankinen H, Lehväslaiho H, Apekina N, Myasnikov Y, Kallio-Kokko H, Henttonen H, Lundkvist A, et al. Tula virus: a newly detected hantavirus carried by European common voles. J Virol. 1994a;68:7833–7839. doi: 10.1128/jvi.68.12.7833-7839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Ulfves K, Lehvaslaiho H, Apekina N, Gavrilovskaya I, Blinov V, Vaheri A. Sequences of wild Puumala virus genes show a correlation of genetic variation with geographic origin of the strains. J Gen Virol. 1994b;75:405–409. doi: 10.1099/0022-1317-75-2-405. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Lehvaslaiho H, Apekina N, Mikhailova T, Gavrilovskaya I, Laakkonen J, Niemimaa J, Henttonen H, et al. Genetic variation of wild Puumala viruses within the serotype, local rodent populations and individual animal. Virus Res. 1995;38:25–41. doi: 10.1016/0168-1702(95)00038-r. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Reip A, Haring B, Sibold C, Stohwasser R, Bautz EK, Darai G, Meisel H, Krüger DH. Coding strategy of the S and M genomic segments of a hantavirus representing a new subtype of the Puumala serotype. Arch Virol. 1995;140:2011–2026. doi: 10.1007/BF01322689. [DOI] [PubMed] [Google Scholar]
- Sachar DS, Narayan R, Song JW, Lee HC, Klein TA. Hantavirus infection in an active duty U.S. Army soldier stationed in Seoul, Korea. Mil Med. 2003;168:231–233. [PubMed] [Google Scholar]
- Sironen T, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus. J Virol. 2001;75:11803–11810. doi: 10.1128/JVI.75.23.11803-11810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MF, Patton JL. Variation in mitochondrial cytochrome b sequence in natural populations of South American Akodontine rodents (Muridae: Sigmodontinae) Mol Biol Evol. 1991;8:85–103. doi: 10.1093/oxfordjournals.molbev.a040638. [DOI] [PubMed] [Google Scholar]
- Song JW, Baek LJ, Kim SH, Kho EY, Kim JH, Yanagihara R, Song KJ. Genetic diversity of Apodemus agrariusborne Hantaan virus in Korea. Virus Genes. 2000;21:227–232. doi: 10.1023/a:1008199800011. [DOI] [PubMed] [Google Scholar]
- Song JW, Gligic A, Yanagihara R. Identification of Tula hantavirus in Pitymys subterraneus captured in the Cacak region of Serbia-Yugoslavia. Int J Infect Dis. 2002;6:31–36. doi: 10.1016/s1201-9712(02)90133-5. [DOI] [PubMed] [Google Scholar]
- Song JW, Baek LJ, Song KJ, Skrok A, Markowski J, Bratosiewicz J, Kordek R, Liberski PP, Yanagihara R. Characterization of Tula virus from common voles (Microtus arvalis) in Poland: evidence for geographic-specific phylogenetic clustering. Virus Genes. 2004;29:239–247. doi: 10.1023/B:VIRU.0000036384.50102.cf. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Vapalahti O, Mustonen J, Lundkvist A, Henttonen H, Plyusnin A, Vaheri A. Hantavirus infections in Europe. Lancet Infect Dis. 2003;3:653–661. doi: 10.1016/s1473-3099(03)00774-6. [DOI] [PubMed] [Google Scholar]
- Xiao SY, Spik KW, Li D, Schmaljohn CS. Nucleotide and deduced amino acid sequences of the M and S genome segments of two Puumala virus isolates from Russia. Virus Res. 1993;30:97–103. doi: 10.1016/0168-1702(93)90019-j. [DOI] [PubMed] [Google Scholar]
- Xiao SY, LeDuc JW, Chu YK, Schmaljohn CS. Phylogenetic analyses of virus isolates in the genus Hantavirus, family Bunyaviridae. Virology. 1994;198:205–217. doi: 10.1006/viro.1994.1023. [DOI] [PubMed] [Google Scholar]
- Yanagihara R, Svedmyr A, Amyx HL, Lee PW, Goldgaber D, Gajdusek DC, Gibbs CJ, Jr, Nyström K. Isolation and propagation of nephropathia epidemica virus in bank voles. Scand J Infect Dis. 1984;16:225–228. doi: 10.3109/00365548409070393. [DOI] [PubMed] [Google Scholar]
- Yanagihara R, Goldgaber D, Gajdusek DC. Propagation of nephropathia epidemica virus in Mongolian gerbils. J Virol. 1985;53:973–975. doi: 10.1128/jvi.53.3.973-975.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]