Abstract
Systemic lupus erythematosus (SLE) is a predominantly female autoimmune disease that affects multiple organ systems. Herein, we report on an X-chromosome gene association with SLE. Methyl-CpG-binding protein 2 (MECP2) is located on chromosome Xq28 and encodes for a protein that plays a critical role in epigenetic transcriptional regulation of methylation-sensitive genes. Utilizing a candidate gene association approach, we genotyped 21 SNPs within and around MECP2 in SLE patients and controls. We identify and replicate association between SLE and the genomic element containing MECP2 in two independent SLE cohorts from two ethnically divergent populations. These findings are potentially related to the overexpression of methylation-sensitive genes in SLE.
Introduction
Systemic lupus erythematosus (SLE) is a debilitating autoimmune disease that affects multiple organs and is associated with significant morbidity and mortality. The disease predominantly affects females with a female to male ratio ranging between 4.3–13.6 to 1 [1]. The etiology of SLE remains incompletely understood, although a number of genetic and environmental factors have been implicated. Strong evidence supports an important role for abnormal T cell DNA methylation in the pathogenesis of SLE [2]. The expression of methylation sensitive genes, such as ITGAL (CD11a), TNFSF7 (CD70), PRF1 (perforin) and CD40LG (CD40L), is increased in T cells from SLE patients, similar to normal T cells treated with DNA methylation inhibitors such as 5-azacytidine [3], [4], [5], [6]. Indeed, 5-azacytidine treated T cells are autoreactive in vitro [7], and produce a SLE-like disease upon adoptive transfer into mice [8]. In active SLE T cells, the expression of DNA methyltransferase 1 (DNMT1), the main enzyme that maintains DNA methylation during cell division, is reduced [9], and the promoter sequences of the aforementioned methylation-sensitive genes are hypomethylated [2], [6]. DNA methylation suppresses gene expression via several mechanisms including the inability of transcription factors to bind methylated promoter sequences [10]. Methyl-CpG-binding protein 2 (MECP2) plays a critical role in this process. MECP2 binds methylcytosine residues and recruits histone deacetylase enzymes, which by deacetylating histone residues, increase the charge attraction between DNA and histone proteins and induce a chromatin configuration that is inaccessible for the transcriptional machinery [11]. Further, DNMT1 associates with and seems to require MECP2 in order to maintain DNA methylation [12].
MECP2 is located on chromosome Xq28 in man and contains four exons. It is ∼76 kb in length and is characterized by the presence of a very large intron 2 (∼60 kb) and a highly conserved 3′ UTR (∼8.5 kb) [13]. The gene encodes a 486 amino acid chromatin-associated protein that consists of three domains; a methyl-binding domain, a transcription repression domain, and a third domain on the C-terminal region that has not been fully functionally characterized [13]. The facts that DNA methylation sensitive genes are overexpressed in SLE [2], and that MECP2 is critical in the transcriptional suppression of methylation sensitive genes [11], make MECP2 an attractive candidate gene for SLE. Using a candidate gene approach and a case-control genetic association study, we report herein on the association of the MECP2 gene region with SLE in two independent cohorts of SLE patients and controls.
Results
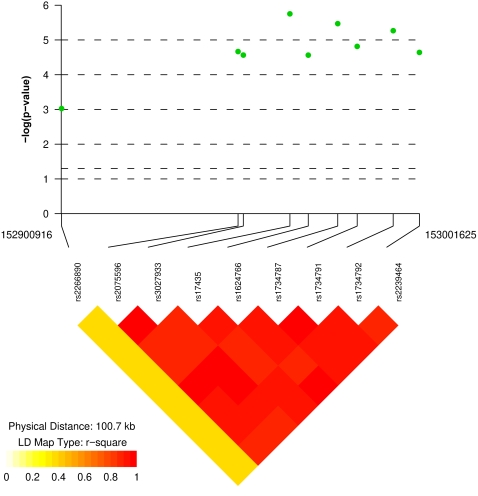
We initially genotyped 628 Korean female SLE patients and 736 healthy female Korean controls across 21 single nucleotide polymorphisms (SNPs) located within or around MECP2 (Table 1). Nine SNPs had a minor allele frequency of more than 5% in our Korean cohort and were used for further analysis. All 9 SNPs were within expected Hardy-Weinberg proportions in both cases and controls (Table 2). Eight out of the nine SNPs are within the MECP2 gene and showed significant association with SLE (Table 2 and Fig. 1). The SNP having the strongest association in the Korean SLE patients is rs17435 (Chi2 = 22.83, OR = 1.58, p = 0.0000018) followed by rs1734787 (Chi2 = 21.58, OR = 1.55, p = 0.0000034), rs1734792 (Chi2 = 20.68, OR = 1.53, p = 0.0000054) and rs1734791 (Chi2 = 18.70, OR = 1.51, p = 0.000015). The SNPs rs1734787, rs1734792, and rs1734791 are all in linkage disequilibrium with rs17435 (r2 = 0.88, 0.92, and 0.86, respectively). We next performed haplotype-based association test using Haploview 3.32 software [14] and WHAP [15]. Three haplotypes (with a frequency of >1%) were identified (Table 3). The haplotype “ACTGCAAA” was identified as a disease risk haplotype with a frequency of 82.3% in SLE patients compared to 75.3% in normal healthy controls (OR = 1.53, p = 0.000013). On the other hand, the haplotype “GGAAATCG” is a protective haplotype with a frequency of 16.8% in SLE patients and 23.4% in normal healthy controls (OR = 0.66, p = 0.000027) (Table 3). Frequencies of the homozygous risk genotypes in the haplotype-forming SNPs were analyzed and summarized in Table 4.
Table 1. SNPs genotyped in the MECP2 region in SLE patients and controls.
| SNP | Position (bp) | Alleles | Gene |
| rs2266890 | 152900916 | A/G | TMEM187 |
| rs10127175 | 152937366 | A/T | IRAK1 |
| rs2075596 | 152950586 | A/G | MECP2 |
| rs3027933 | 152952068 | C/G | MECP2 |
| rs3027935 | 152957662 | A/G | MECP2 |
| rs3027939 | 152963772 | T/C | MECP2 |
| rs17435 | 152965174 | T/A | MECP2 |
| rs7050901 | 152967504 | T/C | MECP2 |
| rs7059306 | 152968184 | A/G | MECP2 |
| rs1624766 | 152970348 | A/G | MECP2 |
| rs7884370 | 152976075 | T/C | MECP2 |
| rs1734787 | 152978640 | A/C | MECP2 |
| rs5987201 | 152983236 | A/G | MECP2 |
| rs1734791 | 152984114 | T/A | MECP2 |
| rs1734792 | 152994254 | A/C | MECP2 |
| rs11156611 | 152997368 | A/G | MECP2 |
| rs5987204 | 153000782 | A/G | MECP2 |
| rs2239464 | 153001625 | A/G | MECP2 |
| rs5986954 | 153002489 | T/C | MECP2 |
| rs5945175 | 153011951 | T/C | LOC728653,MECP2 |
| rs34371500 | 153850987 | C/A | - |
Table 2. Allele frequencies of SNPs in the MECP2 gene region genotyped in Korean SLE patients and that have a minor allele frequency of >0.05.
| SNP | Risk allele | Risk allele frequency* | Chi2 | OR (95% CI) | p value | Permutation p value | HWE (p exact) | ||
| Cases n (%) | Controls n (%) | Cases | Controls | ||||||
| rs2266890 | A | 804 (72.8) | 848 (66.6) | 10.94 | 1.35 (1.13–1.61) | 0.0009 | 0.0029 | 1.00 | 0.24 |
| rs2075596 | A | 1026 (81.7) | 1103 (74.9) | 18.05 | 1.49 (1.24–1.80) | 0.000022 | 0.0001 | 0.59 | 0.56 |
| rs3027933 | C | 1025 (81.6) | 1103 (74.9) | 17.61 | 1.48 (1.23–1.79) | 0.000027 | 0.0001 | 0.35 | 0.56 |
| rs17435 | T | 1036 (82.5) | 1103 (74.9) | 22.83 | 1.58 (1.31–1.90) | 0.0000018 | 0.00001 | 0.33 | 0.85 |
| rs1624766 | G | 1044 (83.1) | 1128 (76.6) | 17.59 | 1.50 (1.24–1.82) | 0.000027 | 0.0001 | 0.15 | 0.31 |
| rs1734787 | C | 1032 (82.2) | 1101 (74.8) | 21.58 | 1.55 (1.29–1.87) | 0.0000034 | 0.00003 | 0.28 | 0.50 |
| rs1734791 | A | 1032 (82.2) | 1109 (75.3) | 18.70 | 1.51 (1.25–1.82) | 0.000015 | 0.00009 | 0.28 | 0.28 |
| rs1734792 | A | 1018 (81.1) | 1085 (73.7) | 20.68 | 1.53 (1.27–1.83) | 0.0000054 | 0.00003 | 0.52 | 0.78 |
| rs2239464 | A | 1044 (83.1) | 1127 (76.6) | 17.94 | 1.51 (1.25–1.82) | 0.000023 | 0.0001 | 0.09 | 0.30 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
A total of 628 independent female SLE patients and 736 healthy unrelated female controls were genotyped
Figure 1. Allelic association results and linkage disequilibrium (LD) plot of the chromosome Xq28 region around the MECP2 gene.
The allelic association p values of the SNPs analyzed are shown in the Korean cohort included in this study.
Table 3. MECP2 haplotype frequencies in the Korean SLE patients and controls.
| Frequency (%) | ||||||||||||
| rs2075596 | rs3027933 | rs17435 | rs1624766 | rs1734787 | rs1734791 | rs1734792 | rs2239464 | Case | Control | OR (95% CI) | p value | |
| Haplotype1 | A | C | T | G | C | A | A | A | 82.3 | 75.3 | 1.53 (1.26–1.85) | 0.000013 |
| Haplotype2 | G | G | A | A | A | T | C | G | 16.8 | 23.4 | 0.66 (0.54–0.80) | 0.000027 |
| Haplotype3 | G | G | T | G | A | T | C | A | 0.9 | 1.3 | 0.74 (0.36–1.50) | 0.39 |
OR, odds ratio; CI, confidence interval.
Table 4. Frequencies of homozygous risk allele genotypes in Korean SLE patients compared to controls.
| SNP | Risk genotype | Risk genotype frequency* | Chi2 | OR (95% CI) | p value | ||
| Cases n (%) | Controls n (%) | ||||||
| rs2075596 | AA | 421(67.0) | 410 (55.7) | 18.28 | 1.62 | (1.30–2.02) | 0.00002 |
| rs3027933 | CC | 422 (67.2) | 410 (55.7) | 18.81 | 1.63 | (1.31–2.03) | 0.00001 |
| rs17435 | TT | 431 (68.6) | 412 (56.0) | 22.98 | 1.72 | (1.38–2.15) | 0.0000016 |
| rs1624766 | GG | 439 (69.9) | 427 (58.0) | 20.66 | 1.68 | (1.34–2.11) | 0.0000055 |
| rs1734787 | CC | 428 (68.2) | 408 (55.4) | 23.10 | 1.72 | (1.38–2.15) | 0.0000015 |
| rs1734791 | AA | 428 (68.2) | 412 (56.0) | 21.23 | 1.68 | (1.35–2.10) | 0.0000041 |
| rs1734792 | AA | 415 (66.1) | 398 (54.1) | 20.29 | 1.66 | (1.33–2.06) | 0.0000067 |
| rs2239464 | AA | 440 (70.1) | 426 (57.9)) | 21.70 | 1.70 | (1.36–2.13) | 0.0000032 |
OR, odds ratio; CI, confidence interval.
A total of 628 independent female SLE patients and 736 healthy unrelated female controls were genotyped
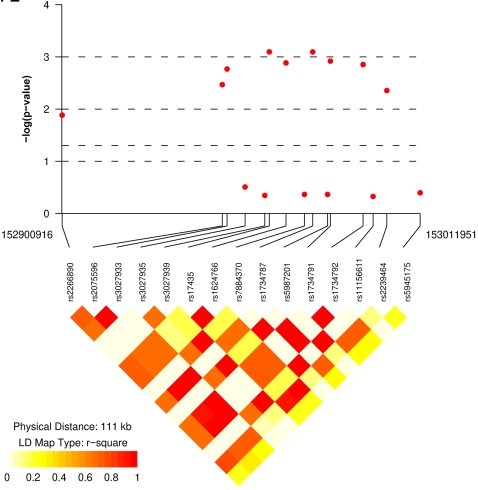
To replicate our initial results, we next genotyped 1080 European-derived independent SLE patients and 1080 healthy unrelated controls matched for sex and race using the same 21 SNPs in the MECP2 region (Table 1). Fifteen SNPs had a minor allele frequency of more than 5% in our European-derived SLE patients and controls and were used for subsequent analysis. All SNPs that were associated with SLE in Korean patients showed significant association with the same risk alleles in the European-derived cohort (Table 5 and Fig. 2). Similarly, the association with these SNPs is confirmed when only analyzing females in the European-derived SLE patients and controls, with the strongest association observed in rs1734787, rs17435, rs1734791, and rs1734792 (p values = 0.0016, 0.0017, 0.0020, and 0.0022, respectively). Our haplotype analysis in female European-derived SLE patients and controls also identified 3 haplotypes with the same risk and protective haplotypes as in the Korean cohort (Table 6). Subset analysis of male European-derived SLE patients and controls was not possible due to small sample size.
Table 5. Allele frequencies of MECP2 SNPs in European-derived SLE patients and controls.
| SNP | Risk allele | Risk allele frequency* | Chi2 | OR (95% CI) | p value | Permutation p value | HWE (p value) | |||
| Cases n (%) | Controls n (%) | Cases | Controls | |||||||
| rs2266890 | A | 418 (21.2) | 354 (18.0) | 6.24 | 1.22 | (1.04–1.43) | 0.013 | 0.068 | 0.45 | 0.21 |
| rs2075596 | A | 371(18.5) | 301(15.5) | 8.59 | 1.28 | (1.09–1.52) | 0.0034 | 0.02 | 0.70 | 1 |
| rs3027933 | C | 383 (19.1) | 308 (15.3) | 9.83 | 1.30 | (1.10–1.53) | 0.0017 | 0.01 | 0.72 | 0.92 |
| rs3027935 | G | 1879 (93.8) | 1865 (93.0) | 1.01 | 1.14 | (0.89–1.46) | 0.31 | 0.79 | 0.03 | 0.14 |
| rs3027939 | G | 104 (5.5) | 97 (5.0) | 0.57 | 1.12 | (0.84–1.48) | 0.45 | 0.92 | 0.02 | 0.68 |
| rs17435 | T | 500 (24.9) | 411 (20.5) | 11.25 | 1.29 | (1.11–1.49) | 0.0008 | 0.0047 | 0.27 | 0.36 |
| rs1624766 | G | 497 (24.8) | 412 (20.5) | 10.39 | 1.28 | (1.10–1.48) | 0.0013 | 0.0074 | 0.48 | 0.36 |
| rs7884370 | G | 109 (5.4) | 98 (4.9) | 0.63 | 1.12 | (0.85–1.48) | 0.43 | 0.91 | 0.02 | 0.68 |
| rs1734787 | C | 387 (19.3) | 307 (15.3) | 11.15 | 1.32 | (1.12–1.56) | 0.0008 | 0.0048 | 1 | 0.92 |
| rs5987201 | A | 109 (5.4) | 98 (4.9) | 0.62 | 1.12 | (0.85–1.48) | 0.43 | 0.91 | 0.02 | 0.68 |
| rs1734791 | A | 389 (19.4) | 311 (15.5) | 10.43 | 1.31 | (1.11–1.54) | 0.0012 | 0.0072 | 0.91 | 1 |
| rs1734792 | A | 388 (19.3) | 311 (15.5) | 10.27 | 1.31 | (1.11–1.54) | 0.0014 | 0.0078 | 0.88 | 1 |
| rs11156611 | A | 108 (5.4) | 98 (4.9) | 0.51 | 1.11 | (0.84–1.47) | 0.47 | 0.94 | 0.02 | 0.68 |
| rs2239464 | A | 475 (23.7) | 400 (19.9) | 8.13 | 1.24 | (1.07–1.45) | 0.0044 | 0.026 | 0.72 | 0.27 |
| rs5945175 | G | 115 (5.7) | 103 (5.1) | 0.70 | 1.12 | (0.86–1.48) | 0.40 | 0.89 | 0.85 | 0.45 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
A total of 1080 independent cases and 1080 healthy unrelated controls matched for race and sex were genotyped
Figure 2. Allelic association results and linkage disequilibrium (LD) plot of the chromosome Xq28 region around the MECP2 gene.
The allelic association p values of the SNPs analyzed are shown in the European-derived cohort included in this study.
Table 6. MECP2 haplotype frequencies in the European-derived female SLE patients and controls.
| rs2075596 | rs3027933 | rs17435 | rs1624766 | rs1734787 | rs1734791 | rs1734792 | rs2239464 | Frequency (%) | OR (95% CI) | p value | ||
| Case | Control | |||||||||||
| Haplotype1 | A | C | T | G | C | A | A | A | 19.0 | 15.4 | 1.29 (1.08–1.53) | 0.0044 |
| Haplotype2 | G | G | A | A | A | T | C | G | 77.0 | 81.0 | 0.81 (0.69–0.95) | 0.012 |
| Haplotype3 | G | G | T | G | A | T | C | A | 4.0 | 4.2 | 0.96 (0.70–1.34) | 0.82 |
OR, odds ratio; CI, confidence interval.
The impact of hidden population stratification on our association study was assessed by genomic control (GC) by estimating the inflation factor (λ) in the samples. We estimated λ = 1.06 in Korean and 1.08 in European-derived samples, hence no significant population stratification was detected. These results are also corroborated with our population structure estimates; one-population model (homogeneous population) fit better than a two-population model (admixture) for both cohorts.
Discussion
DNA methylation plays a critical role in tissue differentiation, imprinting, transcriptional suppression of parasitic DNA, silencing of transcriptional “noise”, and X-chromosome inactivation [16]. Utilizing a candidate gene approach, we first identified significant association with MECP2 SNPs and SLE in a cohort of Korean SLE patients and controls. We next replicated the association with MECP2 SNPs in an independent cohort of SLE patients and controls of European descent. Indeed, the disease-associated alleles in rs17435, rs1734787, rs1734792, and rs1734791 (T, C, A, and A respectively) have meta-analysis p values of 1.2×10−8, 1.6×10−8, 3.3×10−8, and 7.2×10−8 respectively ( Table 7 ). Interestingly, the disease associated alleles in these four MECP2 SNPs are ∼4 times more common in Korean as compared to European-derived controls. This might suggest a possible explanation for the higher frequency of SLE in people of Asian descent as compared to Europeans.
Table 7. Meta-analysis for risk alleles in SLE-associated MECP2 SNPs in Korean and European-derived cohorts.
| SNP | Risk allele | Heterogeneity P value | Meta-analysis OR (95%CI) | Meta-analysis P value | |
| rs2075596 | A | 0.25 | 1.38 | (1.22–1.55) | 2.8×10-07 |
| rs3027933 | C | 0.31 | 1.38 | (1.22–1.56) | 1.6×10-07 |
| rs17435 | T | 0.10 | 1.39 | (1.24–1.56) | 1.2×10-08 |
| rs1624766 | G | 0.18 | 1.36 | (1.21–1.52) | 1.9×10-07 |
| rs1734787 | C | 0.2 | 1.42 | (1.26–1.60) | 1.6×10-08 |
| rs1734791 | A | 0.28 | 1.39 | (1.24–1.57) | 7.2×10-08 |
| rs1734792 | A | 0.23 | 1.40 | (1.24–1.58) | 3.3×10-08 |
| rs2239464 | A | 0.13 | 1.34 | (1.20–1.51) | 6.0×10-07 |
OR, odds ratio; CI, confidence interval
MECP2 has been extensively studied in the setting of mental retardation and, particularly, Rett syndrome, an X-linked neurodevelopmental disease that has a cumulative incidence of ∼1/10,000 females by the age of 12 years [17]. In the majority of cases, this syndrome is caused by mutations in the MECP2 gene [18]. MECP2-deficient mice demonstrate clinical neurological findings similar to those observed in patients with Rett syndrome [19], [20], which can be reversed by MECP2 expression [21]. More recently, mutations in the MECP2 gene have been recognized in a number of other neuropsychiatric illnesses as well [22]. Identifying MECP2 regulated genes had been a challenge in patients with Rett syndrome [23]. Recent studies suggest that MECP2 binding to DNA is selective and requires A/T sequences adjacent to methylated CG sites [24]. In addition to its role in transcriptional regulation, MECP2 interacts with the RNA-binding protein Y box-binding protein 1 (YB-1) and plays a role in RNA splicing [25].
Another interesting gene that is in close proximity to MECP2 is IRAK1 (Interleukin-1 receptor-associated kinase1). Both MECP2 and IRAK1 are on the same haplotype block in combined Japanese and Chinese individuals genotyped in the International HapMap Project (www.hapmap.org). Moreover, this haplotype block harbors only MECP2 and IRAK1 genes. The pivotal role of IRAK1 in Toll-like receptor signaling and innate immune response [26] makes this an important candidate gene for SLE.
We report on an X-chromosome association in SLE. A role for an X-chromosome gene in this predominantly female disease has long been anticipated. Male patients with Klinefelter's syndrome (47,XXY) have similar risk to develop SLE compared to females (46,XX) (Scofield RH, et al Arthritis and Rheumatism 2003; 48:S383) . Possible explanations for the suggested gene-dose effect are the presence of a SLE susceptibility gene(s) on the X-chromosome, or the overexpression of an X-chromosome gene as a result of loss of random X-chromosome inactivation, or both. X-chromosome inactivation is largely mediated by DNA methylation [27], and DNA methylation is defective in SLE T cells [2]. Hence, X-chromosome genes in SLE female patients and SLE male patients with Klinefelter's syndrome are available for transcription from both copies on the two X-chromosomes. This mechanism is suggested to explain the observed overexpression of the X-chromosome gene CD40L in T cells from female SLE patients [6]. Our findings provide evidence for SLE association with an X-chromosome region harboring two genes that are intimately involved in regulating the expression of methylation-sensitive genes and in innate immune response.
In summary, the finding of strong association between the X-chromosome region harboring MECP2 and SLE suggests an important role for genetic and epigenetic interactions in the pathogenesis of this disease. This might provide more insight for the predominance of SLE in females and suggests a novel mechanism to explain the observed overexpression of methylation-sensitive genes in SLE T cells and the resulting T cell autoreactivity in SLE patients.
Materials and Methods
SLE patients and controls
Korean SLE patients and controls were recruited at the Hospital for Rheumatic Diseases, Hanyang University, Seoul, Korea. All patients were of Korean descent and met the 1997 American College of Rheumatology SLE classification criteria. A total of 628 independent female SLE patients and 736 healthy unrelated female controls were studied. No two SLE patients or two controls are blood relatives to avoid intrafamilial correlation bias.
Another independent cohort of SLE patients of European descent was studied. This cohort consisted of 1080 independent cases and 1080 healthy unrelated controls matched for race and sex and recruited in the SLE genetics studies at the Oklahoma Medical Research Foundation as well as from collaborators in the United States, United Kingdom, and Sweden. This cohort included 928 females and 152 males in each of the case and control groups. All SLE patients met the 1997 American College of Rheumatology SLE classification criteria.
The study was approved by the Institutional Review Boards of Hanyang University Medical Center, University of Oklahoma Health Sciences Center, and the Oklahoma Medical Research Foundation. Participants in the study gave written informed consent for the genotyping.
Genotyping
Twenty one SNPs within or around the MECP2 gene (Table 1) were genotyped on an Illumina BeadStation 500GX instrument using Illumina Infinum II genotyping assays following manufacturer's recommendations. The SNPs were selected to cover the entire length of MECP2 and the immediate genetic regions in both the 5′ and 3′ ends of MECP2. SNPs were selected from the published SNP databases (http://www.ncbi.nlm.nih.gov/projects/SNP/). In general, we selected SNPs that have been validated by at least two groups, that have a minor allele frequency of ≥5%, and that had been tested successfully on the Illumina genotyping platform that we used in our study. Genotyping data were only used from samples with a call rate greater than 90% of the SNPs screened (98.05% of the samples). The average call rate for all samples was 97.18%.
Data analysis
A population-based case-control statistical design was employed. For each tested SNP, the quality of the genotyping data was assessed by predetermined quality control inclusion criteria (MAF>5%, SNP call rate>90%, and HWE p value>0.01 among the controls). A Pearson's Chi square was calculated for the frequency of allele associations in cases and controls. P values of <0.05 were considered statistically significant. Odds ratios were calculated under the assumption of normality. Fisher's exact test was used to test for deviation from Hardy-Weinberg equilibrium in the genotyped SNPs in the cases and controls. Permutation p values were calculated to correct for multiple testing using Haploview 3.32 [14]. Haplotype frequencies were estimated using the expectation–maximization algorithm implemented in Haploview 3.32 [14] and WHAP [15]. Haplotype-based association analysis was used to perform regression-based omnibus haplotype frequency tests and haplotype-specific tests, implemented in WHAP. We also used pair-wise SNP correlation (r2) structure between the two populations to identify the minimum haplotype length which could carry the risk of SLE development. To control for possible confounding due to population stratification, we used genomic control (GC) as well as structured association analysis. A panel of 63 randomly chosen “null” SNPs genotyped on the same Illumina SNP platform was used for GC and for estimating hidden population structure.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This publication was made possible by NIH Grant Number P20-RR015577 from the National Center for Research Resources and by funding from the University of Oklahoma College of Medicine (AHS), NIH Grant Number AI63622 (SKN), and NIH Grants Number AR42460, AI024717, AI31584, AR62277, AR048940, AR0490084, Kirkland Scholar award, Alliance for Lupus Research, and the U.S. Department of Veterans Affairs (JBH).
References
- 1.Petri M. Epidemiology of systemic lupus erythematosus. Best Pract Res Clin Rheumatol. 2002;16:847–858. doi: 10.1053/berh.2002.0259. [DOI] [PubMed] [Google Scholar]
- 2.Sawalha AH, Richardson BC. DNA methylation in the pathogenesis of systemic lupus erythematosus. Current Pharmacogenomics. 2005;3:73–78. [Google Scholar]
- 3.Richardson BC, Strahler JR, Pivirotto TS, Quddus J, Bayliss GE, et al. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35:647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 4.Oelke K, Lu Q, Richardson D, Wu A, Deng C, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 6.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. Journal of Immunology. In press doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 7.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum Immunol. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 8.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–3035. [PubMed] [Google Scholar]
- 9.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 11.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 13.Miltenberger-Miltenyi G, Laccone F. Mutations and polymorphisms in the human methyl CpG-binding protein MECP2. Hum Mutat. 2003;22:107–115. doi: 10.1002/humu.10243. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 16.Bird AP. Functions for DNA methylation in vertebrates. Cold Spring Harb Symp Quant Biol. 1993;58:281–285. doi: 10.1101/sqb.1993.058.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Leonard H, Bower C, English D. The prevalence and incidence of Rett syndrome in Australia. Eur Child Adolesc Psychiatry. 1997;6(Suppl 1):8–10. [PubMed] [Google Scholar]
- 18.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 19.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 20.Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 21.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibayama A, Cook EH, Jr, Feng J, Glanzmann C, Yan J, et al. MECP2 structural and 3′-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. Am J Med Genet B Neuropsychiatr Genet. 2004;128:50–53. doi: 10.1002/ajmg.b.30016. [DOI] [PubMed] [Google Scholar]
- 23.Francke U. Mechanisms of disease: neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol. 2006;2:212–221. doi: 10.1038/ncpneuro0148. [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, et al. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 25.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan L, Li L. Regulations and roles of the interleukin-1 receptor associated kinases (IRAKs) in innate and adaptive immunity. Immunol Res. 2006;35:295–302. doi: 10.1385/IR:35:3:295. [DOI] [PubMed] [Google Scholar]
- 27.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]