Abstract
Scavenger receptor BI (SR-BI) is a cell surface receptor that binds high density lipoproteins (HDL) and mediates selective uptake of HDL cholesteryl esters (CE) in transfected cells. To address the physiological role of SR-BI in HDL cholesterol homeostasis, mice were generated bearing an SR-BI promoter mutation that resulted in decreased expression of the receptor in homozygous mutant (designated SR-BI att) mice. Hepatic expression of the receptor was reduced by 53% with a corresponding increase in total plasma cholesterol levels of 50–70% in SR-BI att mice, attributable almost exclusively to elevated plasma HDL. In addition to increased HDL-CE, HDL phospholipids and apo A-1 levels were elevated, and there was an increase in HDL particle size in mutant mice. Metabolic studies using HDL bearing nondegradable radiolabels in both the protein and lipid components demonstrated that reducing hepatic SR-BI expression by half was associated with a decrease of 47% in selective uptake of CE by the liver, and a corresponding reduction of 53% in selective removal of HDL-CE from plasma. Taken together, these findings strongly support a pivotal role for hepatic SR-BI expression in regulating plasma HDL levels and indicate that SR-BI is the major molecule mediating selective CE uptake by the liver. The inverse correlation between plasma HDL levels and atherosclerosis further suggests that SR-BI may influence the development of coronary artery disease.
It is well established that plasma concentrations of high density lipoprotein (HDL) cholesterol are inversely proportional to the risk of developing atherosclerosis and coronary artery disease (1). Although the protective mechanism is not known, HDL is thought to reduce plaque formation by removing cholesterol from arterial cells and delivering it to the liver as cholesteryl ester (CE) for bile acid synthesis and secretion, a process referred to as reverse cholesterol transport (2). HDL also delivers CE to steroidogenic tissues (adrenal gland, ovary, and testis), where it serves as substrate for steroid hormone synthesis (reviewed in ref. 3). The uptake of HDL cholesterol by cells involves selective transfer of CE to the cell without uptake and degradation of HDL proteins, a process known as selective lipid uptake (4, 5). This is markedly different from the mechanism of clearance of low density lipoproteins (LDL), which involves receptor-mediated endocytosis and intracellular degradation of the entire lipoprotein particle (6).
Whereas the receptor that mediates LDL clearance was identified well over a decade ago (6), a functional HDL receptor has only recently been identified. Acton et al. (7) demonstrated that scavenger receptor BI (SR-BI), a multiligand cell surface receptor isolated from Chinese hamster ovary cells by expression cloning (8), binds HDL with high affinity and mediates selective cholesterol uptake in transfected cells. Furthermore, the receptor is primarily expressed in those tissues exhibiting selective lipid uptake in vivo: liver, adrenal gland, ovary, and testis (7, 9, 10).
A number of recent studies have supported a role for SR-BI in vivo in providing cholesterol for steroid hormone synthesis via selective cholesterol uptake in steroidogenic tissues. SR-BI expression was found to be up-regulated under conditions where adrenal stores of cholesterol were depleted (9), or under conditions where cholesterol utilization was increased because of stimulation of hormone synthesis in the adrenal gland, testis, or ovary (9–11). These studies strongly suggest that SR-BI is an in vivo HDL receptor that provides CE for steroid hormone synthesis, but the physiological role of SR-BI in regulating plasma cholesterol metabolism remains uncertain. Although selective uptake by the liver represents the major route of catabolism of plasma HDL cholesteryl ester in rodents (4, 5), the importance of SR-BI in this process is unclear. Adenovirus-mediated overexpression of SR-BI in the liver of mice resulted in a marked decrease of plasma HDL and a significant increase in cholesterol secretion in the bile (12). These data suggest an important role for SR-BI in hepatic HDL metabolism but are based on expression levels that are nonphysiological. To more directly address the role of SR-BI in regulating HDL cholesterol homeostasis, we have generated mice bearing a targeted mutation of the SR-BI gene. Our data indicate that SR-BI plays a central role in the hepatic clearance of HDL cholesterol by the previously described selective uptake pathway (4, 5) and in the regulation of plasma HDL levels.
MATERIALS AND METHODS
Targeting of SR-B1.
Murine SR-BI gene sequences were isolated from a mouse strain 129/Sv genomic phage library (Stratagene). The targeting construct was generated by subcloning the lacZ coding sequence, containing the simian virus 40 large T antigen nuclear localization signal at its 5′ end and the murine protamine-1 first intron and poly(A) sequence at its 3′ end (13), immediately upstream of the PGK-neo expression cassette from the plasmid pKJ1 (14). The 2-kb XbaI-PstI genomic SR-BI fragment upstream of exon 1 was subcloned 5′ of the lacZ coding sequence, and a 3.1-kb EcoRI-SacI gene fragment downstream of exon 2 was subcloned 3′ of the PGK-neo cassette to generate the SR-BI targeting vector. The vector was linearized with NotI digestion before electroporation.
RF-8 ES cells (15) were cultured on SNL76/7 mitotically inactive feeder cells as described (16) and electroporated with the linearized targeting vector as described (17).
For Southern blot analyses, genomic DNA was digested with the indicated restriction endonucleases, electrophoresed through a 0.8% agarose gel, transferred to Hybond N+ membrane (Amersham), and hybridized with the 32P-radiolabeled probes indicated in the text.
Generation of Mutant Mice.
The targeted ES clone was injected into BALB/cByJ blastocysts as described (18) to generate chimeras. Male chimeras were bred with BALB/cByJ females, and offspring heterozygous for the mutation were interbred to generate F2 progeny for analysis. Mice designated as controls are wild-type F2 mice; “SR-BI att” indicates mice that are homozygous for the targeted mutation. Mice were maintained on a 12-h light/12-h dark cycle, fed PMI 5021 chow (PMI Feeds, St. Louis) containing 9% fat, and provided with water ad libitum.
Western Blot Analysis.
Organ homogenates were prepared as described (19). Postnuclear supernatants were prepared by centrifugation of homogenates at 3,000 × g for 10 min at 4°C. Samples were reduced with 2-mercaptomethanol in gel-loading buffer, fractionated on a 8% Tris-glycine gel (Novex), and transferred to 0.45-μM nitrocellulose membranes. Immunoblot analysis was performed by using a rabbit polyclonal antipeptide antibody (Research Genetics, Huntsville, AL) raised against amino acid residues 495–509 of the long form of the human SR-BI sequence (20). Ponceau S staining was used to control for loading and transfer efficiency. Densitometry was performed on scanned autoradiograms by using nih image 1.54 software.
Analysis of Plasma HDL Cholesterol.
Blood was collected from the retroorbital plexus in heparinized capillary tubes, and total plasma cholesterol was determined by using an enzymatic assay (Sigma #352). For size-exclusion chromatography, plasma samples were centrifuged at 20,800 × g for 10 min. One hundred microliters of supernatant was applied to two Superose-6HR columns (Pharmacia) connected in tandem and eluted isocratically at a constant flow rate of 0.5 ml/min with 1.3 column volume of 10 mM Tris/150 mM NaCl/1 mM EDTA, pH 7.4 [BioCad Chromatography workstation (Perceptive Biosystems) fitted with a Thermoseparation autosampler]. Fractions of 0.5 ml were collected and cholesterol content was determined as described above.
Triglyceride, phospholipid, and lipoprotein cholesterol concentrations were determined by using commercial kits (Wako Biochemicals, Osaka) as described (9). Cholesteryl ester content was derived by subtracting free cholesterol from total cholesterol. VLDL (d < 1.006 g/ml), IDL + LDL (d = 1.006–1.055 g/ml), and HDL (d = 1.055–1.21 g/ml) were separated by sequential density ultracentrifugation of pooled mouse plasma. Denaturing polyacrylamide gel analysis of isolated lipoproteins was performed by using 4–20% SDS/PAGE gradient gels from Bio-Rad. Gels were stained with Coomassie brilliant blue R, and the identity of each apolipoprotein confirmed by Western blot analysis. Nondenaturing PAGE of plasma was performed on 4–20% gradient Lipogel (Zaxis, Hudson, Ohio) stained with Sudan Black. Purified murine VLDL, LDL, and HDL were included as standards; the size of HDL was determined by correlation of migration with that of protein size markers (Pharmacia).
HDL Catabolism.
Mouse HDL was prepared in the density range 1.063–1.21 g/ml from plasma of wild-type 129/Sv × BALB/cBy F2 mice by using sequential preparative ultracentrifugation according to standard procedures. HDL was dialyzed against PBS containing 0.3 mM EDTA and 0.02% NaN3, radiolabeled in the protein moiety with 125I-N-methyl-tyramine cellobiose (125I-NMTC) (21), and thereafter with [3H]cholesteryl oleyl ether ([3H]CEt, Amersham) to trace the CE moiety (22).
Experiments to determine plasma fractional catabolic rates (FCRs) of both HDL tracers and their tissue sites of uptake were carried out as previously described (5, 23). Food was removed from four female control and SR-BI att mice each 4 h before tracer injection, and animals were fasted throughout the 24-h study period but had free access to water. Doubly radiolabeled HDL was injected at 10 a.m. in an iliacal vein, and blood samples were drawn from the tail vein of each animal at 0.08, 0.5, 5.0, 2.0, 9.0, and 24.0 h postinjection. Plasma samples were directly radioassayed for 125I and analyzed for [3H] after lipid extraction (5). Twenty-four hours after tracer injection organs and gut contents were collected, weighed, homogenized, and radioassayed. Tissue content of 125I radioactivity was directly assayed, and that of [3H] was analyzed by liquid scintillation counting after lipid extraction (5). Tracers in the gut contents were attributed to primary uptake by the liver (5, 22).
Based on plasma decay of both HDL tracers, plasma FCRs were calculated by using a two-component model, which is similar to established procedures (23). Organ FCRs, representing the fraction of the plasma pool of the traced HDL component cleared per hour by an organ, were calculated as the plasma FCR × fraction of total tracer (%) recovered in a specific organ.
RESULTS
Disruption of the SR-B1 Gene.
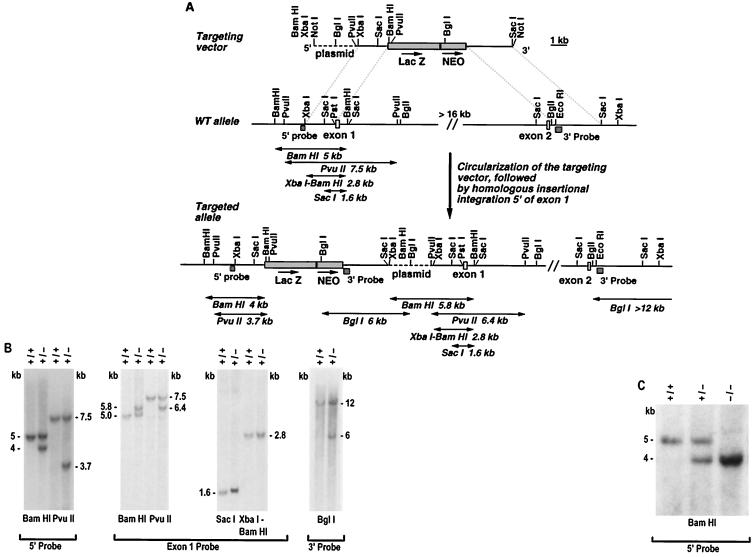
The murine SR-BI locus was disrupted in embryonic stem (ES) cells by homologous recombination with the targeting vector shown in Fig. 1A. A targeted clone was generated in which homologous recombination between the 5′ arm of the vector and genomic sequences upstream of exon 1 was verified by Southern blot hybridization (Fig. 1B). Recombination of the 3′ vector arm with homologous sequences downstream of exon 2 was not detected by a variety of restriction digests (data not shown). Detailed analysis of the targeted allele (as described in the legend to Fig. 1B) demonstrated that the entire targeting vector integrated homologously into genomic DNA upstream of exon 1 as a sequence-insertion event, presumably after circularization within the ES cell nucleus, to generate the genomic structure proposed in Fig. 1A. As a result of this mutation the putative SR-BI promoter region has been disrupted by the insertion of the approximately 13.5-kb targeting vector. However, SRBI coding sequences are intact, as are approximately 2 kb of sequences immediately upstream of exon 1, which include the putative TATA and GC boxes.
Figure 1.
Disruption of the mouse SR-BI locus. (A) Schematic representation and partial restriction maps of the SR-BI targeting vector, the wild-type locus, and the targeted allele. Open boxes represent SR-BI exons 1 and 2, the hatched boxes indicate the 5′ flanking probe and the 3′ probe, the shaded boxes are the lacZ gene and the PGK-neo expression cassette, and the dashed line is plasmid DNA. The arrows above lacZ and neo indicate the direction of transcription. Double-headed arrows indicated approximate predicted restriction fragment sizes. (B) Autoradiogram of Southern blot analysis of ES cell DNA. Genomic DNA from control (+/+) and targeted (+/−) ES cells was digested with the indicated enzymes and hybridized with radiolabeled 5′ probe, 3′ probe, or exon 1 probe, as indicated. When probed with the 5′ flanking probe, BamHI and PvuII digestion of genomic DNA from targeted ES cells produced the expected wild-type 5- and 7.5-kb bands, respectively, and the predicted 4- and 3.7-kb bands diagnostic of homologous recombination with the vector. Hybridization of these digests with an exon 1 probe revealed novel bands of 5.8 (BamHI) and 6.4 kb (PvuII) in the targeted clone, in addition to the wild-type 5- (BamHI) and 7.5-kb (PvuII) bands. Further analysis with the exon 1 probe demonstrated that the wild-type XbaI-BamHI and SacI bands of 2.8 and 1.6 kb were unaltered in the targeted allele. Lastly, hybridization with the 3′ probe detected a control BglI fragment of >12 kb and a novel 6-kb BglI band in the targeted clone. These data are consistent with homologous insertion of the entire targeting vector via single reciprocal recombination (reviewed in ref. 24) as diagrammed. In addition, typical of such sequence insertion recombination events, a genomic duplication was generated (reviewed in refs. 25 and 26), such that in the targeted locus the targeting vector is bracketed by the 2-kb fragment originating from upstream of exon 1. (C) Autoradiogram of Southern blot analysis of genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous mutant (−/−) F2 littermates. Genomic DNA was digested with BamHI and hybridized with the 5′ flanking probe.
SR-B1 Expression in Mutant Mice.
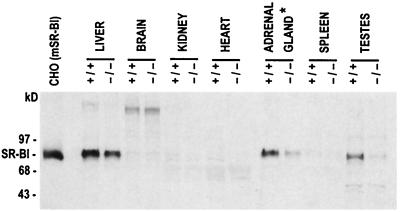
Mice homozygous for the SR-BI promoter mutation were generated (Fig. 1C) with no discernible effect of homozygosity for the SR-BI att mutation upon viability. Mice were assayed for SR-BI expression by Western blot hybridization by using a polyclonal rabbit antiserum. Tissue distribution and size of the SR-BI protein were unaltered by the mutation, but a significant decrease in expression levels was apparent (Fig. 2). In the liver of homozygous mutant mice, SR-BI protein (1117 ± 96.5 arbitrary units) was reduced to 47% of wild-type levels (2,385 ± 345.6 arbitrary units; n = 6 for both wild-type and mutant mice, P = 0.01). In the adrenal glands of homozygous mutant mice, protein expression (427 ± 47 arbitrary units) was further reduced to approximately 13% of wild type (3,266 ± 682 arbitrary units; n = 3 for both wild-type and mutant mice, P = 0.01), as was SR-BI mRNA expression (data not shown). Protein expression in the testis was also dramatically reduced in mutant mice. Based on this attenuated expression, we have designated this mutation SR-BI att. The levels of SR-BI expression in the ovaries of wild-type mice (n = 7) varied significantly, as did expression levels in ovaries of SR-BI att mice (n = 7), with no readily apparent difference between wild-type and control values (data not shown). This variation may reflect cyclical hormonal changes that regulate SR-BI levels coordinately with the demand for CE in the ovary during the 4- to 5-day mouse estrous cycle.
Figure 2.
Immunoblot analysis of SR-BI protein expression in control and homozygous mutant mice. A total of 50 μg of protein from postnuclear supernatants of mSR-BI transfected CHO cells and from liver, spleen, brain, heart, testis, and kidney, and 20 μg from the adrenal gland, of representative control (+/+) and SR-BI att (−/−) mice was analyzed by gel electrophoresis and immunoblotting with a rabbit polyclonal anti-human SR-BI antibody. ∗, 2.5-fold less protein was loaded in this, relative to the other, lanes.
Plasma Cholesterol, HDL, and Lipoproteins.
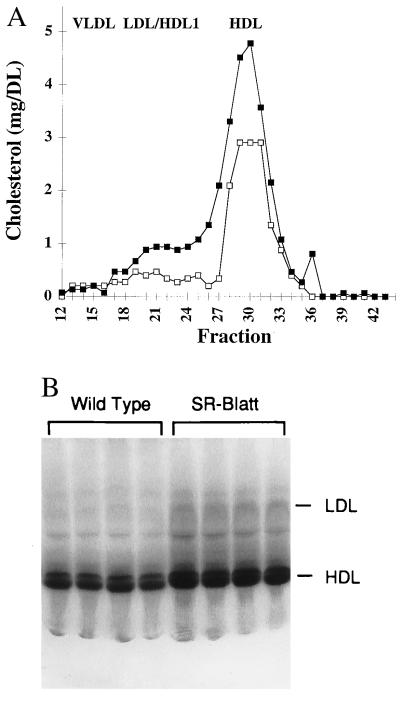
To assess the consequences of the att mutation on plasma cholesterol levels, blood was collected from homozygous mutant and control mice. Attenuated expression of SR-BI was associated with an approximately 50–70% increase in the concentration of total plasma cholesterol (84 ± 2.5 mg/dl control male, 126.6 ± 7.5 mg/dl SR-BI att male, n = 7, P = 0.001; 73.4 ± 6.2 mg/dl control female, 126.5 ± 5.3 mg/dl SR-BI att female, n = 7, P = 0.00003); no alterations of plasma triglyceride levels were observed (data not shown). Fast pressure liquid chromatography (FPLC) revealed a 51% increase in HDL concentration in homozygous mutant mice and a slight increase in cholesterol in the LDL-HDL 1 region (Fig. 3A).
Figure 3.
Plasma cholesterol and HDL analysis. (A) FPLC analysis of plasma lipoproteins. Plasma was pooled (n = 4) from homozygous mutant (▪) and control (□) female mice (ages 10, 11.5, and 15.5 weeks, and 11.5 and 15.5 weeks, respectively) and subjected to FPLC Superose-6 gel filtration, and the cholesterol content of the fractions was determined. (B) Nondenaturing PAGE of plasma from control and mutant mice. Ten microliters of plasma from control and SR-BI att male mice were analyzed.
Analysis of HDL size distribution on nondenaturing polyacrylamide gels revealed an increased staining intensity of HDL lipids and a slight increase in LDL staining (Fig. 3B). HDL migration was slightly retarded in homozygous mutant mice, indicating an increase in mean diameter of the HDL particle from 10.9 ± 0.09 nm to 11.2 ± 0.09 nm (n = 4, P = 0.00085). A similar change in HDL size was observed when plasma samples were normalized for HDL concentration before loading (data not shown). Separation of plasma lipoproteins by ultracentrifugation showed an increase in cholesterol (72%) and phospholipids (29%) in HDL (d = 1.055–1.21 g/ml), with no significant changes in the VLDL (d < 1.006 g/ml) and IDL/LDL (d = 1.006–1.055 g/ml) fractions (Table 1). This suggests that the increase in cholesterol in the LDL/HDL 1 region of the FPLC profile (Fig. 3A) is predominantly due to an increase in HDL 1, indicating that the increased plasma cholesterol in mutant mice is attributable almost exclusively to elevated plasma HDL.
Table 1.
Plasma lipoproteins
| Mouse type | VLDL, mg/dl
|
IDL/LDL, mg/dl
|
HDL, mg/dl
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TC | PL | TG | TC | PL | TG | TC | PL | TG | |
| Control | 16.8 ± 4.8 | 35.6 ± 12.2 | 65.7 ± 17.6 | 5.7 ± 1.7 | 10.5 ± 2.6 | 10.2 ± 2.3 | 62.5 ± 15.6 | 90.2 ± 23.1 | 4.9 ± 1.2 |
| SR-BI att | 14.4 ± 3.9 | 34.7 ± 15.1 | 59.9 ± 15.9 | 6.9 ± 1.9 | 11.6 ± 3.1 | 11.5 ± 2.8 | 107.4 ± 14.9 | 116.7 ± 27.1 | 5.8 ± 2.4 |
| (P < 0.01) | (P < 0.05) | ||||||||
SR-BI att (n = 8) and control females (n = 8) were bled between 16–20 weeks of age. The values are means ± SEM. Significance was determined by Student’s t test. TC, total cholesterol; PL, phospholipids; TG, triglycerides.
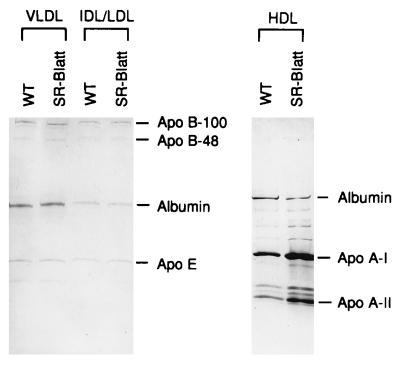
Analysis of HDL percentage composition showed a slight increase in CE content with a corresponding decrease in protein content (Table 2). Characterization of apoprotein composition by denaturing gel electrophoresis showed no significant differences in apo B-100, apo B-48, and apo E composition of VLDL and IDL/LDL in control and mutant mice (Fig. 4). However, the plasma HDL of SR-BI att mice did show a significant increase in apo A-1 levels of approximately 1.4-fold (n = 4, P = 0.045), and a nonsignificant increase in apo A-II levels. The increase in apo AI was confirmed upon additional testing of a separate set of control and mutant animals (n = 4).
Table 2.
HDL chemical composition
| FC, % | CE, % | PL, % | TG, % | Protein, % | |
|---|---|---|---|---|---|
| Control | 1.6 | 18 | 26.2 | 3.7 | 50.6 |
| SR-BI att | 2.3 | 20.4 | 27.4 | 2.7 | 47.2 |
Data shown are the average of two assays, each containing pooled plasma from four male mice. FC, free cholesterol; CE, cholesteryl ester; PL, phospholipids; TG, triglycerides.
Figure 4.
SDS/PAGE of plasma lipoproteins. Lipoprotein fractions were separated by ultracentrifugation of pooled plasma (n = 4) from control and SR-BI att male mice and analyzed on a 4–20% gradient gel.
HDL Metabolism.
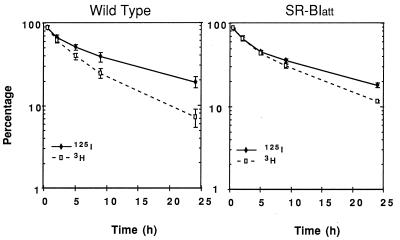
To determine whether the increased plasma HDL cholesterol in homozygous mutant mice reflected differences in HDL clearance between mutant and wild-type animals, female mice were injected with HDL that had been radiolabeled in the protein moiety with 125I-NMTC and in the CE component with [3H]CEt (5). These radiolabels are not degraded in the plasma or tissue compartments. Plasma decay curves for both tracers analyzed during a 24-h period postinjection showed that although the decay of 125I-NMTC-labeled protein was identical in control and mutant mice, in the mutant animals the rate of clearance of [3H]CEt was significantly decreased (Fig. 5). The plasma FCRs calculated from these decay curves similarly showed a significant decrease in lipid tracer catabolism but no change in the protein catabolic rate (Table 3). The selective removal of CE from plasma, calculated as the difference between CE and protein FCRs, was reduced by 53% in mice homozygous for the att mutation (Table 3, P < 0.007).
Figure 5.
Plasma decay curves for 125I-NMTC/[3H]CEt-labeled HDL. Mice were injected with labeled HDL, blood was collected periodically over 24 h, and plasma content of both tracers was determined as described in Materials and Methods. The values are means ± SEM of each of the n = 4 control and SR-BI att mice.
Table 3.
Plasma fractional catabolic rates for 125I-NMTC/[3H]cholesteryl oleyl ether-labeled HDL in mice
| Mice | 125I-NMTC-protein, pools/h | [3H]cholesteryl oleyl ether, pools/h | [3H] − 125I, pools/h |
|---|---|---|---|
| Control | 0.080 ± 0.009 | 0.146 ± 0.01 | 0.066 ± 0.009 |
| SR-BI att | 0.081 ± 0.006 | 0.112 ± 0.006 | 0.031 ± 0.004 |
| (P = 0.47) | (P = 0.048) | (P = 0.007) |
Blood was collected periodically from four female control and four female SR-BI att mice over 24 h after injection of labeled HDL. The values are means ± SEM, and significance was determined by Student’s t test.
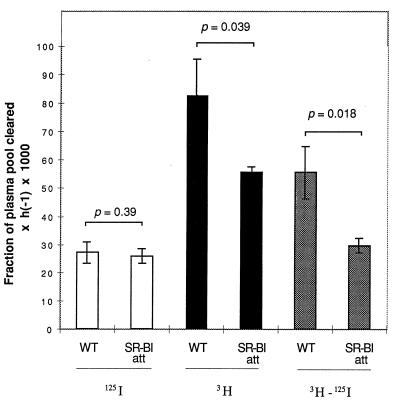
Tissue sites of tracer uptake from doubly radiolabeled HDL were determined, and organ FCRs were calculated (5, 22). The liver was the predominant site of HDL catabolism, and the majority of hepatic CE uptake was mediated by selective uptake as indicated by a higher liver FCR for the lipid, relative to the protein, tracer (Fig. 6). The hepatic FCR for [3H]CEt was decreased in mutant mice by approximately 40%. More importantly, selective CE uptake (lipid–protein FCRs) by the liver of female mutant mice was approximately 50% of control values (Fig. 6), correlating closely with the 53% reduction observed in SR-BI expression in the liver (Fig. 2) and the 53% reduction in selective removal of CE from the plasma (Table 3).
Figure 6.
Uptake of labeled HDL by the liver. Liver FCRs, calculated as described in Materials and Methods, are shown for uptake of labeled protein (125I, open bars) and cholesteryl ether (3H, solid bars). The shaded bars represent selective CE uptake (3H − 125I). Significance was determined by Student’s one-tailed t test for unpaired data.
The selective uptake of HDL CE by the adrenal glands of SR-BI att mice was decreased by about 20%, but the change was not significant (FCR of 0.743 ± 0.17 in control mice, 0.596 ± 0.16 in mutant mice; P = 0.28), despite the substantial reduction of basal SR-BI expression in this organ (Fig. 2). Because att is a promoter mutation, it is possible that the conditions of the HDL clearance study, which involved periodic bleedings over a 24-h period, up-regulated SR-BI expression. In this regard, it has been documented that stress, and the resulting induction of adrenocorticotropic hormone plasma levels, markedly induces adrenal SR-BI expression (9, 11). To determine whether the clearance study itself had stimulated adrenal SR-BI expression, adrenal glands were assayed from control and mutant mice that had been exposed to periodic anesthetization, as in the HDL tracer analyses. This protocol resulted in up-regulation of adrenal SR-BI expression in mutant mice to 77% of control levels (n = 3, no significant difference between control and mutant mice; data not shown), as contrasted with untreated SR-BI att mice in which adrenal expression is 13% of control levels (Fig. 2). In other organs (kidney, lungs, heart, spleen) where SR-BI expression is low or undetectable (Fig. 2), there was no effect of the SR-BI mutation on tissue uptake of [3H]CEt (data not shown).
DISCUSSION
In this study we have addressed the physiological role of SR-BI in the regulation of HDL homeostasis. Mice were generated bearing an SR-BI promoter mutation, which resulted in decreased expression of the receptor in homozygous mutant mice (designated SR-BI att mice). While this manuscript was in preparation, Rigotti et al. (27) reported generation of an SR-BI null mutation that resulted in increased concentrations of plasma HDL and decreased adrenal gland cholesterol content. The present study extends these findings by demonstrating a central role for SR-BI in the selective uptake of HDL CE in the liver.
Previous studies in rodents have shown that the liver is the major organ regulating HDL metabolism (4, 5). HDL cholesterol catabolism by the liver occurs primarily by the process of selective CE uptake, whereby CE is transferred from the HDL particle to hepatic cells without transfer and degradation of HDL protein (4, 5). Although this process has been known for several years at the physiological level, the molecular identification of the receptor mediating selective CE uptake has proven difficult. The recent characterization of SR-BI as a functional HDL receptor that can mediate selective uptake in vitro (7) raised the possibility that SR-BI is the physiologically relevant hepatic receptor for HDL CE. In the present study we present compelling evidence in support of this hypothesis. In SR-BI att mice, hepatic expression of SR-BI was reduced by 53% with a corresponding increase in total plasma cholesterol levels of 50–70%. Metabolic studies using HDL radiolabeled in both the protein and lipid components demonstrated that reducing hepatic SR-BI expression by half was associated with a decrease of 47% in selective uptake of CE by the liver and a corresponding reduction of 53% in selective removal of HDL CE from plasma. Taken together, these findings strongly support a central role for hepatic SR-BI expression in regulating plasma HDL levels and indicate that SR-BI is the major molecule mediating selective CE uptake by the liver.
Other factors, such as hepatic lipase activity or probucol treatment of rodents, have been shown to increase selective uptake in the liver or in cultured hepatocytes (28–30). The present finding, that hepatic SR-BI expression levels closely parallel changes in plasma and hepatic CE selective uptake, suggests that these factors may not be directly responsible for selective uptake but rather may modulate SR-BI activity, for example, by modifying interaction with HDL or by affecting SR-BI expression. The quantitative importance of SR-BI in selective uptake in vivo suggests that it may be possible to modulate HDL plasma levels by therapies aimed at modulating SR-BI expression.
The delayed rate of clearance of HDL CE in the plasma of SR-BI att mice was associated with a slight increase in HDL particle size, which was largely attributable to increased CE content as well as a small increase in phospholipids. These observations suggest that SR-BI plays a role in regulating the size and composition of HDL particles. In addition to an increase in HDL lipids, there was also an increase in HDL apo AI in SR-BI att mice, not noted by Rigotti et al. (27). This difference could reflect methodological differences in the quantitation of apo AI. Detailed analysis of HDL composition indicates that the proportional increase in lipid was greater than in protein, but a 1.4-fold increase in the absolute level of apo AI was observed in HDL from SR-BI att mutants. Because the protein catabolic rate is unchanged in mutant mice, the increase in apo AI levels must be at the level of synthesis. This raises the intriguing possibility that the hepatic synthesis of apo AI may at least in part be regulated by selective uptake of HDL CE.
The SR-BI att mutation was generated by homologous insertion of an approximately 13.5-kb targeting vector approximately 2 kb upstream of exon 1. Neither SR-BI coding sequences nor the putative SR-BI TATA box or GC box situated within 2 kb upstream of exon 1 was disrupted, but sequences further upstream were displaced by insertion of the targeting vector. The mutation had no discernible effect on the size of the SR-BI protein or RNA transcript (data not shown) in any of the tissues tested, nor was the tissue distribution of SR-BI expression altered. Only quantitative effects on gene expression were observed, which were differentially manifested in various tissues. Whereas hepatic SR-BI expression was reduced by approximately half in SR-BI att mice, basal expression was even more dramatically reduced in adrenal glands to approximately 10% of wild type and was also significantly reduced in testis. Interestingly, although basal adrenal SR-BI expression was substantially reduced, inducible regulation by stress was maintained. The differential effect of the promoter mutation may be mediated by disruption of tissue-specific regulatory elements by insertion of the targeting vector or may be the result of a transcriptional silencer effect of the neo gene within the targeting vector (31), which exhibits tissue specificity.
Targeted mutation has demonstrated a critical role for murine SR-BI in HDL CE uptake and thus presumably in reverse cholesterol transport, the process by which cholesterol is transported from extrahepatic tissues to the liver. The reverse cholesterol pathway has been implicated in mediating the atheroprotective effect of HDL via the proposed hepatic clearance of HDL cholesterol originating from atherosclerotic plaques. Because extraction of CE by the liver is thought to be the final step in this process, SR-BI may play an important role in determining susceptibility to atherosclerosis. In addition, SR-BI also mediates cellular cholesterol efflux and is expressed in atheroma (32). Thus the receptor may be involved in the initial steps of reverse cholesterol transport, i.e., the removal of cholesterol from arterial foam cells, as well as in the final clearance stage of the process.
In humans, but not rodents, the reverse cholesterol transport pathway also involves cholesteryl ester transfer protein (CETP), which transfers HDL CE to other lipoproteins with subsequent clearance in the liver. Human genetic deficiency of CETP results in increased HDL levels but paradoxically also increases coronary heart disease susceptibility (33). By analogy, the relationship of SR-BI expression to atherosclerosis may also be complex. The SR-BI att mutation described in this study should prove a useful tool for assessing the consequences of SR-BI down-regulation on the development of atherosclerotic lesions in atherosclerosis-susceptible mouse models.
Acknowledgments
We thank Sharon Squazzo, Pei Shu, Doreen Osgood, Roslyn Feeley, and Michael Donovan for technical assistance, Jingya Ma and Betty Woolf for sequence information, Anne Tsimboukis and Nash Mason for help with graphics, and Geoffrey S. Ginsburg for reviewing the manuscript. This work was financially supported by Eli Lilly, and by Grants HL22682 and HL58033 from the National Institutes of Health.
ABBREVIATIONS
- HDL and LDL
high and low density lipoproteins
- CE
cholesteryl esters
- SR-BI
scavenger receptor BI
- FCR
fractional catabolic rates
- ES cell
embryonic stem cell
Footnotes
A commentary on this article begins on page 4077.
References
- 1.Gordon D J, Rifkind B M. N Engl J Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 2.Glomset J A. J Lipid Res. 1968;9:155–167. [PubMed] [Google Scholar]
- 3.Gwynne J T, Strauss J F. Endocr Rev. 1982;3:299–329. doi: 10.1210/edrv-3-3-299. [DOI] [PubMed] [Google Scholar]
- 4.Glass C, Pittman R C, Weinstein D W, Steinberg D. Proc Natl Acad Sci USA. 1983;80:5435–5439. doi: 10.1073/pnas.80.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glass C, Pittman R C, Civen M, Steinberg D. J Biol Chem. 1985;260:744–750. [PubMed] [Google Scholar]
- 6.Brown M S, Goldstein J L. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 7.Acton S, Rigotti A, Landschultz K T, Xu S, Hobbs H H, Krieger M. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 8.Acton S, Scherer P E, Lodish H F, Krieger M. J Biol Chem. 1994;269:21003–21009. [PubMed] [Google Scholar]
- 9.Wang N, Weng W, Breslow J L, Tall A R. J Biol Chem. 1996;271:21001–21004. doi: 10.1074/jbc.271.35.21001. [DOI] [PubMed] [Google Scholar]
- 10.Landschulz K T, Pathak R K, Rigotti A, Krieger M, Hobbs H H. J Clin Invest. 1996;98:984–995. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigotti A, Edelman E R, Seifert P, Iqbal S N, DeMattos R B, Temel R E, Krieger M, Williams D L. J Biol Chem. 1996;271:33545–33549. doi: 10.1074/jbc.271.52.33545. [DOI] [PubMed] [Google Scholar]
- 12.Kozarsky K F, Donahee M H, Rigotti A, Iqbal S N, Edelman E R, Krieger M. Nature (London) 1997;387:414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 13.Mercer E H, Hoyle G W, Kapur R P, Brinster R L, Palmiter R D. Neuron. 1991;7:703–716. doi: 10.1016/0896-6273(91)90274-4. [DOI] [PubMed] [Google Scholar]
- 14.McBurney M W, Sutherland L C, Adra C N, Leclair B, Rudnicki M A, Jardine K. Nucleic Acids Res. 1991;19:5755–5761. doi: 10.1093/nar/19.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X-Z, Wu J F, Cass D, Erle D J, Corry D, Young S G, Farese R V, Sheppard D. J Cell Biol. 1996;133:921–928. doi: 10.1083/jcb.133.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson E J. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL; 1987. pp. 71–112. [Google Scholar]
- 17.Huszar D, Lynch C A, Fairchild-Huntress V, Dunmore J H, Fang Q, Berkemeier L R, Gu W, Kesterson R A, Boston B A, Cone R D, et al. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 18.Bradley A. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL; 1987. pp. 113–151. [Google Scholar]
- 19.Jokinen E V, Landschulz K T, Wyne K L, Ho Y K, Frykman P K, Hobbs H H. J Biol Chem. 1994;269:26411–26418. [PubMed] [Google Scholar]
- 20.Calvo D, Vega M A. J Biol Chem. 1993;268:18929–18935. [PubMed] [Google Scholar]
- 21.Pittman R C, Taylor C A., Jr Methods Enzymol. 1986;129:612–628. doi: 10.1016/0076-6879(86)29094-1. [DOI] [PubMed] [Google Scholar]
- 22.Rinninger F, Pittman R C. J Lipid Res. 1987;28:1313–1325. [PubMed] [Google Scholar]
- 23.Le N A, Ramakrishnan R, Dell R B, Ginsberg H N, Brown W V. Methods Enzymol. 1986;129:384–395. doi: 10.1016/0076-6879(86)29081-3. [DOI] [PubMed] [Google Scholar]
- 24.Sedivy J M, Joyner A L. Gene Targeting. New York: Freeman; 1992. pp. 77–121. [Google Scholar]
- 25.Koller B H, Smithies O. Annu Rev Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 26.Morrow B, Kucherlapati R. Current Opin Biotechnol. 1993;4:577–582. doi: 10.1016/0958-1669(93)90080-g. [DOI] [PubMed] [Google Scholar]
- 27.Rigotti A, Trigatti B L, Penman M, Rayburn H, Herz J, Krieger M. Proc Natl Acad Sci USA. 1997;94:12610–12615. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamberger M, Glick J M, Rothblat G H. J Lipid Res. 1983;24:869–876. [PubMed] [Google Scholar]
- 29.Komaromy M, Azhar S, Cooper A D. J Biol Chem. 1996;271:16906–16915. doi: 10.1074/jbc.271.28.16906. [DOI] [PubMed] [Google Scholar]
- 30.Richard B M, Pfeuffer M A, Pittman R C. Arterioscler Thromb. 1992;12:862–869. doi: 10.1161/01.atv.12.7.862. [DOI] [PubMed] [Google Scholar]
- 31.Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H. Gene. 1991;99:249–254. doi: 10.1016/0378-1119(91)90134-w. [DOI] [PubMed] [Google Scholar]
- 32.Ji Y, Jian B, Wang N, Sun Y, de la Llera Moya M, Phillips M C, Rothblat G H, Swaney J B, Tall A R. J Biol Chem. 1997;272:20982–20985. doi: 10.1074/jbc.272.34.20982. [DOI] [PubMed] [Google Scholar]
- 33.Zhong S, Sharp D S, Grove J S, Bruce C, Yano K, Curb J D, Tall A R. J Clin Invest. 1996;97:2917–2923. doi: 10.1172/JCI118751. [DOI] [PMC free article] [PubMed] [Google Scholar]