Abstract
Loss-of-function siz1 mutations caused early flowering under short days. siz1 plants have elevated salicylic acid (SA) levels, which are restored to wild-type levels by expressing nahG, bacterial salicylate hydroxylase. The early flowering of siz1 was suppressed by expressing nahG, indicating that SIZ1 represses the transition to flowering mainly through suppressing SA-dependent floral promotion signaling under short days. Previous results have shown that exogenous SA treatment does not suppress late flowering of autonomous pathway mutants. However, the siz1 mutation accelerated flowering time of an autonomous pathway mutant, luminidependens, by reducing the expression of FLOWERING LOCUS C (FLC), a floral repressor. This result suggests that SIZ1 promotes FLC expression, possibly through an SA-independent pathway. Evidence indicates that SIZ1 is required for the full activation of FLC expression in the late-flowering FRIGIDA background. Interestingly, increased FLC expression and late flowering of an autonomous pathway mutant, flowering locus d (fld), was not suppressed by siz1, suggesting that SIZ1 promotes FLC expression by repressing FLD. Consistent with this, SIZ1 facilitates sumoylation of FLD that can be suppressed by mutations in three predicted sumoylation motifs in FLD (i.e. FLDK3R). Furthermore, expression of FLDK3R in fld protoplasts strongly reduced FLC transcription compared with expression of FLD, and this affect was linked to reduced acetylation of histone 4 in FLC chromatin. Taken together, the results suggest that SIZ1 is a floral repressor that not only represses the SA-dependent pathway, but also promotes FLC expression by repressing FLD activity through sumoylation, which is required for full FLC expression in a FRIGIDA background.
Keywords: SIZ1, SA, flowering, SUMO, FLD, FLC
Introduction
Sumoylation is a post-translational regulatory process that conjugates small ubiquitin modifier peptides (SUMO) to protein substrates (Matunis et al., 1996; Mahajan et al., 1997). Like ubiquitination, SUMO attachment to a target substrate involves a series of steps referred to as activation (E1), conjugation (E2) and ligation (E3) (Seeler and Dejean, 2003; Johnson, 2004). SUMO E3 ligases of the PIAS/SIZ family facilitate SUMO conjugation to lysine (K) residues in the SUMO consensus motif, ΨKXE/D (Ψ, a large hydrophobic residue; K, the acceptor lysine; X, any amino acid; E/D, glutamate or aspartate), located in protein substrates (Schmidt and Muller, 2003). SUMO modification of target proteins in yeast and metazoans has been implicated in the regulation of innate immunity, cell-cycle progression and mitosis, DNA repair, chromatin stability, nucleocytoplasmic trafficking, subnuclear targeting, ubiquitination antagonism and transcriptional regulation (Johnson, 2004; Gill, 2005). Sumoylation in plants is reported to be involved in biotic and abiotic stress responses, flowering and development (Chosed et al., 2006; Downes and Vierstra, 2005; Kurepa et al., 2003; Lee et al., 2007; Miura et al., 2005, 2007; Novatchkova et al., 2004; Yoo et al., 2006). The Arabidopsis PIAS-type SUMO E3 ligase, AtSIZ1, facilitates SUMO modification of transcription factors, PHR1 and ICE1, which regulate phosphate-starvation signaling and low-temperature response, respectively (Miura et al., 2005, 2007). A SUMO protease, AtESD4 (EARLY SHORT DAY FLOWERING4), and its interacting protein NUA (NUCLEAR PORE ANCHOR) negatively regulate transition to flowering, suggesting that SUMO homeostasis is important for flowering time regulation (Murtas et al., 2003; Reeves et al., 2002; Xu et al., 2007).
Flowering is the result of a plant developmental process that controls the transition from vegetative maturity to the reproductive stage (Baurle and Dean, 2006). Floral transition is regulated by day length, light quality and temperature, and this responsive capacity is thought to optimize the environmental fitness of plants (Ballare, 1999; Corbesier et al., 1996; Michaels and Amasino, 2001). The vegetative to floral transition of Arabidopsis, and other rosette-type plants, is characterized by the rapid proliferation of an extended floral shoot that is the result of internodal expansion (Blazquez et al., 1997). In Arabidopsis, signal regulatory cascades, such as the photoperiodic- (or long-day), vernalization, autonomous and gibberellin (GA)-dependent pathways, control floral transition (He and Amasino, 2005).
Photoperiodic-pathway genes promote transition to flowering in response to a long-day photoperiod (Koornneef et al., 1991). Mutations to photoperiodic-pathway genes, such as GIGANTEA (GI), CONSTANS (CO) and FLOWERING LOCUS T (FT), cause significantly delayed flowering under long days (Koornneef et al., 1991). Rhythmic expression of CO transcript is regulated by circadian clock oscillators [e.g. CIRCADIAN CLOCK-ASSOCIATED PROTEIN 1 (CCA1)] (Green and Tobin, 1999) and clock- and light-regulated genes (i.e. GI) (Fowler et al., 1999). Under long days (LD) CO protein accumulates to levels that promote floral transition, mainly through activation of FLOWERING LOCUS T (FT) expression. FT in turn activates expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and floral identity genes such as APETALA1 (AP1) (Yoo et al., 2005; Wigge et al., 2005).
In contrast to the photoperiodic pathway, which directly activates the floral transition, the vernalization and autonomous pathways indirectly promote transition to flowering through repression of the central floral repressor, FLOWERING LOCUS C (FLC) (Michaels and Amasino, 1999). FLC encodes a MADS box-containing transcription factor that antagonizes floral transition facilitated by the photoperiodic pathways, by repressing FT and SOC1 expression (Michaels and Amasino, 1999; Searle et al., 2006). The presence of an active allele of FRIGIDA (FRI, an activator of FLC) in winter-annual ecotypes causes increased expression of FLC and delayed flowering, which is reversed by lesions in FLC or by vernalization treatment (Michaels and Amasino, 1999; Sheldon et al., 1999). To date, eight autonomous pathway genes have been identified from screening for late-flowering mutants that are responsive to daylength and vernalization treatment (Koornneef et al., 1991; Schmitz and Amasino, 2007). Among the autonomous pathway genes, FLOWERING LOCUS D (FLD), a plant ortholog of the human protein KIAA0601/LYSINE-SPECIFIC HISTONE DEMETHYLASE1 (LSD1), represses FLC expression by facilitating deacetylation of histone H4 in FLC chromatin, but how this process is regulated remains to be elucidated (He et al., 2003; Shi et al., 2004).
In addition to light and temperatures, the plant hormones GA and salicylic acid (SA) are implicated in the regulation of floral transition (Cleland and Ben-Tal, 1982; Wilson et al., 1992). GA promotes flowering through the activation of SOC1 and LEAFY expression, and is considered to be involved in the main floral-inducing cascade under short days (SD) (Blazquez et al., 1998; Langridge, 1957; Moon et al., 2004). Exogenous SA treatment or UV-C light stress, which induces accumulation of SA, accelerates the transition to flowering (Martinez et al., 2004). SA-deficient nahG-expressing plants, or eds5/sid1 and sid2 mutants, exhibit a delayed flowering phenotype that is evident under SD (Martinez et al., 2004). SA control of the transition to flowering appears to be complex, and the extent of its role remains to be elucidated (Martinez et al., 2004).
Although sumoylation/desumoylation has been implicated in flowering-time regulation, target proteins that are involved in flowering remain unknown (Murtas et al., 2003; Xu et al., 2007). In this report, evidence indicates that FLD is a sumoylation target for SIZ1. SUMO conjugation to FLD inhibits its activity to repress FLC expression, which is required for full activation of FLC expression in a FRI background. Our results also demonstrate that the early flowering of siz1 in SD is mainly the result of an elevated SA level. This and other results (Lee et al., 2007) indicate that sumoylation has an important role in the regulation of SA accumulation, although the sumoylation targets involved in SA accumulation are unidentified.
Results
The SUMO E3 ligase SIZ1 regulates flowering time
Flowering time of Col-0 wild-type and siz1 (siz1-2 and siz1-3) loss-of-function mutant plants (Miura et al., 2005) under LD (16-h light and 8-h dark) or SD (8-h light and 16-h dark) conditions (Lee et al., 1994a) indicated that SIZ1 negatively regulates the transition to flowering (Figure 1a–c). Flowering time of siz1 plants relative to wild type was slightly earlier under LD, and was substantially earlier under SD (Figure 1a–c). Rosette leaf numbers at flowering under LD and SD were 10 and 13, respectively, for siz1 plants, and were 13 and 49, respectively, for wild type (Figure 1c). Thus, the floral transition of siz1 plants is much less photoperiodic responsive than that of wild type.
Figure1.
SIZ1 represses transition to flowering.
(a) Wild-type (Col-0) and siz1 (siz1-2 and siz1-3) plants were grown under long days (LD) for 32 days.
(b) Wild-type (Col-0) and siz1 (siz1-2 and siz1-3) plants were grown under short days (SD) for 113 and 50 days, respectively.
(c) The number of rosette leaves at flowering of wild-type and siz1 plants. Plants were grown under LD or SD.
(d) Phenotypic comparison of plants: wild-type (Col-0), siz1-2 and transgenic ProSIZ1:SIZ1:GFP expressing siz1-2 (SSG1,8,12,14,15) and ProSIZ1:GUS:GFP expressing siz1-2 (SGG) plants.
(e) The number of rosette leaves at the flowering of plants, as in (d), which were grown under SD. Data illustrated in (c) and (e) are means ± SE of 15–20 plants per analysis.
The role of SIZ1 in flowering-time regulation was confirmed by genetic complementation of the siz1 mutation with the wild-type SIZ1 allele. Expression of ProSIZ1:SIZ1:GFP (SSGs) in siz1-2 plants suppressed the dwarf and early flowering phenotypes of plants from multiple, independent transformed lines (Figure 1d,e). Expression of ProSIZ1:GUS:GFP (SGG) in siz1-2 failed to complement these siz1-2 phenotypes in the same experiments (Figure 1d,e).
SIZ1 regulates flowering, independent of the photoperiodic- and GA-dependent pathways
Mutations in circadian oscillator genes cause a short-period phenotype, which results in early flowering under SD (Mizoguchi et al., 2002). To test whether siz1 affects the circadian clock, the rhythmic expression patterns of CCA1 and COLD CIRCADIAN RHYTHM RNA BINDING 2 (CCR2) were determined (Green and Tobin, 1999; Kreps and Simon, 1997). The diurnal rhythmic expression of CCA1 and CCR2::LUCIFERASE (CCR2::LUC) was not altered by siz1 mutations, suggesting that SIZ1 does not regulate the circadian clock (Figures S1A,B, respectively).
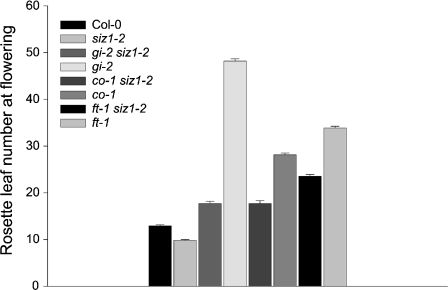
To determine if there is an interaction between SIZ1 and the photoperiodic-pathway genes GI, CO and FT, the double mutants gi-2 siz1-2, co-1 siz1-2 and ft-1 siz1-2 were produced. Flowering times of the double mutants were intermediate to that of siz1-2 and the corresponding late-flowering parental mutant plants under LD, suggesting that SIZ1 may function independently of the photoperiodic pathway (Figure 2). Consistent with this hypothesis, expression patterns of GI, CO and FT were not altered by siz1 (data not shown). To determine whether siz1 mutations affect the GA-dependent floral promotion pathway, exogenous GA was applied to wild-type, siz1-2 and siz1-3 plants, and flowering times were determined under LD and SD (Figure S2). Exogenous GA treatment accelerated the flowering time of both wild-type and siz1 plants to an equivalent extent (Figure S2), indicating that the GA floral promotion pathway is not impaired in siz1 plants.
Figure2.
siz1 partially suppresses late-flowering photoperiodic-pathway mutants. The flowering times of wild-type (Col-0), siz1-2, gi-2 siz1-2, gi-2, co-1 siz1-2, co-1, ft-1 siz1-2 and ft-1 plants were estimated under long days (LD). Data are means ± SE of 15–20 plants per analysis.
SIZ1 regulates flowering mainly through an SA-dependent pathway in the Columbia background
Compared with wild-type Col-0 plants, siz1 plants accumulate higher levels of SA, which causes increased plant innate immunity (Lee et al., 2007). The bacterial gene nahG encodes a protein that rapidly and efficiently converts SA to inactive catechol in planta (Delaney et al., 1994). To elucidate SA effects on the siz1 early flowering phenotype, flowering times of wild-type, siz1-2, nahG siz1-2 and nahG plants were compared (Figure 3a,b). In a previous report, we have shown that nahG and nahG siz1-2 plants accumulate SA at levels similar to wild-type plants (Lee et al., 2007). nahG siz1-2 plants also exhibit nearly normal leaf morphology and rosette plant size (Figure 3a; Lee et al., 2007). Flowering time of nahG siz1-2 plants was similar to that of siz1-2 plants under LD, but similar to nahG and wild-type plants under SD (Figure 3b). These results indicate that in the Columbia genetic background, the early flowering phenotype of siz1 is mainly dependent on an SA-dependent pathway under SD, but not under LD.
Figure3.
Early flowering of siz1 under short days (SD) is mainly caused by elevated salicylic acid (SA) levels.
(a) Wild-type (Col-0), siz1-2, nahG siz1-2 and nahG plants were grown under long days (LD).
(b) Number of rosette leaves at flowering of wild-type, siz1-2, nahG siz1-2 and nahG plants. Plants were grown under LD or short days (SD). Data are means ± SE of 15–20 plants per analysis.
The siz1 early flowering phenotype is in part linked to reduced MAF4 expression and elevated SOC1 expression
To determine if SIZ1 regulates floral repressors, the expression of FLC, FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING1 (MAF1), MAF2, MAF3, MAF4 and MAF5, and SHORT VEGETATIVE PHASE (SVP) were analyzed (Hartmann et al., 2000; Ratcliffe et al., 2001, 2003; Scortecci et al., 2001). FLC transcript abundance in siz1 seedlings was slightly reduced compared with wild-type seedlings under SD (Figure 5b). MAF4 mRNA abundance was substantially lower in siz1 plants compared with wild type under both LD and SD, whereas, expression of the other floral repressors FLM/MAF1, SVP, MAF2, MAF3 and MAF5 was not affected in siz1 plants. Together, these results indicate that SIZ1 positively regulates FLC and MAF4 gene expression (Figures 4a and 5b).
Figure5.
siz1 partially suppresses the late flowering of FRI and ld-1, but not of fld-6.
(a) The flowering times of wild-type (Col-0), siz1-2, ld-1 siz1-2, ld-1, fld-6 siz1-2 and fld-6 plants were analyzed under short days (SD). Stars indicate that flowering had not occurred after producing more than 100 rosette leaves in fld-6 and fld-6 siz1-2 plants.
(b) Relative FLC mRNA levels were determined in 14-day-old SD-grown wild-type (Col-0), siz1-2, ld-1 siz1-2, ld-1, fld-6 siz1-2 and fld-6 seedlings by quantitative PCR.
(c) Flowering times of wild-type (Col-0), siz1-2, FRI siz1-2 and FRI plants were estimated under long days (LD).
(d) Relative FLC mRNA levels in wild-type (Col-0), siz1-2, FRI siz1-2 and FRI seedlings were determined by quantitative PCR. RNA was isolated from 10-day-old seedlings grown under long days (LD). Data illustrated in (a) and (c) are means ± SE of 15–20 plants per analysis.
Figure4.
SIZ1 represses SOC1 expression but activates MAF4 expression.
(a) FLM/MAF1, SVP, MAF2, MAF3, MAF4, MAF5 and SOC1 mRNA levels in wild-type (Col-0) and siz1-2 plants were determined by RT-PCR. RNA was isolated from 14-day-old seedlings grown under long days (LD) or short days (SD). TUBULIN was used as a control for loading.
(b, c) Flowering time of wild-type (Col-0), siz1-2, flc-3 siz1-2, flc-3, soc1-2 siz1-2 and soc1-2 plants were estimated under LD and SD. Data are means ± SE of 15–20 plants per analysis.
To test whether reduced FLC expression levels contribute to early flowering of siz1, a double mutant was made containing siz1-2 and an FLC null allele (flc-3) (Michaels and Amasino, 1999). Under LD, flc-3 flowered slightly earlier than wild-type plants (Figure 4b). The flowering time of flc-3 siz1-2 was earlier than that of flc-3 or siz1-2 plants (Figure 4b). Consistent with a previous report (Michaels and Amasino, 2001), the flowering time of flc-3 was slightly earlier than in wild-type plants under SD (Figure 4b). Double-mutant flc-3 siz1-2 plants flowered at the same time as siz1-2 plants, but flowered much later than flc-3 plants under SD (Figure 4b). These results indicate that reduced FLC expression does not contribute to the early flowering of siz1 plants. Thus, in the Columbia background, SIZ1 regulates the transition to flowering mainly through an FLC-independent pathway(s).
SOC1 transcript levels were greater in siz1 compared with wild-type plants under both LD and SD (Figure 4a). Flowering time of soc1-2 siz1-2 was similar to that of soc1-2 plants under LD, suggesting that the early flowering of siz1 under LD is mainly dependent on elevated SOC1 expression levels (Figure 4c). The flowering time of soc1-2 siz1-2 was intermediate to that of siz1-2 and soc1-2 under SD. It appears that the early flowering of siz1 under SD was only partially dependent on elevated SOC1 expression (Figure 4c).
The interaction of SIZ1 with FRI and the autonomous pathway genes
Although reduction of FLC transcript abundance by the siz1 mutation does not affect flowering time in the Columbia background (i.e. fri null, basal FLC expression), SIZ1 appears to be required for full FLC expression in late-flowering autonomous pathway mutants and a FRI background. Double mutants were made between siz1-2 and dysfunctional alleles of late-flowering autonomous pathway mutants, such as luminidependens (ld-1) and fld-6 (Lee et al., 1994b; Sanda and Amasino, 1996). Also, an active FRI allele was introduced into siz1-2 plants by crossing with FRI-Col (Michaels and Amasino, 1999; Sheldon et al., 1999). As established in previous studies (Lee et al., 1994b; Michaels and Amasino, 1999), ld-1 and FRI-Col plants flowered substantially later than Col-0 or siz1-2 plants, which results from greater FLC transcript abundance relative to wild-type plants (Figure 5). The siz1-2 mutation partially suppressed the late-flowering phenotype of ld-1 and FRI-Col plants (Figure 5a,c). Consistent with the flowering time phenotype, FLC expression levels were reduced in ld-1 siz1-2 and FRI siz1-2 compared with that of the late-flowering parental plants ld-1 and FRI-Col, respectively (Figure 5b,d). Note that exogenous SA treatment does not accelerate the flowering time of autonomous pathway mutants (Martinez et al., 2004). Together, these results suggest that SIZ1 activates FLC expression, possibly through an SA-independent pathway, which is required for full FLC activation by ld-1 and FRI. However, interestingly, fld-6 and fld-6 siz1-2 plants did not flower after producing more than 100 rosette leaves under SD (Figure 5a). Moreover, increased FLC expression in fld-6 was not suppressed by the siz1 mutation (Figure 5b). These results suggest that FLD may be required for SIZ1 to promote FLC expression. No difference in FLD transcript abundance was observed between wild-type and siz1 plants (data not shown), indicating that SIZ1 does not regulate expression of FLD.
SIZ1 facilitates SUMO1 modification of FLD
Three potential sumoylation motifs (IK287VE, PK693AD and IK770AE) in FLD were identified by SUMOplot (http://www.abgent.com/tool/sumoplot) analyses. Thus, FLD could be a potential SUMO target protein (Schmidt and Muller, 2003). To test this possibility, human influenza hemagglutinin (HA)-tagged FLD (HA:FLD) was transiently expressed in wild-type or siz1-2 protoplasts (Jin et al., 2001). Proteins from wild-type and siz1-2 protoplasts were separated by SDS-PAGE gel (Figure 6a, left panel). Interestingly, a single anti-HA reactive protein was detected after expressing HA:FLD in wild-type protoplasts, even though two anti-HA reactive proteins bands are predicted (sumoylated and unsumoylated) (Figure 6a, lane 1). Presumably SUMO modification of FLD in planta is very efficient, and only the sumoylated protein is observed. The molecular mass of AtSUMO1 is 12 kDa. Thus, it is expected that sumoylated FLD should have a molecular mass that is 12 kDa larger than unsumoylated FLD. However, the HA:FLD band resulting from wild type (Figure 6a, lane 1) appeared to be only 3.5–4 kDa larger than the corresponding band from siz1-2 protoplasts (Figure 6a, lane 3). There are probably two possibilities that cause this aberrant protein mobility on the SDS-PAGE. First, SUMO modification of proteins induces conformational change in some proteins (Steinacher and Schar, 2005). The sumoylated FLD may undergo a conformational change that may be insensitive to SDS treatment. Second, sumoylation and other post-translational modification processes (e.g. phosphorylation) that coordinately regulate protein function, etc. are often interdependent (Vu et al., 2007). Perhaps the smaller molecular mass differences between sumoylated and unsumoylated FLD are the consequence of additional post-translational processes.
Figure6.
SIZ1 mediates SUMO modification of FLD. (a) HA:FLD or HA:FLDK3R (K287R, K693R and K770R) and T7:AtSUMO1 translational fusions were co-expressed in wild-type (Col-0) or siz1-2 protoplasts (Jin et al., 2001). T7:AtSUMO1 and HA:FLD or HA:FLDK3R were co-immunoprecipitated (IP) from extracts, and were then detected on the western blot (WB) with anti-HA. No IP is protoplast lysate before IP. The star indicates the position of sumoylated FLD proteins (right panel). HA:FLD and HA:FLDK3R transient expression levels were similar in siz1 and wild-type (Col-0) plants (left, No IP panel); vector, total protein extract from protoplasts transformed with the empty vector. (b) A WB with anti-T7 was used to determine the expression level of T7:AtSUMO1 in the No-IP samples from (a). Free T7:AtSUMO1 transient expression levels were similar in siz1 and wild-type (Col-0) plants (lower arrow). T7:AtSUMO1 conjugates (upper arrow) were nearly undetectable in siz1-2 (lanes 11 and 12) compared with wild-type (Col-0) (lanes 9 and 10) protoplasts. No IP and vector are as in (a).
To determine if the slightly larger molecular mass of the protein in wild type is indeed the result of sumoylation, HA:FLD and T7:AtSUMO1 were transiently co-expressed in wild-type and siz1-2 protoplasts (Jin et al., 2001). Total protein was isolated from protoplasts under denaturing conditions to minimize desumoylation of the conjugated peptides by SUMO proteases during extraction. Protoplast lysates were then diluted with immunoprecipitation buffer, and T7:AtSUMO1 was immunoprecipitated with anti-T7. A single peptide band was detected with anti-HA from immunoprecipitated proteins of wild-type protoplasts, indicating that HA:FLD physically interacts with T7:AtSUMO1 (Figure 6a, lane 5). However, the anti-HA reactive band was almost non-detectable on the immunoblot of proteins from siz1 protoplasts (Figure 6a, lane 7). These results indicate that SIZ1 facilitates SUMO1 conjugation to FLD.
K to R mutations in sumoylation motifs block SUMO conjugation to protein substrates (Hilgarth et al., 2004; Miura et al., 2005). Consequently, K residues in the three predicted sumoylation motifs of FLD were substituted with R residues (HA:FLDK3R), and Co-IP analysis was performed as described above (Figure 6a). T7:AtSUMO1 conjugation to HA:FLDK3R was not detected on the immunoblot of protein isolated from wild-type or siz1-2 protoplasts (Figure 6a, compare lanes 6 and 8). These results indicate that the K residues in one or more of the three sumoylation motifs are necessary for SUMO1 modification of FLD, and that the capacity for AtSUMO1 conjugation to FLD is impaired in siz1.
SIZ1-mediated SUMO modification of FLD represses H4 deacetylation of FLC chromatin
Effects of FLD and FLDK3R (constitutively unsumoylated) on FLC expression were evaluated to determine if SIZ1-mediated sumoylation of FLD alters its activity (Figure 7a). HA:FLD, HA:FLDK3R or empty vector were transiently expressed in fld-6 protoplasts that were isolated from pre-flowering plants grown under SD. After a 40-h incubation, protoplasts were harvested and the FLC mRNA level was determined by quantitative real-time PCR (Figure 7a). Transient, but equivalent, expression in protoplasts of HA:FLDK3R (unsumoylated) reduced FLC expression to a greater extent than expression of HA:FLD (sumoylated) relative to vector control (Figure 7a,c).
Figure7.
SIZ1-mediated SUMO modification of FLD represses deacetylation of histone H4 in FLC chromatin. (a) Relative FLC mRNA levels were determined by quantitative PCR in fld-6 protoplasts expressing vector, HA:FLD or HA:FLDK3R. Data are means ± SD (n = 4).
(b) The acetylation state of H4 in FLC chromatin was assessed by chromatin immunoprecipitation (ChIP) analysis in fld-6 protoplasts expressing FLD or FLDK3R. Input is fld-6 chromatin before immunoprecipitation that was isolated from protoplasts transformed with empty vector; No AB and ACTIN are as in (b). The fold enrichment in H4 acetylation of fld-6 protoplasts expressing FLD or FLDK3R overexpressing the empty vector is shown. (c) HA:FLD and HA:FLDK3R were expressed equally in fld-6 protoplasts in this experiment.
FLD mutations, which disturb protein function, cause FLC transcript accumulation and late flowering that is linked to hyperacetylation of histone 4 (H4) in chromatin associated with the first intron of FLC (He et al., 2003). Consequently, the H4 acetylation status of FLC chromatin in the protoplast samples used in the experiments presented in Figure 7(a) were evaluated using a chromatin immunoprecipitation (ChIP) assay (Figure 7b). Consistent with the FLC mRNA level seen in Figure 7(a), H4 in FLC chromatin was less acetylated in protoplasts expressing HA:FLDK3R compared with those expressing HA:FLD (Figure 7b). These results suggest that SIZ1-mediated SUMO modification of FLD inhibits its ability to repress FLC expression by reducing acetylation of H4 in FLC chromatin. This SUMO-mediated regulatory mechanism appears to be required for full FLC activation by FRI.
Discussion
Mechanisms for SIZ1 regulation of flowering in the Columbia background
Under LD, the flowering time of soc1-2 siz1-2 plants was simililar to that of soc1-2, suggesting that the slightly earlier flowering of siz1 under LD is mainly the result of elevated SOC1 expression (Figure 4). The photoperiodic pathway activates SOC1 expression through FT, which is repressed by FLC (Searle et al., 2006; Wigge et al., 2005). However, expression of FT and FLC was nearly unaffected by siz1 under LD (data not shown and Figure 5d, respectively). Thus, increased SOC1 expression in siz1 plants is not the result of elevated expression of FT or of reduced expression of FLC. If the activation of SOC1 expression is the result of increased SA levels caused by siz1, then the flowering time of nahG siz1-2 plants should be similar to that of nahG plants under LD. However, the flowering time of nahG siz1-2 plants was similar to that of siz1-2 under LD (Figure 3b), suggesting that SA does not activate SOC1 expression, which is consistent with a previous report that the UV-C light-induced accumulation of SA does not affect SOC1 expression (Martinez et al., 2004). Therefore, SIZ1 represses SOC1 expression through FT- and FLC-independent pathways, and through an unknown SA-independent pathway. The siz1 mutation substaintially reduced the expression of the floral repressor MAF4, which may also contribute to early flowering of siz1 under LD.
Under SD, the substaintial early flowering phenotype of siz1 is mainly the result of elevated SA levels (i.e. the flowering time of nahG siz1-2 was similar to that of nahG plants). Under SD, SIZ1 function on flowering time also showed little dependence on FLC (i.e. the flowering time of flc-3 siz1-2 was similar to that of siz1-2 plants), indicating that under SD, SA promotes transition to flowering mainly through FLC-independent pathway(s). These results further indicate that SA accelerates flowering through pathways that are independent of the vernalization and the autonomous pathways, as these two pathways promote transition to flowering through repression of FLC expression. SA could facilitate transition to flowering by shortening the circadian period (Mizoguchi et al., 2002). However, rhythmic expression of CCA1 and CCR2 were not altered in siz1 plants (Figure S1), suggesting that SA does not regulate the circadian clock. Moreover, GA-dependent floral promotion is operative in siz1 plants that contain a high level of SA (Figure S2). Thus, it is likely that SA accelerates flowering through pathways that are independent of photoperiodic- and vernalization-dependent pathways, and the autonomous and GA-dependent pathways. Despite the major role of SA in the early flowering of siz1 under SD, the flowering time of nahG siz1-2 was slightly earlier than that of nahG plants under SD (Figure 3b), indicating that siz1 also accelerates flowering through SA-independent machanisms under SD. This may include activation of SOC1 and/or repression of MAF4 expression. Although SIZ1 is now strongly implicated in SA accumulation, and in its subsequent affect on flowering time under SD, the sumoylation targets of SIZ1 that affect SA accumulation remain to be discovered. SA-mediated flowering control has escaped major attention, perhaps because it is dependent on the interaction of particular environmental and genetic background conditions. However, the other major flowering signal pathways are also similarly affected, and the roles of SA and sumoylation in flowering time control may prove as important as other major signal systems.
Possible mechanism for FLC activation by SIZ1
Interestingly, activated FLC expression in fld-6 was not affected by siz1, wheareas siz1 caused partial suppression of FLC expression in FRI and ld-1 plants. The first interpretation of these results is that SIZ1 promotes FLC expression by inhibiting FLD activity, which is required for the full activation of FLC expression in FRI and ld-1 plants. Alternatively, SIZ1 and FLD may function in independent pathways, and activation of FLC expression by fld-6 could overcome repression of FLC expression caused by siz1. However, we also found that SIZ1 facilitates sumoylation of FLD, which represses FLD activity. This strongly supports the first interpretation. In addition, partial suppression of FLC expression in FRI and ld-1 plants by siz1 is difficult to explain by the alternative interpretation, as FRI or ld-1 also causes strong FLC activation. Therefore, it is likely that the partial suppression of FLC expression in an FRI and ld-1 background by siz1 mutation is, at least in part, caused by the inhibition of FLD activity by SIZ1-mediated SUMO modification. However, we must consider the possibility that SIZ1 also activates FLC expression through an FLD-independent mechanism.
Possible mechanisms by which SUMO modification of FLD affects HDAC activity
We have found that sumoylation/desumoylation of FLD controls histone acetylation/deacetylation, but the mechanism(s) by which SUMO modification of FLD affects HDAC activity remains unelucidated. The FLD homolog, KIAA0601/LSD1, has lysine-specific demethylase activity that is associated with numerous co-repressor complexes, such as CoREST, BHC80 and HDAC in humans (Humphrey et al., 2001; Shi et al., 2003, 2004). LSD1 and HDAC1 function cooperatively in a co-repressor complex (Lee et al., 2006). CoREST induces LSD1 demethylation activity, but BHC80 negatively regulates LSD1 demethylation activity (Shi et al., 2005). Interestingly, LSD1 undergoes some unknown post-translational modification (You et al., 2001). LSD1 contains three potential sumoylation sites, which are likely targets for SUMO modification. Although the biochemical function of FLD remains unknown, it is possible that the activity and regulatory mechanisms of FLD are similar to that of LSD1. It is possible that SUMO modification of FLD enhances and/or inhibits interaction with an FLD repressor and/or activator, respectively, and consequently inhibits HDAC function in the repressor complex. Identification and characterization of FLD interacting partners will help us to understand mechanisms by which AtSIZ1-mediated sumoylation affects FLD activity, and that understanding will have broad scientific relevance to both plant and animal systems.
Experimental procedures
Plant materials
Genotypes used in all experiments were in the Columbia genetic background. The siz1-2 (SALK_065397) and siz1-3 (SALK_034008) lines were obtained from ABRC at Ohio State University (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm; Miura et al., 2005). Early and late flowering mutants, flc-3, ft-1, soc1-2, gi-2, co-1, ld-1 and FRI-SF2 in the Columbia background (FRI-Col) were described previously (Fowler et al., 1999; Kardailsky et al., 1999; Kobayashi et al., 1999; Koornneef et al., 1991; Lee et al., 1994b, 2000; Michaels and Amasino, 1999). fld-6 (SAIL_642 C05) was isolated from T-DNA mutant SAIL lines, which were kindly provided by Dr R.M. Amasino (University of Wisconsin, http://www.wisc.edu). Homozygous double mutants were obtained by crossing various flowering-time mutants with siz1-2. The presence of siz1-2 and fld-6 mutations were analyzed by diagnostic PCR analysis according to the SALK T-DNA verification protocol (http://signal.salk.edu), and the presence of the FRI-SF2, ld-1, flc-3, gi-2, co-1, ft-1 and soc1-2 mutations was analyzed according to a previous report (Lee et al., 2000; Moon et al., 2005).
Growth conditions
To break seed dormancy, seeds were stratified on soil for 4 days at 4°C before transfer to normal growth conditions. Plants were grown at 23°C in a greenhouse under LD (16-h light/8-h dark), whereas for SD (8-h light/16-h dark), plants were grown at 22°C under fluorescent lights (100 μmol m−2 sec−1) in growth chambers, which were equipped with seven ALTO PLUS T8 fluorescent lamps (#F32T8/TL735/PLUS/ALTO; Philips, http://www.philips.com) and one PLANT & AQUARIUM 40W lamp (GE; http://www.ge.com). For exogenous GA treatment, 100 μM GA3 was sprayed twice onto 7- or 14-day-old seedlings grown under either LD or SD. The flowering time is estimated based on the number of rosette leaves formed by the primary shoot apical meristem prior to flowering under LD and SD, as described above. At least 15–20 plants were used to determine the flowering time of each genotype.
RT-PCR
Total RNA was isolated using the PureLink Micro-to-Midi Total RNA Purification System (#12183-018; Invitrogen, http://www.invitrogen.com) according to the manufacturer’s protocol. A 200-ng sample of RNA was used as the template for first-strand cDNA synthesis with the ThermoScript RT-PCR System (#11146-016; Invitrogen) and an oligo (dT21) primer. Specific gene expression levels were analyzed by semiquantitative RT-PCR or real-time PCR (Miura et al., 2005). Primer sequences for gene amplifications are listed in Table S2.
In vivo sumoylation assay
In vivo sumoylation was assayed as described previously by Hilgarth et al. (2004). HA:FLD or HA:FLDK3R was transiently co-expressed with T7:AtSUMO1 in protoplasts prepared from 14-day-old wild-type and siz1-2 seedlings by polyethylene glycol-mediated transformation (Jin et al., 2001). After 40-h incubation, immunoprecipitation was performed using T7 tag monoclonal antibody (#69522-3; Novagen, http://www.emdbiosciences.com) with protein-A-sepharose CL-4B (#17-0963-03; Amersham, http://www.amersham.com). Immunoprecipitated proteins were released in 2× SDS sample buffer, separated by SDS- PAGE, and detected by western blotting using anti-HA monoclonal antibodies (#sc-7392; Santa Cruz Biotechnology, Inc., http://www.scbt.com) (Jin et al., 2001).
Chromatin immunoprecipitation analysis
The chromatin immunoprecipitation experiments were performed as described previously (He et al., 2003). The primer pair CH1/H12 and JP1595/JP1596 were used to amplify the first intron region of FLC chromatin and ACTIN 2/7, respectively (see Table S2 for primer sequence) (He et al., 2003). SD-grown 20-day-old wild-type, siz1-2 and fld-6 plants and anti-acetylated H4 antibody were used for ChIP analyses. To check the activity of the sumoylation-deficient mutant, FLDK3R, HA:FLD or HA:FLDK3R were transiently expressed in SD-grown 20-day-old fld-6 protoplasts. After 40 h of incubation, ChIP analysis was performed to determine the acetylation status of H4 in FLC chromatin as described above. The fold enrichment in H4 acetylation was calculated as follows: FLC was first normalized to ACTIN in each sample, and these values were normalized against their respective wild type or vector controls.
Rhythm analysis
Rhythm analysis was performed as described by Kim et al. (2005).
Acknowledgments
We are grateful to Dr Rick M. Amasino for providing FRI-Col and fld-6 seeds, and for critical advice. This work was supported by the National Science Foundation Plant Genome Award (DBI–98–13360), by the Basic Science Project of Korea Science and Engineering Foundation (grant no. RO1–2006–000–10123–0), by the Environmental Biotechnology National Core Research Center Project of Korea Science and Engineering Foundation (grant no. R15–2003–012–01002–00), by the Biogreen 21 Program of the Rural Development Administration, Korea (grant no. 20070301034030), and by the Korea Research Foundation (MOEHRD, KRF-2006-352-F00008). This work is Purdue University Agricultural Research Program Paper 2007-18230.
Supplementary material
The following supplementary material is available for this article online:
Lesions in SIZ1 do not impair clock-controlled gene expression.
Exogenous gibberellin (GA) treatment accelerates flowering in both wild-type and siz1 plants.
Period of circadian rhythms estimation in different light conditions at 21°C.
Primers for RT-PCR, real-time PCR or chromatin immunoprecipitation (ChIP) analysis.
Primers for subcloning.
Experimental procedures: plasmid construction.
This material is available as part of the online article from http://www.blackwell-synergy.com
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ballare CL. Keeping up with the neighbours: phytochrome sensing and other signaling mechanisms. Trends Plant Sci. 1999;4:201. doi: 10.1016/s1360-1385(99)01408-9. [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Mukherjee S, Lois LM, Orth K. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem. J. 2006;398:521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland CF, Ben-Tal Y. Influence of giving salicylic acid for different time periods on flowering and growth in the long-day plant Lemna gibba G3. Plant Physiol. 1982;70:287–290. doi: 10.1104/pp.70.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Gadisseur I, Silvestre G, Jacqmard A, Bernier G. Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J. 1996;9:947–952. doi: 10.1046/j.1365-313x.1996.9060947.x. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, et al. A central role of salicylic acid in plant resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Downes B, Vierstra RD. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem. Soc. Trans. 2005;33:393–399. doi: 10.1042/BST0330393. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl Acad. Sci. USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- Hilgarth RS, Murphy LA, O’Connor CM, Clark JA, Park-Sarge OK, Sarge KD. Identification of Xenopus heat shock transcription factor-2: conserved role of sumoylation in regulating deoxyribonucleic acid-binding activity of heat shock transcription factor-2 proteins. Cell Stress Chaperones. 2004;9:214–220. doi: 10.1379/CSC-8R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kim WY, Hicks KA, Somers DE. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 2005;139:1557–1569. doi: 10.1104/pp.105.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Simon AE. Environmental and genetic effects on circadian clock-regulated gene expression in Arabidopsis. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- Langridge J. Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature. 1957;180:36–37. [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell. 1994a;6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994b;6:903–909. [Google Scholar]
- Lee H, Suh S, Park E, Cho E, Ahn JH, Kim S, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Martinez C, Pons E, Prats G, Leon J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 2004;37:209–217. doi: 10.1046/j.1365-313x.2003.01954.x. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavass E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl Acad. Sci. USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun D-J, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2004;35:613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005;46:292–299. doi: 10.1093/pcp/pci024. [DOI] [PubMed] [Google Scholar]
- Murtas G, Reeves PH, Fu Y-F, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A. SUMO conjugation in plants. Planta. 2004;220:1–8. doi: 10.1007/s00425-004-1370-y. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Murtas G, Dash S, Coupland G. early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. 2002;129:5349–5361. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- Sanda SL, Amasino RM. Ecotype-specific expression of a flowering mutant phenotype in Arabidopsis thaliana. Plant Physiol. 1996;111:641–644. doi: 10.1104/pp.111.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Amasino RM. Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta. 2007;1769:269–275. doi: 10.1016/j.bbaexp.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J-S, Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y-J, Matson C, Lan F, Iwase S, Bada T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- Vu EH, Kraus RJ, Mertz JE. Phosphorylation-dependent sumoylation of estrogen-related receptor alpha1. Biochemistry. 2007;46:9795–9804. doi: 10.1021/bi700316g. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992;100:403–408. doi: 10.1104/pp.100.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl Acad. Sci. USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lesions in SIZ1 do not impair clock-controlled gene expression.
Exogenous gibberellin (GA) treatment accelerates flowering in both wild-type and siz1 plants.
Period of circadian rhythms estimation in different light conditions at 21°C.
Primers for RT-PCR, real-time PCR or chromatin immunoprecipitation (ChIP) analysis.
Primers for subcloning.
Experimental procedures: plasmid construction.