Abstract
Clinical isolates of the periodontopathogen Aggregatibacter actinomycetemcomitans form matrix-encased biofilms on abiotic surfaces in vitro. A major component of the A. actinomycetemcomitans biofilm matrix is PGA, a hexosamine-containing polysaccharide that mediates intercellular adhesion. In this report we describe studies on the purification, structure, genetics and function of A. actinomycetemcomitans PGA. We found that PGA was very tightly attached to A. actinomycetemcomitans biofilm cells and could be efficiently separated from the cells only by phenol extraction. A. actinomycetemcomitans PGA copurified with LPS on a gel filtration column. 1H-NMR spectra of purified A. actinomycetemcomitans PGA were consistent with a structure containing a linear chain of N-acetyl-D-glucosamine residues in β(1,6) linkage. Genetic analyses indicated that all four genes of the pgaABCD locus were required for PGA production in A. actinomycetemcomitans. PGA mutant strains still formed biofilms in vitro. Unlike wild-type biofilms, however, PGA mutant biofilms were sensitive to detachment by DNase I and proteinase K. Treatment of A. actinomycetemcomitans biofilms with the PGA-hydrolyzing enzyme dispersin B made them 3 log units more sensitive to killing by the cationic detergent cetylpyridinium chloride. Our findings suggest that PGA, extracellular DNA and proteinaceous adhesins all contribute to the structural integrity of the A. actinomycetemcomitans biofilm matrix.
Keywords: Congo red, Crystal violet, DspB
1. Introduction
Aggregatibacter actinomycetemcomitans (formerly Actinobacillus actinomycetemcomitans) is a member of the Pasteurellaceae, a family of Gram-negative bacteria that includes many important human and animal pathogens. A. actinomycetemcomitans colonizes the human oral cavity and causes periodontitis and nonoral infections including endocarditis [1]. Clinical isolates of A. actinomycetemcomitans are known for their ability to form extremely tenacious biofilms on abiotic surfaces in vitro [2, 3]. Mutants that fail to form biofilms in vitro are unable to colonize the oral cavity of the rat or cause bone loss in a rat model of periodontitis [4, 5]. These findings suggest that biofilm formation is an important virulence factor in A. actinomycetemcomitans.
Tenacious biofilm formation by A. actinomycetemcomitans requires the production of adhesive, bundled, type IV pili (known as Flp-pili) that form on the surface of the cell [3, 6, 7]. Mutants that lack Flp-pili are deficient in surface attachment, autoaggregation, biofilm formation and colonization of the rat oral cavity [3, 5–7]. However, Flp-pili mutants still form weak biofilms on abiotic surfaces in vitro [6]. These findings indicate that additional adhesins mediate cohesion in A. actinomycetemcomitans biofilm colonies. Interestingly, biofilms produced by a Flp-pili mutant strain were sensitive to detachment by DNase I [6], which suggests that extracellular DNA (eDNA) may function as a biofilm matrix adhesin in A. actinomycetemcomitans.
Another major component of the A. actinomycetemcomitans biofilm matrix is a hexosamine-rich polysaccharide that is functionally and genetically related to extracellular polysaccharide adhesins produced by Staphylococcus aureus, S. epidermidis, Escherichia coli and Actinobacillus pleuropneumoniae [8]. These polysaccharides, usually referred to as PNAG, PIA (polysaccharide intercellular adhesin), or PGA, consist of linear chains of N-acetyl-D-glucosamine (GlcNAc) residues in β(1,6) linkage (hereafter referred to as PGA). Various forms of PGA appear to differ in their molecular weight, in the degree of N-deacetylation of the GlcNAc residues, and in the presence of O-succinate substituents [9–12]. PGA has been shown to play a role in abiotic surface attachment and intercellular adhesion [11–15], protection from killing by antibiotics, antimicrobial peptides and phagocytes [12, 16], and virulence [17]. In A. actinomycetemcomitans, PGA has been shown to mediate intercellular adhesion and resistance to killing by the anionic detergent sodium dodecyl sulfate (SDS) [8, 18].
The purpose of the present study was to gain better insight into the structure, synthesis and function of A. actinomycetemcomitans PGA. We purified PGA polysaccharide from a biofilm-producing clinical strain of A. actinomycetemcomitans and analyzed its chemical structure by using NMR spectroscopy. We also investigated the genetics of PGA production and the role of PGA in biofilm cohesion and detergent resistance in A. actinomycetemcomitans. In this report we present evidence that A. actinomycetemcomitans PGA is a β(1,6)-linked GlcNAc polymer that mediates intercellular adhesion and detergent resistance in A. actinomycetemcomitans biofilm colonies.
2. Results
2.1 Purification and structural analysis of A. actinomycetemcomitans PGA
We attempted to extract PGA polysaccharide from biofilms produced by A. actinomycetemcomitans strain JK1044 by sonication. This method has been successfully used to extract PGA adhesins from biofilm cells of Staphylococcus epidermidis [10] and Actinobacillus pleuropneumoniae [12]. Surprisingly, only trace amounts of PGA were liberated from JK1044 biofilm cells after this treatment. We therefore subjected JK1044 biofilm cells to a hot phenol-water extraction [19], which is a harsher extraction method that disintegrates the outer membrane of Gram-negative bacteria and is widely used for LPS extraction. The hexosamine-enriched aqueous phase was further deproteinated and fractionated on a Sephacryl S-300 column (Fig. 1A). Unlike sonic extracts from S. epidermidis or A. pleuropneumoniae biofilms (Figs. 1B and 1C, respectively), the high molecular weight fraction of the A. actinomycetemcomitans phenol extract corresponded to a substance containing both neutral and aminosugars (Fig. 1A).
Fig. 1.
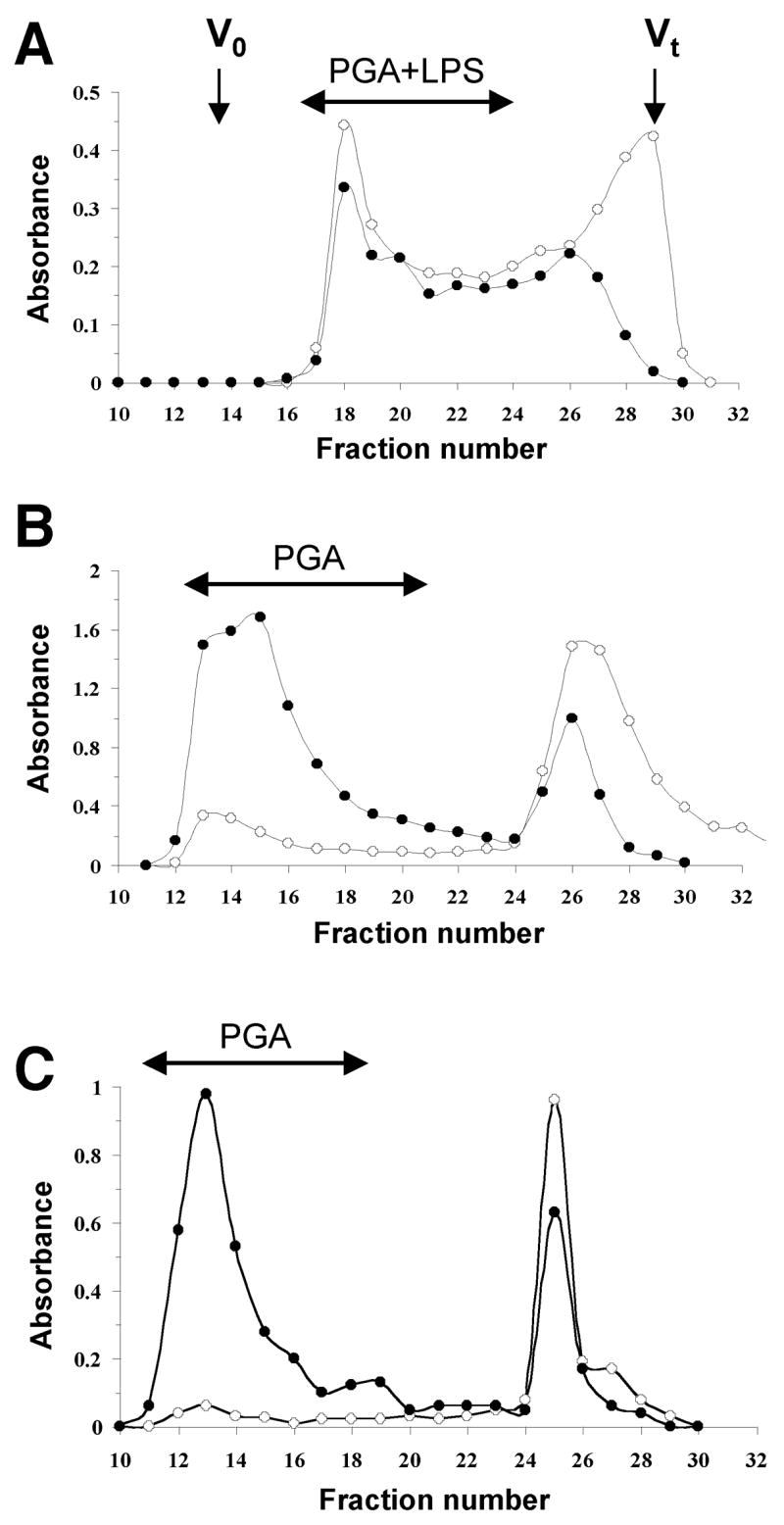
Purification of PGA polysaccharides by gel filtration chromatography. (A) Elution profile of the aqueous layer of the hot-phenol extract of A. actinomycetemcomitans JK1044 biofilms on a Sephacryl S-300 column irrigated with water. Aliquots (200 μL) of each 5-mL fraction were assayed for aminosugars (●, A485) and neutral sugars (○, A530). Void (Vo) and total (Vt) volumes of the column are indicated with arrows. (B and C) Elution profiles of sonic extracts from biofilms produced by A. pleuropneumoniae strain IA1 [12] (B) and S. epidermidis strain RP62A [35] (C).
The high molecular fractions from the A. actinomycetemcomitans phenol extract were pooled and lyophilized. The sample was analyzed by 1D and 2D NMR spectroscopy, DOC-PAGE and monosaccharide analysis. DOC-PAGE analysis indicated that this preparation contained LPS (data not shown). Monosaccharide analysis indicated the presence of rhamnose and glucosamine, consistent with the composition of O-polysaccharide (O-PS) of A. actinomycetemcomitans serotype e LPS [20], as well as heptose, a monosaccharide typical of the core oligosaccharides of LPS. Examination of the 1H-NMR spectrum of the water-soluble fraction indicated the presence of resonances corresponding to H1 and H6 of α-Rha of the O-PS, and to H1 of α-GlcNAc (Fig. 2). In addition, the H1 of the β-GlcNAc of PGA could be clearly identified (Fig. 2B, arrow). 2D NMR analysis resulted in 1H and 13C chemical shifts closely corresponding to the →4)-α-GlcpNAc-(1–3)-α-Rhap-(1→ structure described in the literature for A. actinomycetemcomitans serotype e O-PS [20] mixed with PGA (data not shown). The 1H-NMR spectrum of the HCl-soluble fraction closely corresponded to the spectrum of A. pleuropneumoniae PGA, which is highly characteristic of a β(1, 6)-linked GlcNAc backbone (Fig. 3). Chemical analyses indicated that the soluble mixture of A. actinomycetemcomitans PGA and LPS contained 10% N-deacetylated GlcNAc residues. Since there is no report of N-deacetylated GlcNAc in serotype e O-PS [20], it is likely that these N-deacetylated GlcNAc residues were from PGA.
Fig. 2.
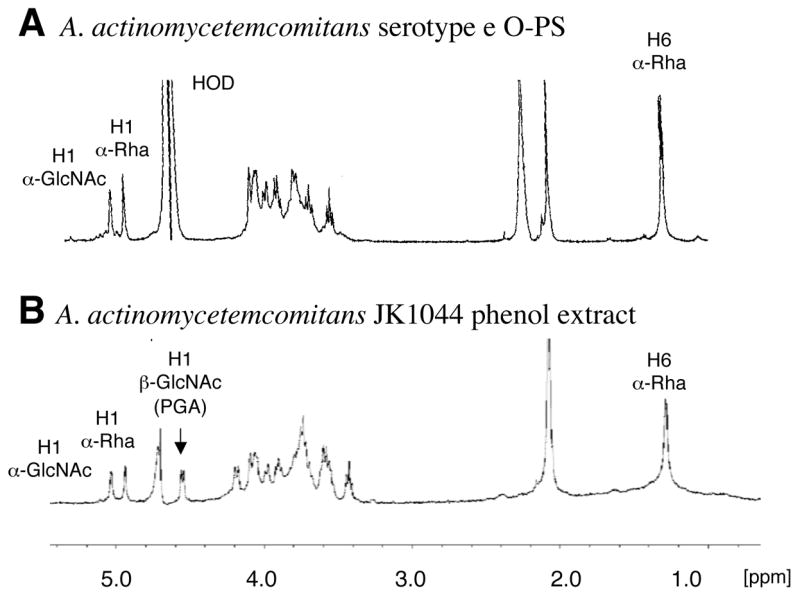
1H NMR spectra of A. actinomycetemcomitans serotype e O-PS [20] (A) and the high molecular weight fraction extracted from biofilms of A. actinomycetemcomitans JK1044 (B).
Fig. 3.
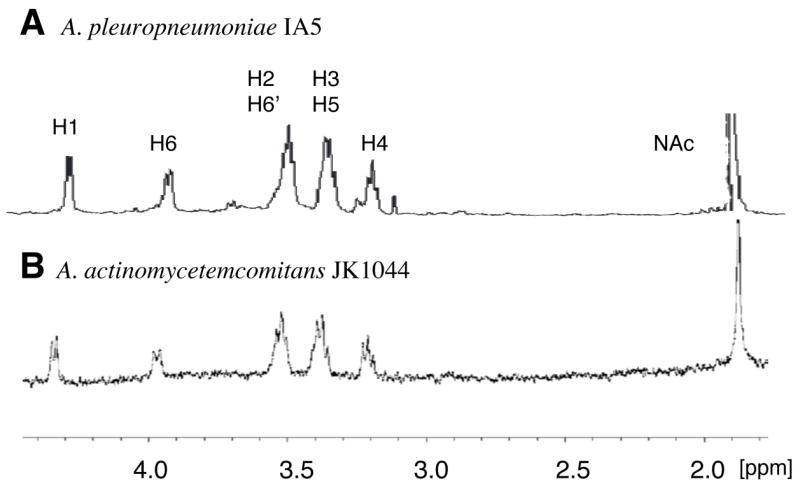
1H NMR spectra of PGA from A. pleuropneumoniae strain IA5 [12] (A) and A. actinomycetemcomitans strain JK1044 (B).
2.2 The pgaABCD gene cluster is required for PGA synthesis in A. actinomycetemcomitans
Previous studies showed that all four genes of the pgaABCD gene cluster are required for PGA production in E. coli [11] and Yersinia pestis [21]. The A. actinomycetemcomitans genome contains a pgaABCD locus that is homologous, gene-for-gene, to the pgaABCD loci of E. coli and Y. pestis [8]. To test whether all four pga genes were required for PGA production in A. actinomycetemcomitans, we isolated and characterized a series of random transposon mutants that were deficient in PGA production (Fig. 4A). These mutants were isolated by selecting ones that formed white colonies on Congo red agar [8]. Transposon insertions in all four pga genes were obtained (Fig. 4A). We also constructed a series of targeted deletion mutants in the A. actinomycetemcomitans pga locus (Fig. 4B). All of the pga insertion and deletion mutants were deficient in PGA production as determined by using a Congo red binding assay (Fig. 5A). Genetic complementation experiments confirmed that all four pga genes were required for PGA production in A. actinomycetemcomitans (Fig. 5B).
Fig. 4.
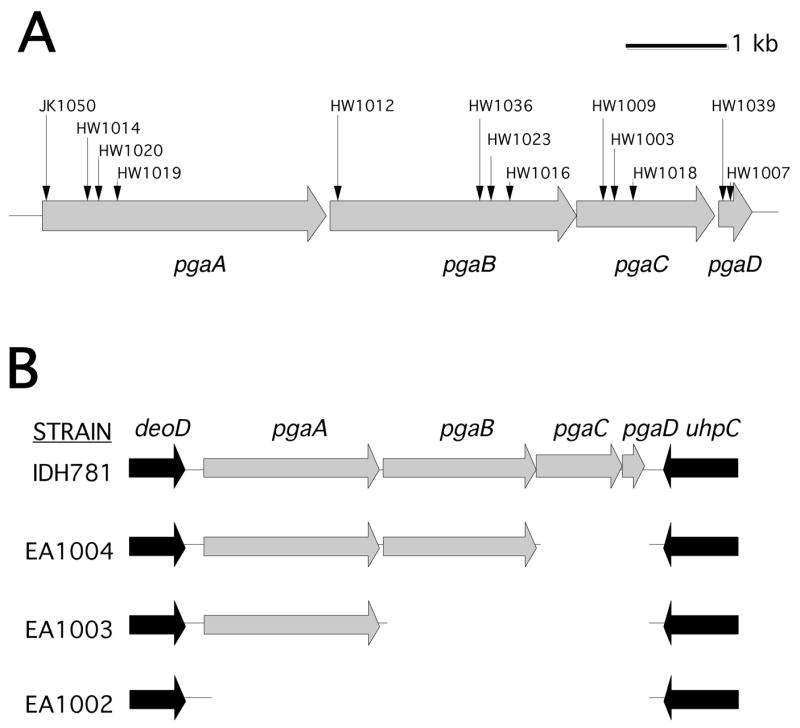
Genetic maps of the pgaABCD gene clusters in wild-type and mutant A. actinomycetemcomitans strains. (A) Map of pgaABCD in strain CU1000. Horizontal arrows indicate open reading frames and direction of transcription. Vertical arrows indicate the insertion sites of transposon IS903φkan in 13 mutant strains. All 13 transposons inserted in the same transcriptional orientation as the pga genes. (B) Map of pgaABCD and flanking genes in strain IDH781 and three deletion mutants. In each mutant, the deleted region was replaced by a 879 bp spectinomycin-resistance gene that was transcribed in the same orientation as the pga genes.
Fig. 5.
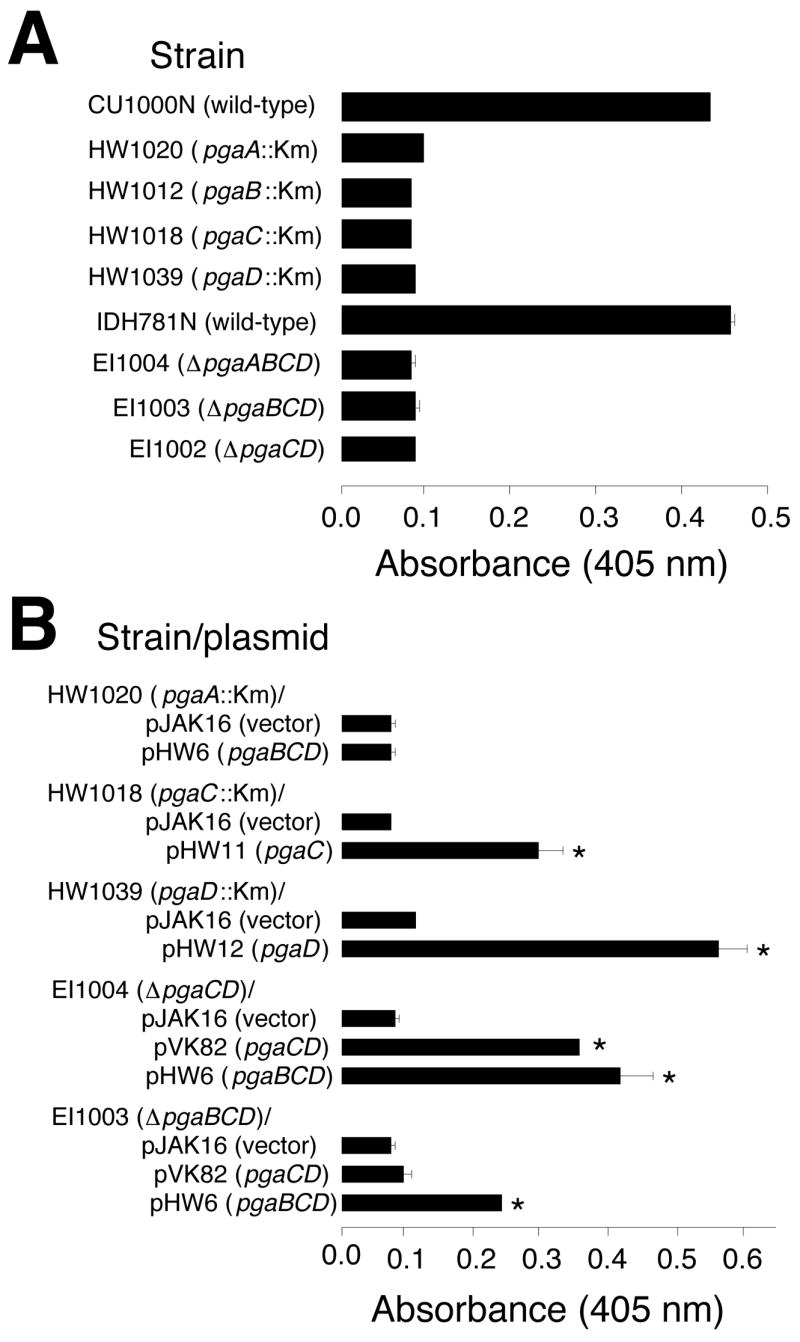
Congo red binding assay. Biofilms grown in microtiter plates were stained with Congo red dye and the amount of bound dye was quantitated by measuring its absorbance at 405 nm. The amount of bound Congo red is proportional the amount of PGA. Values indicate the mean absorbance for duplicate wells. Error bars indicate range. Background levels were <0.1 absorbance unit. Km, transposon IS903φkan. (A) Congo red binding by wild-type and mutant strains. (B) Congo red binding by mutant strains harboring complementary plasmids. Asterisks indicate values that were significantly greater than those of the vector control (P <0.05; Student’s t-test).
2.3 PGA, Flp-pili and eDNA mediate cohesion in A. actinomycetemcomitans biofilms
To investigate the role of PGA, Flp-pili and eDNA in A. actinomycetemcomitans biofilm cohesion, we treated wild-type, PGA mutant, and Flp-pili mutant biofilms with various matrix degrading enzymes (Fig. 6). Wild-type biofilms were resistant to detachment by dispersin B, a PGA-hydrolyzing enzyme, as well as by proteinase K (Fig. 6A). PGA mutant biofilms, as well as wild-type biofilms pretreated with dispersin B, were sensitive to detachment by proteinase K. In addition, biofilms produced by two different Flp-pili mutants were sensitive to detachment by dispersin B (Fig. 6B). Previous studies showed biofilms produced by a Flp-pili mutant strain [6], but not by a PGA mutant strain [8], were sensitive to detachment by the carbohydrate-modifying agent sodium metaperiodate. Also, biofilms produced by a Flp-pili mutant strain were resistant to detachment by pronase [6]. Taken together, these findings suggest that PGA and Flp-pili are the major polysaccharide and proteinaceous adhesins in the A. actinomycetemcomitans biofilm matrix. Wild-type biofilms were resistant to detachment by DNase I (Fig. 6A), whereas PGA mutant biofilms (Fig. 6A) and Flp-pili mutant biofilms [6] were sensitive to detachment by DNase I. These findings indicate that eDNA also mediates cohesion in A. actinomycetemcomitans biofilms.
Fig. 6.
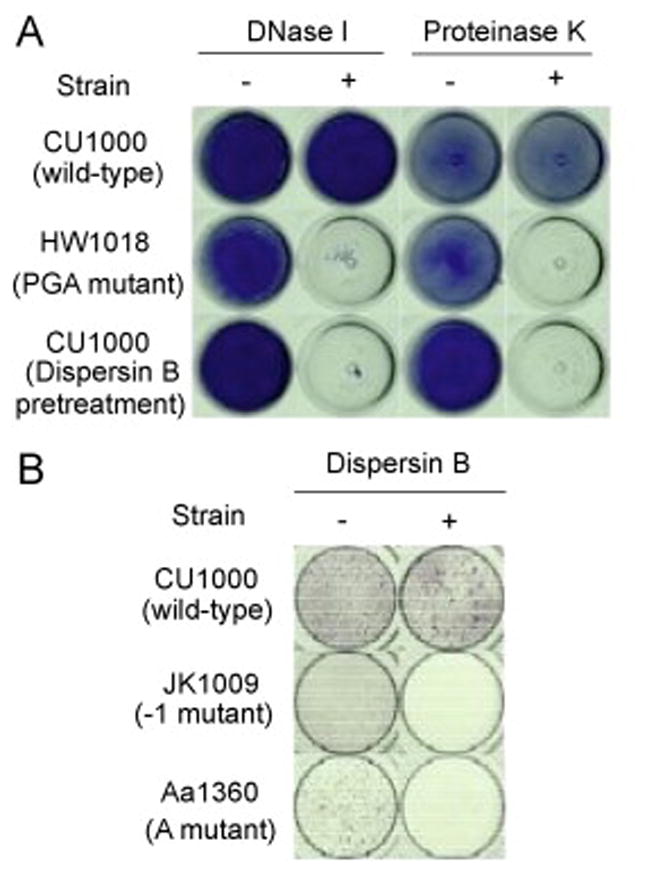
Growth and detachment of A. actinomycetemcomitans biofilms in polystyrene tubes and 96-well microtiter plates. (A) Biofilms produced by parental strain CU1000N and isogenic PGA mutant strain HW1018 in polystyrene tubes. Tubes were stained with crystal violet and photographed from the bottom. Biofilms were treated for 1 h with 500 μg/mL of DNase I or 100 μg/mL of proteinase K (or the appropriate buffer alone as a control) prior to staining. The bottom row shows CU1000N biofilms pretreated with 20 μg/mL of dispersin B prior to the DNase I and proteinase K treatments. (B) Microtiter plate biofilm formation by CU1000N and isogenic Flp-pili mutants JK1009 and Aa1360, which contain transposon insertion in flp-1 (the Flp-pilus structural gene [3]) and tadA (an ATPase required for secretion of Flp-pili [36], respectively. Biofilms were rinsed, treated with PBS or PBS plus dispersin B, and then rinsed and stained with crystal violet.
2.4 Depolymerization of A. actinomycetemcomitans PGA increases detergent sensitivity
We tested the sensitivity of A. actinomycetemcomitans biofilms to killing by 0.02% cetylpyridinium chloride (CPC), which corresponds to 10 times the MIC against A. actinomycetemcomitans planktonic cells, but which is below the concentration required for biofilm detachment (data not shown). Biofilms grown in tubes were pretreated for 30 min with phosphate buffer saline (PBS; mock pretreatment) or PBS with 20 μg/mL of dispersin B, and then treated with 0.02% CPC for 5 min. Biofilms treated with dispersin B or CPC alone exhibited little or no reduction in the number of CFUs/tube compared to the mock-treated controls (Fig. 7). Biofilms treated with dispersin B and then CPC, however, exhibited an approximately 3 log unit decrease in the number of CFUs/tube compared to biofilms treated with dispersin B or CPC alone.
Fig. 7.
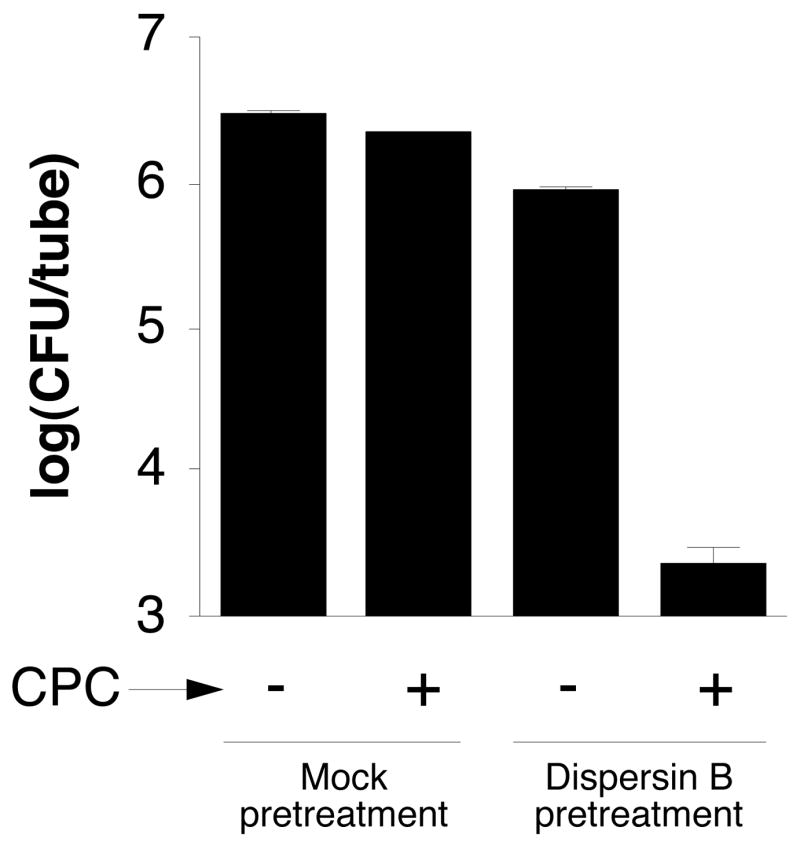
Pretreatment of A. actinomycetemcomitans CU1000 biofilms with dispersin B increases their sensitivity to killing by CPC. Biofilms grown in polystyrene tubes were rinsed with PBS and treated for 30 min with PBS (mock pretreatment) or PBS containing 20 μg/mL of dispersin B, and then treated for 5 min with 0.02% CPC. CFUs were enumerated by dilution plating. Values indicate the log10 of the mean number of CFUs/tube for duplicate tubes. Error bars indicate range.
3. Discussion
Although PGA polysaccharides produced by S. epidermidis and A. pleuropneumoniae were readily released from biofilm cells by sonication, we found that sonication alone was not sufficient to release PGA from A. actinomycetemcomitans biofilm cells. These findings suggest that PGA may be more tightly bound to A. actinomycetemcomitans cells than to cells of other PGA-producing bacteria. A. actinomycetemcomitans PGA co-eluted with LPS on a gel filtration column. It is not known, however, whether PGA binds directly to LPS. Structural analyses suggested that A. actinomycetemcomitans PGA contains a linear polymer of GlcNAc residues in β(1,6) linkage, and that some of the GlcNAc residues may be N-deacetylated. This structure is nearly identical to those of PGA polysaccharides produced by S. aureus, S. epidermidis, E. coli and A. pleuropneumoniae [9–12].
Previous studies showed that all four pga genes are required for PGA production in E. coli [11] and Y. pestis [21]. Among the four Pga proteins, PgaC and PgaD are subunits of an integral membrane glycosyltransferase that synthesizes PGA from UDP-GlcNAc monomers [22], and PgaB is a putative PGA-specific N-deacetylase [23]. The function of PgaA is unknown. Our findings confirm that all four genes of the pgaABCD gene cluster were required for PGA production in A. actinomycetemcomitans. PCR assays using pgaC-specific primers showed that pgaC was present on the chromosome of 16 out of 16 phylogenetically diverse strains of A. actinomycetemcomitans (J.B. Kaplan, unpublished), indicating that the pga genes are widespread among A. actinomycetemcomitans isolates.
Our findings indicate that Flp-pili, PGA and eDNA contribute to cohesion in A. actinomycetemcomitans biofilm colonies. Flp-pili appear to be the major biofilm matrix adhesin because Flp-pili mutants are severely defective in biofilm formation, whereas PGA mutants still form tenacious biofilms. Biofilms produced by Flp-pili mutants were resistant to detachment by pronase [6], which suggests that Flp-pili are the major proteinaceous adhesin in A. actinomycetemcomitans biofilms. However, biofilms produced by Flp-pili mutants were readily detached by dispersin B or DNase I, which indicates that PGA and eDNA also mediate cohesion in A. actinomycetemcomitans biofilm colonies. PGA is the major biofilm matrix adhesin in S. epidermidis [24] and A. pleuropneumoniae [12], and a major virulence factor in S. aureus [17]. eDNA has been shown to be a structural component of the biofilm matrix in several bacterial species including Haemophilus influenzae, Pseudomonas aeruginosa, S. aureus, Streptococcus pneumoniae and S. mutans [25–29].
Our findings indicate that depolymerization of PGA by dispersin B renders A. actinomycetemcomitans biofilm cells sensitive to killing by the cationic detergent CPC. Previous studies showed that depolymerization of PGA by dispersin B renders A. actinomycetemcomitans biofilm cells sensitive to killing by the anionic detergent SDS [18], and A. pleuropneumoniae biofilm cells more sensitive to killing by ampicillin [12]. In addition, a PGA mutant of S. epidermidis exhibited increased sensitivity to killing by the cationic antimicrobial peptides β-defensin 3 and LL-37, and by the anionic peptide dermcidin [16]. These findings suggest that the matrix formed by PGA polysaccharide may act as a general diffusion barrier that prevents penetration of various antimicrobial agents into the biofilm.
4. Materials and methods
4.1. Bacterial strains
The A. actinomycetemcomitans strains used in this study are listed in Table 1. A. actinomycetemcomitans strain JK1044, a dispersin B mutant, was constructed by transferring the kanamycin resistance gene from strain JK1023 to strain IDH1705N using natural transformation as previously described [8]. Strains were maintained on Trypticase Soy agar (TSA) supplemented with 6 g of yeast extract and 8 g of glucose per liter. Plates were incubated at 37°C in 10% CO2 and passaged twice weekly.
Table 1.
A. actinomycetemcomitans strains
| Strain | Relevant characteristicsa | Source or reference |
|---|---|---|
| IDH1705 | Wild-type (serotype e) | 37 |
| IDH1705N | Spontaneous Nalr variant of IDH1705 | D. Figurski |
| JK1044 | IDH1705N dspB::IS903φkan (Kmr) | This study |
| CU1000 | Wild-type (serotype f) | 38 |
| CU1000N | Spontaneous Nalr variant of CU1000 | D. Figurski |
| JK1023 | CU1000N dspB::IS903φkan (Kmr) | 34 |
| JK1050 | CU1000N pgaA::IS903φkan (Kmr) | This study |
| HW1014 | CU1000N pgaA::IS903φkan (Kmr) | This study |
| HW1020 | CU1000N pgaA::IS903φkan (Kmr) | This study |
| HW1019 | CU1000N pgaA::IS903φkan (Kmr) | This study |
| HW1012 | CU1000N pgaB::IS903φkan (Kmr) | This study |
| HW1036 | CU1000N pgaB::IS903φkan (Kmr) | This study |
| HW1023 | CU1000N pgaB::IS903φkan (Kmr) | This study |
| HW1016 | CU1000N pgaB::IS903φkan (Kmr) | This study |
| HW1009 | CU1000N pgaC::IS903φkan (Kmr) | This study |
| HW1003 | CU1000N pgaC::IS903φkan (Kmr) | This study |
| HW1018 | CU1000N pgaC::IS903φkan (Kmr) | 18 |
| HW1039 | CU1000N pgaD::IS903φkan (Kmr) | This study |
| HW1007 | CU1000N pgaD::IS903φkan (Kmr) | This study |
| JK1009 | CU1000N flp-1::IS903φkan (Kmr) | 3 |
| Aa1360 | CU1000N tadA::IS903φkan (Kmr) | 32 |
| IDH781 | Wild-type (serotype d) | 37 |
| IDH781N | Spontaneous Nalr variant of IDH781 | D. Figurski |
| EI1004 | IDH781N ΔpgaCD (Smr) | This study |
| EI1003 | IDH781N ΔpgaBCD (Smr) | This study |
| EI1002 | IDH781N ΔpgaABCD (Smr) | This study |
Nalr, nalidixic acid resistant; Kmr, kanamycin resistant; Smr, spectinomycin resistant.
4.2. Purification of A. actinomycetemcomitans PGA
Biofilms of A. actinomycetemcomitans strain JK1044 were grown in ten 150-mm-diam tissue-culture-treated polystyrene petri dishes (Corning no. 430199). The medium was Tryptic soy broth supplemented with yeast extract and glucose as described above (TSB). Dishes were filled with 60 mL of TSB containing 105–106 CFUs/mL and incubated for 24–48 h at 37°C in 10% CO2. Biofilms were washed with 0.9% (w/v) NaCl, detached from the surface using a cell scraper, and harvested by centrifugation (2,200 g, 4°C, 15 min). The cell pellet (approx. 1.0 g of wet cells) was resuspended in 40 mL of water. An equal volume of 90% phenol was added and the suspension was stirred for 45 min at 75°C and then cooled on ice. The layers were separated by centrifugation (3,100 g, 4°C, 20 min). Aqueous and phenol layers were dialyzed separately against deionized water and the quantity of amino sugars in each extract was assayed as described below. TCA was added to a concentration of 5%, and the precipitated proteins and nucleic acids were removed by centrifugation (8,700 g, 4°C, 10 min). The supernatant was neutralized with 1 M NaOH, concentrated to a volume of 15 mL using an Amicon ultrafiltration cell (Millipore) with a 10-kDa molecular weight cutoff membrane, and applied to a Sephacryl S-300 column (1-cm × 90-cm; Pharmacia) irrigated with water. High molecular weight fractions that were positive for both neutral sugars [30] and amino sugars [31] were pooled and lyophilized.
4.3. Structural analyses
Polysaccharides were converted to alditol acetates by conventional methods. GLC identification of monosaccharides and colorimetric measurement of N-deacetylated GlcNAc residues were carried out as previously described [12]. For NMR analysis, 3 mg of lyophilized sample was suspended in 1 mL of D2O. Insoluble material was collected by centrifugation, dissolved in 30 μL of 20% (w/v) DCl in D2O, and the total volume was adjusted to 1 mL with D2O (HCl-soluble fraction). The D2O and HCl-soluble fractions were analyzed separately (Figs. 2 and 3, respectively). NMR spectra were recorded at 25°C with a Varian Unity Inova 500 MHz spectrometer using acetone as internal reference (1H, δ 2.225 ppm; 13C, δ 31.5 ppm). Variant standard programs COSY and HSQC were employed.
4.4. Transposon mutagenesis
A. actinomycetemcomitans strain CU1000N was mutagenized with transposon IS903φkan as previously described [8]. Mutants were plated on TSA plates containing 20 μg/mL of kanamycin and 0.1% Congo red dye. Thirteen mutants that produced white colonies (from approx. 10,000 total mutants) were selected. The location of the IS903φkan insertion in each mutant was mapped by PCR using primers 5-GTTTCCCGTTGAATATGGCTGGG-3 and 5-GCAGTTTCATTTGATGCTCGA-3, which hybridize to the ends of the kanamycin-resistance gene in IS903φkan and are oriented outward, and pga-specific primers P1–P6 (Table 2). To confirm that each mutant contained a single IS903φkan insertion, PCR primer pairs P1/P2, P3/P4 and P5/P6 were employed. The conditions for PCR were as previously described [8]. The precise location of each insertion was determined by DNA sequence analysis of the PCR products.
Table 2.
A. actinomycetemcomitans pga-specific PCR primers
| Name | Sequence (5′ → 3′)a | Orientationb | GenBank coordinatesc |
|---|---|---|---|
| P1 | GATGGTATTTATGAGTGG | Forward | 1012–1030 |
| P2 | CGCAGCTGCTCTTGACGG | Reverse | 2883–2900 |
| P3 | CCGTCAAGAGCAGCTGCG | Forward | 2883–2900 |
| P4 | TGTGCAAACCATTCTTCCG | Reverse | 4972–4990 |
| P5 | AGCGGAAGAATGGTTTGC | Forward | 4970–4987 |
| P6 | GTGTTGATAGCGTTAAACC | Reverse | 6909–6927 |
| P7 | TCTGTCGACGGTCGATTATTTCAGCAGG | Forward | 6839–6857 |
| P8 | TGTAAGCTTCCATAGGTTTGCCGACCGTGG | Reverse | 7677–7697 |
| P9 | CATGAGCTCTTTCGGTTTAACTCAGGCGG | Forward | 390–409 |
| P10 | GATGGATCCAAATTGCGTTGGTGTGCAGG | Forward | 1258–1277 |
| P11 | CATGAGCTCTGAGTTAAATCAGCGTTGGG | Forward | 2825–2844 |
| P12 | GATGGATCCGGCGGAAATGGTTTGCGGG | Reverse | 3572–3590 |
| P13 | CATGAGCTCTATGCAAAATTTGACGG | Forward | 4737–4753 |
| P14 | GATGGATCCTCAGCTGAAGCAAGTGCGG | Reverse | 5561–5579 |
| P15 | ACCGGATCCCTTTCTCACTTTATTATTAGTACTCGG | Forward | 3391–3417 |
| P16 | CCGCTGCAGTTATTTTTTCTTTTTTCTCC | Reverse | 6863–6882 |
| P17 | ACCGGATCCTCAAGCAGGTAAACCATAG | Forward | 5324–5342 |
| P18 | CCGCTGCAGGTTATACTCCTCTATCCG | Reverse | 6570–6587 |
| P19 | ACCGGATCCTATGGACCAGCCCGGATAG | Forward | 6558–6576 |
Restriction sites used for cloning are underlined.
With respect to transcription of the pga genes.
GenBank accession no. EF535005.
4.5. Construction of pga deletion mutants
A plasmid for deleting the entire pgaABCD locus from the A. actinomycetemcomitans chromosome (Fig. 4B) was constructed as follows. First, the region containing the 3′ end of pgaD, the pgaD/uhpC intergenic region, and the 3′ end of uhpC (corresponding to bp 6839–7697 in GenBank accession no. EF535005) was amplified by PCR using genomic DNA isolated from strain CU1000 and PCR primers P7 and P8. The PCR product was digested with SalI and HindIII and ligated into the SalI/HindIII sites of pUC18 (New England Biolabs), resulting in plasmid pEI1. Next, the region containing the 3′ end of deoD, the deoD/pgaA intergenic region, and the 5′ end of pgaA (bp 390–1267 in accession no. EF535005) was amplified by PCR using primers P9 and P10. The PCR product was digested with SacI and BamHI and ligated into the SacI/BamHI sites of pEI1, resulting in pEI2. Finally the spectinomycin-resistance gene and promoter from transposon Tn21 (provided by D. Figurski) was amplified using PCR primers 5-GTCATCGATCCTTGACCGAACGCAGCGG-3 and 5-GCTATCGATGCGCCGCGAAGCGGCGTCGG-3 (ClaI recognition sequences underlined), and the PCR product was digested with ClaI and ligated into the AccI site of pEI2, resulting in pEI5. A plasmid for deleting pgaBCD (pEI6) was constructed in a similar way, except that a DNA fragment containing the pgaA/pgaB intergenic region (PCR primers P11 and P12; bp 2825–3590 in accession no EF535005) was substituted for the deoD/pgaA fragment in pEI5. Similarly, a plasmid for deleting pgaCD (pEI7) was constructed by substituting a fragment containing the pgaB/pgaC intergenic region (PCR primers P13 and P14; bp 4737–5179 in accession no. EF535005) for the deoD/pgaA fragment in pEI5. Plasmids pEI5, pEI6 and pEI7 were linearized with SwaI and transformed into strain IDH781N by using natural transformation as previously described [8]. Transformants were plated on TSA plates containing 20 μg/mL of spectinomycin. Integration of the plasmid into the chromosome by homologous recombination was confirmed by PCR using primers 5-GCGCTTGCTGCTTGGATGCC-3 and 5-GAGATCACCAAGGTAGTCGG-3, which hybridize to the ends of the spectinomycin-resistance gene in Tn21 and are oriented outward, and pga-specific primers P1–P6.
4.6. Congo red binding assay
Biofilms of each test strain were grown for 24 h in 96-well microtiter plates as described below. Biofilms were washed with water and stained with 1% Congo red dye (in water) for 1 min, and then rinsed and dried. The bound dye was solubilized in 200 μL of DMSO for 1 h at room temperature and its absorbance was measured at 405 nm. All assays were preformed in duplicate wells, which exhibited minimal variation, and on several occasions with similar results.
4.7. Genetic complementation
The vector used for genetic complementation experiments was pJAK16, an IncQ expression plasmid that contains a chloramphenicol-resistance gene and an IPTG-inducible tac promoter located upstream from a multiple cloning site [32]. Plasmid pHW6 was constructed by amplifying the pgaBCD genes from strain CU1000 (bp 3391–6882 in accession no. EF535005) using PCR primers P15 and P16, digesting the PCR product with BamHI and PstI, and ligating the fragment into the BamHI/PstI sites of pJAK16. Plasmid pHW11 was constructed by amplifying the pgaC gene (bp 5324–6587 in accession no. EF535005) with PCR primers P17 and P18 and ligating the fragment into the BamHI/PstI sites of pJAK16. Similarly, plasmid pHW12 was constructed by amplifying the pgaD gene (bp 6558–6882 in accession no. EF535005) with PCR primers P19 and P16 and ligating the fragment into the BamHI/PstI sites of pJAK16. Plasmid pVK82, which contains the A. actinomycetemcomitans pgaCD genes in pJAK16, was described previously [8]. Complementary plasmids were conjugated from E. coli into A. actinomycetemcomitans using the RK2 oriT-defective mutant plasmid pRK21761 as previously described [33]. Plasmid-harboring strains were grown in TSB containing 3 μg/mL of chloramphenicol and 1 mM IPTG.
4.8. Biofilm cultures and crystal violet assay
Biofilms were grown in 16-mm × 100-mm polystyrene test tubes (Falcon no. 352051) or tissue-culture-treated 96-well polystyrene microtiter plates (Falcon no. 353072). Tubes were filled with 1 mL of TSB containing 103–105 CFUs/mL and incubated statically at 37°C in 10% CO2 for 18 h (for biofilm detachment assays) or 24 h (for biofilm killing assays). Previous studies showed that 24-h old biofilms grown in polystyrene tubes, but not 18-h old biofilms, were sensitive to detachment by dispersin B [18]. For microtiter plates, wells were filled with 200 μL of inoculum and incubated for 24 h. For crystal violet staining, tubes were rinsed with water and stained with 1 mL of crystal violet (Fisher no. 23255960) for 1 min, and then rinsed and dried. Microtiter plate wells were gently aspirated, rinsed with PBS and dried, and then stained with 100 μL of crystal violet.
4.9 Biofilm detachment assays
Biofilms were grown for 18 h in tubes as described above. Biofilms were rinsed with PBS and then treated with 1 mL of 20 μg/mL of dispersin B (103 units/mg of protein [34]), 200 μg/mL of bovine DNase I (Roche), or 500 μg/mL of proteinase K (Sigma). Control wells were treated with the appropriate buffer alone (PBS for dispersin B; 40 mM Tris [pH 8.0], 1 mM CaCl2, 10 mM MgCl2 for DNase I; and 50 mM Tris [pH 8.0], 1 mM EDTA for proteinase K). In some experiments, biofilms were treated with dispersin B and then with DNase I or proteinase K. After treatment, biofilm were stained with crystal violet as described above. For biofilms grown in microtiter plates, biofilms were treated for 1 h with 200 μL of PBS or PBS containing 20 μg/mL of dispersin B. After treatment, wells were rinsed with water and dried, and then stained with crystal violet as described above.
4.10. Biofilm killing assay
Biofilms were grown for 24 h in polystyrene tubes as described above. Biofilms were washed three times with sterile PBS and then treated with 1 mL of CPC (0.02% in PBS). After 5 min the biofilms were rinsed three times with PBS to remove the CPC and then treated with 1 mL of dispersin B (20 μg/mL in PBS) for 5 min to detach the cells. Tubes were vortexed for 10 sec and the surviving CFUs were enumerated in microtiter plates as previously described [18]. In some assays, biofilms were pretreated with 1 mL of dispersin B (20 μg/mL in PBS) for 5 min prior to the CPC treatment. In these assays, 100 μL of 0.2% CPC in PBS was added directly to the dispersin B-treated cell suspension and mixed. Control tubes received 100 μL of PBS alone. After 5 min, tubes were vortexed briefly and 20 μL aliquots were enumerated as described above. Killing assays were performed in duplicate tubes and on several occasions with similar results.
Acknowledgments
We thank David Figurski (Columbia University) and members of his laboratory for providing some of the bacterial strains and plasmids used in this study. Supported by grants DE15124 and DE16291 from the United States Public Health Service, and by the Regional Council of Nord-Pas-de-Calais (France).
Abbreviations
- CPC
cetylpyridinium chloride
- eDNA
extracellular DNA
- GlcNAc
N-acetyl-D-glucosamine
- PGA
poly-β-1,6-N-acetyl-D-glucosamine
- PBS
phosphate buffered saline
- SDS
sodium dodecyl sulfate
- TSA
Tryptic Soy agar
- TSB
Tryptic Soy broth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 2.Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–76. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 3.Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2001;40:542–54. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- 4.Fine DH, Goncharoff P, Schreiner HC, Chang KM, Furgang D, Figurski DH. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch Oral Biol. 2001;46:1065–78. doi: 10.1016/s0003-9969(01)00067-x. [DOI] [PubMed] [Google Scholar]
- 5.Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Nat Acad Sci USA. 2003;100:7295–300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue T, Shingaki R, Sogawa N, Sogawa CA, Asaumi J, Kokeguchi S, et al. Biofilm formation by a fimbriae-deficient mutant of Actinobacillus actinomycetemcomitans. Microbiol Immunol. 2003;47:877–81. doi: 10.1111/j.1348-0421.2003.tb03454.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Chen C. Mutational analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene. 2005;351:61–71. doi: 10.1016/j.gene.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–20. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyce JG, Abeygunawardana C, Xu Q, Cook JC, Hepler R, Przysiecki CT, et al. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res. 2003;338:903–22. doi: 10.1016/s0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 10.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–83. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–34. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izano EA, Sadovskaya I, Vinogradov E, Mulks MH, Velliyagounder K, Ragunath C, et al. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathogen. 2007;43:1–9. doi: 10.1016/j.micpath.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agladze K, Wang X, Romeo T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol. 2005;187:8237–46. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–91. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 15.McKenney D, Hubner J, Muller E, Wang Y, Goldmann DA, Pier GB. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66:4711–20. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–75. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 17.Kropec A, Maira-Litrán T, Jefferson KK, Grout M, Cramton SE, Götz F, et al. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 2005;73:6868–76. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izano EA, Wang H, Ragunath C, Ramasubbu N, Kaplan JB. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dent Res. doi: 10.1177/154405910708600707. in press. [DOI] [PubMed] [Google Scholar]
- 19.Westphal O, Jann K. Bacterial lipopolysaccharides, extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 20.Perry MB, MacLean LM, Brisson JR, Wilson ME. Structures of the antigenic O-polysaccharides of lipopolysaccharides produced by Actinobacillus actinomycetemcomitans serotypes a, c, d and e. Eur J Biochem. 1996;242:682–8. doi: 10.1111/j.1432-1033.1996.0682r.x. [DOI] [PubMed] [Google Scholar]
- 21.Lillard JW, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) locus of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 22.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–93. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 23.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–6. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48:2633–6. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–75. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 27.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Nat Acad Sci USA. 2007;104:8113–8. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moscoso M, García E, López R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–95. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187:4392–400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubois M, Gilles KA, Hamilton JF, Rebers PA, Smith F. Colorimetric methods for determination of sugars and related substances. Anal Biochem. 1956;28:350–6. [Google Scholar]
- 31.Enghofer E, Kress H. An evaluation of the Morgan-Elson assay for 2-amino-2-deoxy sugars. Carbohydr Res. 1979;76:233–8. doi: 10.1016/0008-6215(79)80022-1. [DOI] [PubMed] [Google Scholar]
- 32.Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J Bacteriol. 2000;182:6169–76. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson VJ, Bhattacharjee MK, Fine DH, Derbyshire KM, Figurski DH. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J Bacteriol. 2003;185:4693–8. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–17. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–36. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haubek D, Poulsen K, Asikainen S, Kilian M. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1995;33:395–401. doi: 10.1128/jcm.33.2.395-401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, et al. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–47. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]


