Abstract
RABEX-5 and other exchange factors with VPS9 domains regulate endocytic trafficking through activation of the Rab family GTPases RAB5, RAB21 and RAB22. Here we report the crystal structure of the RABEX-5 catalytic core in complex with nucleotide-free RAB21, a key intermediate in the exchange reaction pathway. The structure reveals how VPS9 domain exchange factors recognize Rab GTPase substrates, accelerate GDP release and stabilize the nucleotide-free conformation. We further identify an autoinhibitory element in a predicted amphipathic helix located near the C terminus of the VPS9 domain. The autoinhibitory element overlaps with the binding site for the multivalent effector RABAPTIN-5 and potently suppresses the exchange activity of RABEX-5. Autoinhibition can be partially reversed by mutation of conserved residues on the nonpolar face of the predicted amphipathic helix or by assembly of the complex with RABAPTIN-5.
The essential regulatory function of Rab GTPases in membrane trafficking, organelle biogenesis and cell growth depends on their ability to cycle between active (GTP-bound) and inactive (GDP-bound) conformations1,2. In the active conformation, Rab GTPases mediate selective interactions with effectors, including cargo-sorting complexes, motor proteins, tethering factors and lipid kinases, as well as proteins implicated in signal transduction, cytoskeletal dynamics and cytokinesis. The intrinsic rates of activation by nucleotide exchange and deactivation by GTP hydrolysis are slow compared with the timescale of the relevant cellular processes. Consequently, the interconversion between states is tightly regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs).
Null and temperature-sensitive alleles of the yeast VPS9 gene result in mis-sorting of proteins targeted to the vacuole3. Yeast Vps9p has GEF activity for the yeast RAB5 homolog Vps21p (also called Ypt51p)4 and possesses a CUE domain on the C-terminal side of the GEF domain. The CUE domain binds monoubiquitin and promotes autoubiquitination of Vps9p5,6. At least seven mammalian proteins contain VPS9 domains, including RABEX-5 (ref. 7), the RIN family of RAS effectors8–10, the ALSIN protein encoded by the amyotrophic lateral sclerosis type 2 (ALS2) gene11, the homolog of the Caenorhabditis elegans RME-6 protein identified in a screen for receptor-mediated endocytosis12 and the VPS9 domain–ankyrin repeat protein, VARP13.
RABEX-5 was originally identified by copurification of an endogenous complex with RABAPTIN-5 (ref. 7). The RABEX-5–RABAPTIN-5 complex has RAB5 GEF activity and cooperates with other factors to promote endosome fusion7,14–16. Rabex-5−/− mice show a substantial increase in perinatal mortality, severe skin inflammation, increased numbers of skin mast cells and elevated serum levels of IgE and histamine17. Prolonged signaling and enhanced degranulation after activation are evident in cultured mast cells derived from the Rabex-5−/− mice, indicating that Rabex-5 negatively regulates mast cell activation17. Consistent with a role in endocytosis, c-Kit internalization is delayed in cultured Rabex-5−/− mast cells and can be restored by lentiviral expression of Rabex-5 (ref. 18). RABEX-5 also contains two distinct ubiquitin binding sites corresponding to an A20 zinc finger, which promotes intramolecular ubiquitination, and an inverted UIM motif in an adjacent helix19–21. RABEX-5 binds the ubiquitinated epidermal growth factor (EGF) receptor (EGFR), is recruited to the plasma membrane following stimulation with EGF and colocalizes with the EGFR at several stages within the endocytic pathway, suggesting that RABEX-5 may also function in EGFR downregulation21. RABAPTIN-5 is a multivalent effector with distinct binding sites for active RAB4 and RAB5 (ref. 22) as well as the GAE and GAT domains of GGA proteins23,24.
The catalytic core of RABEX-5 consists of a helical bundle (HB)-VPS9 domain tandem (HB-VPS9 tandem) with potent GEF activity for RAB5 and RAB21, weak activity for RAB22 and no detectable activity for other Rab GTPases25. RAB5, RAB21 and RAB22 comprise a subfamily of ubiquitously expressed Rab GTPases that have overlapping subcellular distributions in the endosomal system26,27. RAB5 is well established as a master regulator of early endosome biogenesis, sorting and fusion2. Although less well characterized, RAB21 and RAB22 have recently been implicated in endosomal trafficking28–30. The low nucleotide affinity (“dominant negative”) mutant of RAB21 blocks transferrin and EGFR internalization and interferes with EGFR sorting to late endosomes and lysosomes29. RAB21 also regulates phagocytosis in Dictyostelium discoideum31 and regulates integrin-containing focal adhesions as well as the adhesion and migration of breast and prostrate cancer cell lines32.
Despite the critical function of Rab GTPases in membrane trafficking and the obligatory requirement of GEFs for activation, little is known about the structural bases of Rab GTPase recognition and activation by Rab GEFs or the mechanisms for allosteric regulation of Rab GEF activity. To investigate the structural basis for selective activation of Rab GTPases by VPS9 domain GEFs, we determined the crystal structure of the HB-VPS9 tandem of human RABEX-5 in complex with the nucleotide-free form of human RAB21. The structure reveals how VPS9 domains recognize the GDP-bound form of RAB5-subfamily GTPases, promote GDP release and stabilize the high-energy nucleotide-free intermediate. To gain insight into the mechanism for allosteric regulation of RABEX-5 GEF activity by RABAPTIN-5 (ref. 15), we identified an autoinhibitory element that potently suppresses the GEF activity of RABEX-5, found that the autoinhibitory element overlaps with the binding site for RABAPTIN-5, and determined that autoinhibition can be partially reversed by assembly of the complex with human RABAPTIN-5.
Results
Structure of the nucleotide-free RABEX-5–RAB21 complex
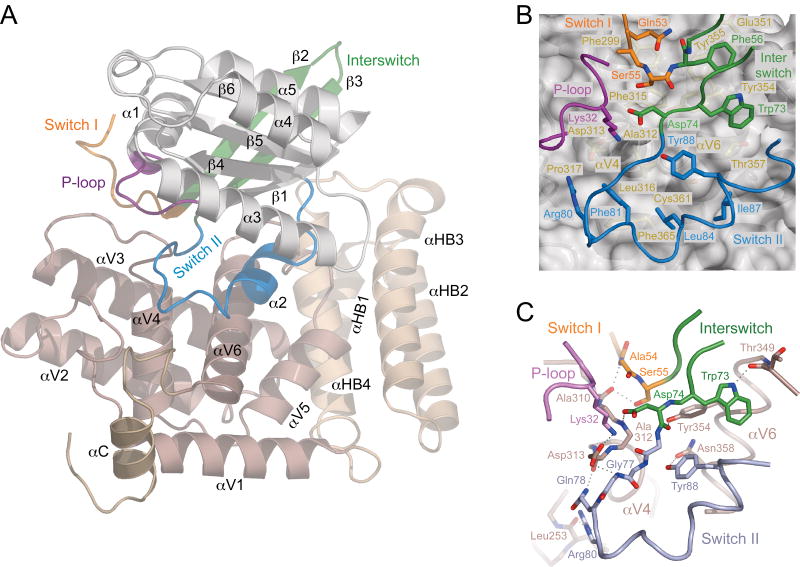
To understand how RABEX-5 and other VPS9 domain GEFs selectively catalyze GDP-GTP exchange for RAB5-subfamily GTPases, we determined the crystal structure of the RABEX-5 catalytic core in complex with nucleotide-free RAB21 (Supplementary Fig. 1 online). The RABEX-5 catalytic core consists of a helical bundle, HB (comprising helices αH1–αH4), and a VPS9 domain (helices αV1–αV6). RAB21 has a canonical small GTPase fold with a central sheet (β1–β6) surrounded by helices (α1–α5). The VPS9 domain engages the P-loop, switch and interswitch regions of RAB21 in a substantial, 2,400 Å2 interface centered on a ‘V’-shaped arrangement of the αV4 and αV6 helices (Fig. 1a). Two invariant aromatic residues (Phe56 and Trp73) anchor the interswitch (β2-β3) hairpin in adjacent nonpolar pockets (Fig. 1b) lined by conserved residues from the N terminus of αV6 (Glu351, Tyr354, Tyr355, Thr357 and backbone of 349–351). Preceding Phe56, switch I residues 51-Thr-Leu-Gln-Ala-Ser-55 are channeled through a notch between the C terminus of αV3 and the αV3-αV4 loop. Here, Gln53 packs against Phe299 from αV3 while Ser55 occupies a nonpolar pocket bounded by the backbone of residues 310 and 311 in the αV3-αV4 loop and the side chains of Phe299 from αV3, Phe315 from αV4 and Tyr354 from αV6. In the switch II region, the conserved Phe81 wedges into a nonpolar crevice between Leu316 and Pro317 from αV4 and Phe365 from αV6 while Leu84, Ile87 and Tyr88 pack against Tyr354, Thr357, Cys361 and Phe365 from αV6.
Figure 1.
Structure of the RABEX-5 HB-VPS9 tandem in complex with nucleotide-free RAB21. (a) Ribbon representation. The catalytic core of RABEX-5 consists of a helical bundle (light brown; αHB1–αHB4), a VPS9 domain (brown; αV1–αV6) and a C-terminal helix (dark brown; αC). RAB21 (α1–α5 and β1–β6) is depicted in gray with the P-loop, switch and interswitch regions colored as indicated. (b) Docking of nonpolar residues in the switch and interswitch regions of Rab21 in the hydrophobic groove between the αV4 and αV6 helices of the VPS9 domain. RABEX-5 is depicted as a gray ribbon with yellow side chains underneath a semitransparent surface. RAB21 is depicted as a tube with side chains. (c) Network of intermolecular polar interactions at the RAB21–VPS9 domain interface. Dotted lines represent hydrogen bonds.
The interswitch, switch and P-loop regions are further secured through a network of polar contacts (Fig. 1c). In the interswitch region, the backbone carbonyl and side chain carboxylate of the invariant Asp74 from the DxxG motif participate in hydrogen bonds with the side chain hydroxyl of Tyr354 from αV6 and backbone NH of Ala312 from the αV3-αV4 loop. In the switch I region, residues 53-Gln-Ala-Ser-55 are pinned down through a pair of hydrogen bonds donated by the backbone NH of Ala54 and side chain hydroxyl of Ser55 to the backbone carbonyl of Ala310 from the αV3-αV4 loop. In the switch II region, the side chains of the invariant Arg80 and conserved Tyr88 mediate hydrogen bonds with the backbone carbonyl of Leu253 from the αV1-αV2 loop and side chain carboxamide of Gln358 from αV4. Finally, the only strictly invariant residue in the VPS9 domain, Asp313, mediates a bipartite interaction with the invariant Lys32 from the P-loop GxxxxGK(S/T) motif and the backbone NH group of the invariant Gly77 from the switch II DxxG motif, thus mimicking canonical interactions with the γ phosphate of GTP.
Conformational changes accompanying GDP-GTP exchange
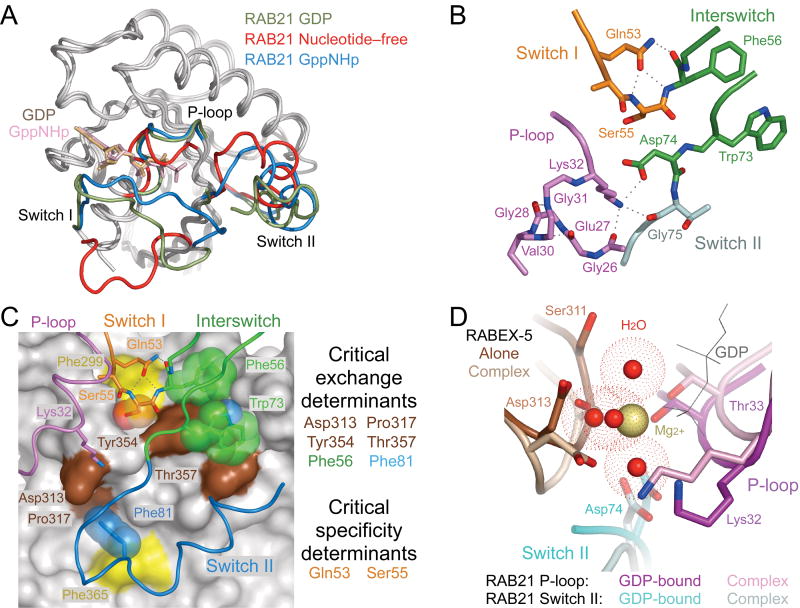
Comparison with nucleotide-bound structures of RAB21 reveals dramatic differences in the conformation of the P-loop and switch regions (Fig. 2a). In the nucleotide-free complex, P-loop residues 26-GEGSVGKT-33 adopt a collapsed conformation stabilized by a network of intramolecular polar contacts, including backbone hydrogen bonds from residues Val30 and Gly31 to residues Glu27 and Gly28 (Fig. 2b). The invariant Lys32, which contacts the β and γ phosphates of bound nucleotides, flips into an extended configuration to mediate polar interactions with Gly26 in the P-loop, Asp74 in the interswitch region and Asp313 in the VPS9 domain. Similar intramolecular interactions between the P-loop lysine and DxxG aspartic acid occur in the nucleotide-free forms of the bacterial signal-recognition particle protein subunit Ffh and the eukaryotic elongation factor eEF-G33,34. The switch I region of RAB21 is propped in an open conformation stabilized by polar interactions with the αV3-αV4 loop and by the side chain carboxamide of Gln53, which participates in intramolecular hydrogen bonds with the backbone NH of Ser54 and backbone NH and carbonyl of Phe56. Following the α1 helix, switch I residues 43–46, which include the highly conserved Phe44 that packs against the guanine base in most nucleotide-bound GTPases, are completely disordered. The switch II region of RAB21, though mobile in both the GDP- and GTP-bound states35, is engaged in an ordered conformation distinct from that of GTP-bound RAB5. In the VPS9 domain, the most substantial differences are in the αV3-αV4 and αV5-αV6 loops (Supplementary Fig. 2 online), which have relatively high B-factors in the isolated structure but are stabilized in well-ordered conformations in the nucleotide-free complex.
Figure 2.
Conformational changes accompanying formation of the nucleotide-free complex. (a) Superposition of GDP-bound (PDB 1Z0I (PDB 1YZU) RAB21 with the nucleotide-free form of RAB21 from the complex with RABEX-5. (b) Stabilization of the nucleotide-free state by intramolecular polar interactions. Dotted lines represent hydrogen bonds. (c) Correlation with critical exchange and specificity determinants identified in an earlier mutational analysis25. VPS9 domain of RABEX-5 is depicted as gray surface with critical exchange determinants highlighted in brown. Two additional residues discussed in the text are highlighted in yellow. RAB21 is depicted as a tube. Side chains with or without semitransparent spheres are shown for critical exchange or specificity determinants as well as other residues discussed in the text. (d) Steric crowding in the Mg2+-binding site following superposition of GDP-bound RAB21 (PDB 1Z0I) with nucleotide-free RAB21 from the complex with RABEX-5. Water molecules coordinated by the Mg2+ ion in GDP-bound RAB21 are depicted as red spheres surrounded by dot surfaces indicating the van der Waals radii.
Correlation with mutational analyses
Mutational analyses have identified four highly conserved residues in the VPS9 domain (Asp313, Pro317, Tyr354 and Thr357) and two in RAB5 (corresponding to Phe56 and Phe81 in RAB21) as crucial determinants of exchange activity25. Asp313 contacts both the invariant P-loop lysine and switch II backbone, whereas Pro317, Tyr354 and Thr357 contribute to binding pockets for conserved aromatic residues in the interswitch (Phe56 and Trp73) and switch II (Phe81) regions (Fig. 2c). Although not included in the mutational analyses, other conserved residues such as Tyr355 and Phe365 in αV6 or Trp73 in the interswitch region are likely to make substantial contributions to the stability of the complex.
A key issue concerns how the RABEX-5 VPS9 domain achieves selective recognition of RAB5-subfamily GTPases and further discriminates RAB5 and RAB21 from RAB22. The most important determinant of RAB5-subfamily selectivity is a strict requirement for a small nonacidic residue preceding the invariant Phe56 in the switch I–interswitch T(I/V)GxxF motif. Whereas substituting the alanine in RAB5 with serine, as found in RAB21 and RAB22, has no effect, substitution with aspartic acid or glutamic acid, which are broadly conserved in nearly all Rab GTPases, results in a 200-fold reduction in catalytic efficiency25. The side chain of Ser55 is buried in a solvent-excluded pocket bounded by nonpolar residues with hydrogen-bonding potential on one side for the hydroxyl group of a serine residue (Fig. 1b,c). This pocket is just large enough to accommodate a serine side chain but too small for the larger, negatively charged side chains of aspartic and glutamic acid (Fig. 2c). The relatively low exchange activity for RAB22 reflects the cumulative effect of weak determinants in the interswitch and switch II regions25, involving residues that are located directly within or proximal to the interface with the VPS9 domain. The equivalent catalytic efficiencies for RAB5 and RAB21, on the other hand, reflect a gain-of-function substitution whereby Gln53 in RAB21 replaces the otherwise invariant glycine in the T(I/V)GxxF motif. Substitution of Gln53 in RAB21 with glycine reduces the catalytic efficiency by 50-fold, whereas the corresponding Gly→Gln substitutions in RAB5 and RAB22 have the opposite effect25. Gln53 packs against Phe299 in the VPS9 domain and stabilizes the open switch I conformation through polar interactions with the backbone of Ser55 and Phe56 (Fig. 2c).
Structural mechanism of nucleotide exchange
The catalytic properties of the RABEX-5 HB-VPS9 tandem can be explained within the framework of the general allosteric competition mechanism in which rapid equilibration to form an initial GDP-bound GTPase-GEF intermediate is followed by rate-limiting release of GDP to form the nucleotide-free intermediate36–38. Excess GTP drives the reverse reaction by competing with the GEF for binding to the nucleotide-free GTPase. The dynamic flexibility of the switch regions in GDP-bound Rab GTPase substrates facilitates formation of the initial GDP-bound intermediate. Superposition of GDP-bound RAB21 with nucleotide-free RAB21 in the complex reveals that a small reorientation would be necessary to alleviate steric conflicts in the Mg2+- and β phosphate–binding sites (Fig. 2d). These observations suggest that VPS9 domains initially accelerate GDP release through disruption of the Mg2+-binding site coupled with electrostatic repulsion between the β phosphate and the invariant Asp313, which further reduces the energetic barrier for formation of the high-energy nucleotide-free intermediate through a favorable electrostatic interaction with the P-loop lysine.
Comparison with Sec7–ARF1 and MSS4–Rab8 complexes
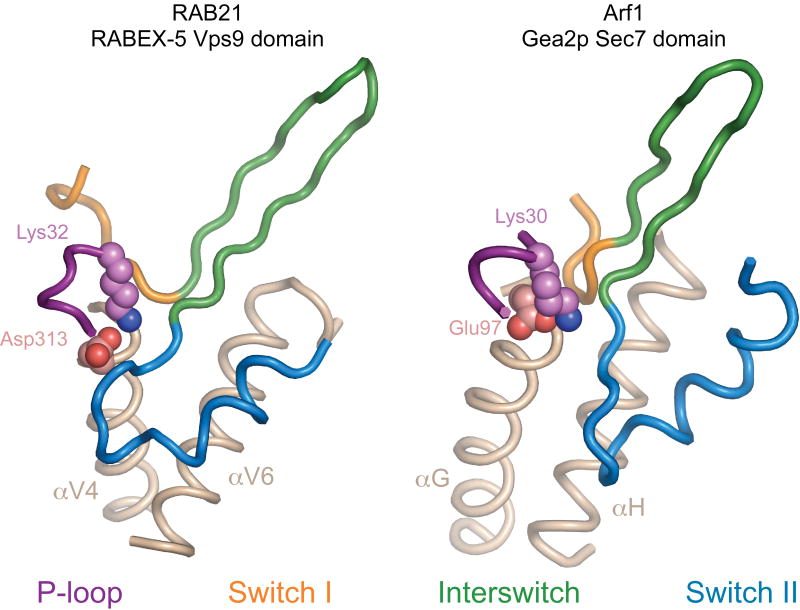
The overall mode of interaction in the VPS9 domain–RAB21 complex is strikingly similar to that in the Sec7 domain–ARF1 complexes (Fig. 3)39,40. Despite unrelated folds, the VPS9 and Sec7 domains engage the switch regions of their respective GTPases through a hydrophobic groove between apposing helices, prop switch I in an open nucleotide-accessible conformation and supply a crucial ‘glutamic acid finger’ (Sec7 domain) or ‘aspartic acid finger’ (VPS9 domain) to stabilize a collapsed P-loop through interaction with the invariant lysine.
Figure 3.
Comparison with the nucleotide-free Sec7–Arf GTPase complex. Left, nucleotide-free RAB21 in complex with the HB-VPS9 tandem of RABEX-5. Right, nucleotide-free ARF1 in complex with the Sec7 domain of Gea2 (ref. 39). The helices that comprise the core of the GTPase binding site in the VPS9 and Sec7 domains are depicted in brown. The invariant P-loop lysine (RAB21 and ARF1), aspartic acid finger (VPS9 domain) and glutamic acid finger (Sec7 domain) are depicted as spheres.
Nevertheless, the orientation of RAB21 is rotated and shifted relative to ARF1, allowing invariant aromatic residues in the interswitch region to dock in the hydrophobic groove while the aspartic acid finger contacts the switch II backbone in addition to the P-loop lysine. In contrast, there is little similarity to the recently characterized MSS4–Rab8 complex41. MSS4 binds tightly to the nucleotide-free form of RAB1, RAB3, RAB8 and RAB10, has weak exchange activity for both the GDP-bound and GTP-bound forms, and has been suggested to function in cells as a molecular chaperone for nucleotide-free and possibly misfolded Rab GTPases42–44. In the complex with Rab8, MSS4 engages residues in the switch I region and β2 strand of the interswitch region. Unlike the VPS9 domain, MSS4 does not contact the switch II region or stabilize a collapsed conformation of the P-loop but instead indirectly promotes the unfolding of the P-loop, α1 helix and other loop regions involved in nucleotide binding41.
Identification of an autoinhibitory element
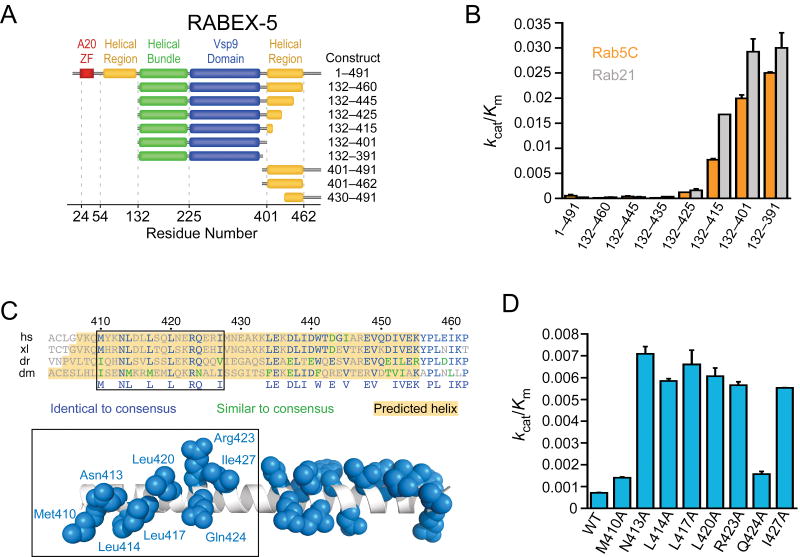
Full-length RABEX-5 has weak exchange activity comparable to that of Dss4p37 and approximately two orders of magnitude lower than that of the isolated HB-VPS9 tandem25. Following purification to homogeneity, we found that full-length RABEX-5 is uniformly monomeric, with high solubility and no evidence of instability. These observations are consistent with autoinhibition by elements outside the HB-VPS9 tandem. To identify the relevant autoinhibitory element(s), we analyzed the catalytic efficiency of N- and C-terminal truncation constructs (Fig. 4a). Notably, a construct (RABEX-5132–460) lacking both the ubiquitin-binding domains at the N terminus19–21 and a proline-rich region at the C terminus is fully autoinhibited, as is a construct (RABEX-5132–445) lacking another 15 residues from the C terminus of a predicted helical/coiled-coil region (Fig. 4b). Elimination of an additional 20 residues (RABEX-5132–425) results in slight activation. Shorter truncation constructs (RABEX-5132–415 and RABEX-5132–401) have high catalytic efficiencies equivalent to that of the isolated HB-VPS9 tandem (RABEX-5132–391). Thus, the exchange activity of RABEX-5 is robustly suppressed by an autoinhibitory element on the C-terminal side of the VPS9 domain that includes critical determinants within residues 415–425. This region is predicted to form an amphipathic helix with a conserved nonpolar surface (Fig. 4c). To determine whether this surface contributes to autoinhibition, the conserved residues were individually mutated to alanine in the context of RABEX-5132–460. Although without effect on the expression level, stability or solubility of the purified protein, the alanine substitutions substantially increased kcat/Km by two- to ten-fold (Fig. 4d).
Figure 4.
Identification of an autoinhibitory element in RABEX-5. (a) RABEX-5 truncation constructs. (b) Catalytic efficiency of RABEX-5 truncation constructs. Values are mean ± s.d. for n = 2. (c) Alignment of RABEX-5 sequences from diverse organisms (Hs, Homo sapiens; Xl, Xenopus laevis; Dr, Danio rerio; Dm, Drosophila melanogaster). Below the alignment is a hypothetical helical model for RABEX-5407–456 with conserved side chains depicted in blue (in the default rotamer conformation). The helix was constructed using PyMOL. (d) Effect of alanine substitutions on the catalytic efficiency of RABEX-5132–460. Values are mean ± s.d. for n = 2.
Partial relief of autoinhibition by RABAPTIN-5
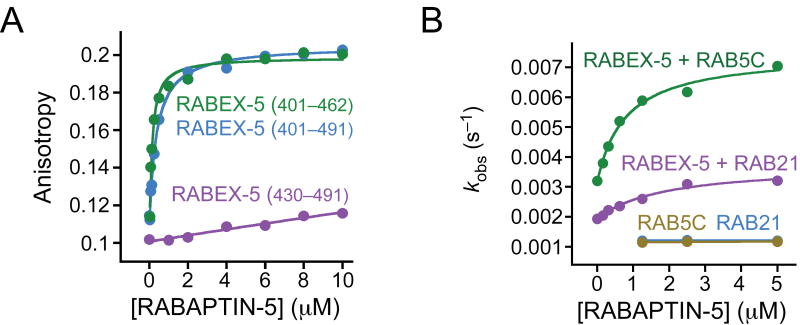
The binding sites for formation of the RABEX-5–RABAPTIN-5 complex have recently been mapped by two-hybrid experiments to residues 401–462 of RABEX-5 and 551–651 of RABAPTIN-5 (ref. 20). Using fluorescence anisotropy (Fig. 5a), we found that RABEX-5401–462 and RABEX-5401–491 bind RABAPTIN-5551–862 with similar dissociation constants (Kd) of 0.13 and 0.37 μM, respectively, whereas RABEX-5430–491 binds only weakly (Kd > 10 μM). Given that the binding site for RABAPTIN-5 evidently overlaps with the autoinhibitory element, we investigated whether autoinhibition can be relieved through formation of the complex with RABAPTIN-5. The exchange activity of RABEX-5 increases as a function of the concentration of RABAPTIN-5551–862 and approaches saturation in the low micromolar range (Fig. 5b). The observed rate constant is well described by a binding isotherm, yielding an apparent Kd of 0.8 μM for RAB5 and 1.8 μM for RAB21 and a maximum increase in exchange activity of threefold for both RAB5 and RAB21. Equivalent results were obtained for RABEX-5132–460, whereas RABAPTIN-5551–862 has no effect on the exchange activity of RABEX-5132–391. Furthermore, the magnitude of the effect is the same as that reported for the complex of the full-length proteins coexpressed in insect cells15. In contrast, ubiquitin does not stimulate RABEX-5 exchange activity, consistent with the observation that the region on the N-terminal side of the HB-VPS9 tandem is not required for autoinhibition (A.D., unpublished data). Likewise, the exchange activity is not enhanced by active H-RAS, which interacts with the region on the C-terminal side of the HB-VPS9 tandem in two-hybrid and coimmunoprecipitation experiments17.
Figure 5.
Partial reversal of autoinhibition by formation of the complex with RABAPTIN-5. (a) Fluorescence anisotropy of RABEX-5 constructs labeled with Alexa 488 as a function of RABAPTIN-5551–862 concentration. Solid lines are fitted model functions for a 1:1 binding isotherm. (b) Dependence of the observed rate constant (kobs) for RABEX-5–catalyzed nucleotide exchange on the concentration of RABAPTIN-5551–862. Solid lines are fitted model functions for a 1:1 binding isotherm.
Discussion
The structure of the RABEX-5–RAB21 complex reveals how VPS9 domains recognize RAB5-subfamily GTPases, accelerate GDP release and stabilize the nucleotide-free intermediate. A substantial mixed-polarity interface involving primarily backbone atoms and conserved side chains provides the driving force for displacement of GDP. The formation of this interface forces the residue preceding the invariant interswitch phenylalanine into a confined pocket that is just large enough to accommodate the small alanine or serine residues of RAB5-subfamily GTPases but not the larger acidic residues of other Rab GTPases. Discrimination within the RAB5 subfamily depends on two distinct factors: (i) overall physiochemical complementarity, which is optimized for RAB5 (ref. 25); and (ii) intramolecular stabilization of the open switch I conformation by a unique glutamine residue in RAB21 that substitutes for the otherwise invariant glycine residue in other Rab GTPases. The general mode of interaction and structural mechanism for acceleration of nucleotide release is notably similar to that used by the Sec7 domain39,40,45. In both cases, the GEFs supply a key acidic finger to initially destabilize the Mg2+-and β phosphate–binding sites and subsequently stabilize the nucleotide-free intermediate through interaction with the invariant P-loop lysine. In contrast, MSS4 uses an indirect local unfolding mechanism to stabilize a conformation of the switch I region that is incompatible with nucleotide binding41.
The HB-VPS9 tandems of both RABEX-5 and RIN1 have equivalently high exchange activity for RAB5 and RAB21 but relatively weak activity for RAB22 (ref. 25). This specificity profile is preserved in full-length RABEX-5 and in the complex with RABAPTIN-5. For other less well-characterized VPS9 domain GEFs, sequence variation in the Rab GTPase binding site could potentially contribute to alternative specificity profiles for the RAB5 subfamily. It is also possible that subcellular localization or other unidentified mechanisms might sharpen the specificity for RAB5 and/or RAB21, or otherwise provide conditions favorable for activation of RAB22. Recent studies indicate that RAB5, RAB21 and RAB22 have overlapping subcellular distributions in the endosomal system and may have distinct functional roles in common or related endocytic processes28–32,46. One hypothesis consistent with the structural observations, specificity profiles and currently available cell-biological data is that RABEX-5, RIN1 and potentially other mammalian VPS9 domain GEFs have coevolved with RAB5-subfamily GTPases from common progenitors in lower ancestral eukaryotes to facilitate and coordinate more elaborate control mechanisms required for regulation of complex endosomal trafficking networks in higher eukaryotic organisms. Thus, we suspect that at least a fraction of RAB5, RAB21 and possibly RAB22 in cells may be activated through common VPS9 domain GEFs, including RABEX-5 and RIN1. Nevertheless, the Gly→Gln substitution that renders RAB21 a good substrate for RABEX-5 also prevents RAB21 from interacting with some, if not all, RAB5 effectors35. Consequently, functional specificity would still be maintained at the level of interactions with effectors. Conversely, RAB22 interacts with several RAB5 effectors28,35 but may well require GEFs other than RABEX-5 or RIN1 for activation.
An investigation of the kinetic properties of Vps9p, RABEX-5 and Dss4p (the yeast homolog of MSS4) has revealed weak exchange activities, several orders of magnitude lower than those of other well-characterized GEFs37. Formation of the RABEX-5–RABAPTIN-5 complex by coexpression in Sf9 cells stimulates the exchange activity of RABEX-5 (ref. 15). Unlike Dss4 and MSS4, which are small single-domain proteins with intrinsically weak exchange activity, the HB-VPS9 tandem of RABEX-5 has a catalytic efficiency that is approximately two orders of magnitude greater than that of the full-length protein25. Here we have identified an autoinhibitory element that overlaps with the RABAPTIN-5–binding site recently mapped by two-hybrid analysis20. These observations explain both the weak exchange activity of full-length RABEX-5 compared with the isolated HB-VPS9 tandem as well as the stimulatory effect of RABAPTIN-5.
Autoregulatory mechanisms have been described for GEFs that catalyze exchange for other GTPase families, including Vav and SOS47–49, both of which have a DH-PH domain tandem with exchange activity for Rho GTPases50. SOS also contains a CDC25 domain with exchange activity for RAS51. The GTPase-binding site in the DH domain of Vav is occluded by an autoinhibitory N-terminal extension. Autoinhibition is reversed by phosphorylation of a tyrosine residue in the N-terminal extension47. The CDC25 domain of SOS is autoinhibited by the DH-PH domain tandem, which occludes the binding site for RAS49. The CDC25 domain of SOS is further regulated by a conformational change induced by binding of activated RAS to an allosteric site distinct from the exchange site48. Although it is possible that the autoinhibitory mechanism in RABEX-5 involves a conformational change in the VPS9 domain, several observations are consistent with direct occlusion of the GTPase-binding site as a likely mechanism. First, the autoinhibitory element coincides with a predicted amphipathic helix with a conserved nonpolar face. Second, the core of the GTPase-binding site in the VPS9 domain consists of a nonpolar groove between the αV4 and αV6 helices. Third, the C terminus of the RABEX-5 HB-VPS9 tandem is located on a surface proximal to the GTPase-binding site. Finally, the 15-residue region linking the N terminus of the amphipathic helix to the C terminus of the HB-VPS9 tandem corresponds to a predicted loop, the length of which is sufficient to allow the conserved nonpolar surface in the amphipathic helix to dock in the nonpolar groove between the αV4 and αV6 helices of the VPS9 domain. This model is further supported by the substantially enhanced exchange activity resulting from mutation of conserved nonpolar residues in the autoinhibitory region.
Even though RABAPTIN-5 stimulates the exchange activity of RABEX-5, the catalytic efficiency of the complex remains more than an order of magnitude lower than that of the isolated HB-VPS9 tandem. It is possible that additional interactions with RAB5, RAB4 and/or the GGA proteins might further relieve autoinhibition. Alternatively, full reversal may be coupled to the assembly of higher-order complexes on membranes, allowing for spatially and temporally limited activation of RAB5 and/or RAB21. Future studies will be necessary to fully elucidate the structural bases, molecular mechanisms and cellular consequences of RABEX-5 autoregulation.
Methods
Constructs
Constructs were amplified with Vent polymerase (NEB). Human RABEX-5 and RABAPTIN-5 constructs were subcloned into a modified pET15b vector containing an N-terminal His6 tag (MGHHHHHHGS). Human RAB21 and mouse RAB5C constructs were subcloned into pGEX-4T1 (Amersham Biosciences) for expression as an N-terminal glutathione S-transferase (GST) fusion. All constructs were verified by sequencing the entire coding region.
Expression and purification
Constructs were expressed in BL21(DE3)-RIL cells (Stratagene) cultured in 2× YT medium supplemented with 100 mg l–1 ampicillin at 25 °C to an A600 of 0.4 and induced with 0.05 mM IPTG for 14 h. Cells were disrupted by sonication in 50 mM Tris (pH 8.0), 100 mM NaCl, 0.1% (v/v) 2-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride and 1 mg ml–1 lysozyme. Following addition of Triton X-100 to 0.5% (v/v), cell lysates were centrifuged at 35,000g for 40 min. For His6 fusions, the supernatants were loaded onto Ni2+–nitrilotriacetic acid-agarose columns (Qiagen), which were washed with 50 mM Tris (pH 8.0), 500 mM NaCl, 10 mM imidazole and 0.1% (v/v) mercaptoethanol and eluted with a gradient of 10–150 mM imidazole. For GST fusions, the supernatants were loaded onto a glutathione-Sepharose column (Amersham Biosciences) equilibrated with 50 mM Tris (pH 8.0), 0.1 M NaCl and 0.1% (v/v) 2-mercaptoethanol. After washing with ten column volumes of the same buffer, the GST fusions were eluted with 10 mM reduced glutathione. To generate the untagged form of RAB21, GST fusions at a concentration of 2–4 mg ml–1 were incubated with 2 μg ml–1 human α-thrombin (Hematologic Technologies) overnight at 4 °C in 50 mM Tris (pH 8.0), 2 mM CaCl2, 0.1% (v/v) mercaptoethanol. Ion exchange using Source Q (Amersham Biosciences) followed by gel filtration over Superdex-75 (Amersham Biosciences) resulted in preparations that were >99% pure as judged by SDS-PAGE. For RAB21, all buffers were supplemented with 2 mM MgCl2.
Exchange kinetics
Exchange kinetics were measured by monitoring the quenching of fluorescence after release of the nucleotide analog 2′-(3′)-bis-O-(N-methylanthraniloyl)-guanosine 5′-diphosphate (mant-GDP; Invitrogen). Rab GTPases were loaded with mant-GDP as described52 and diluted to 1.0 μM in 50 mM Tris (pH 8.0), 150 mM NaCl and 2 mM MgCl2. Samples were excited at 360 nm and the emission monitored at 440 nm. Data were collected using a Saphire multimode microplate spectrophotometer (Tecan). Observed pseudo first-order rate constants (kobs) were extracted from a nonlinear least-squares fit to
where I(t) is the emission intensity at time t, I0 is the initial emission intensity and I∞ is the final emission intensity. Catalytic efficiencies (kcat/Km) were obtained from the slope of a linear least-squares fit to
where kintr is the intrinsic rate constant for nucleotide exchange in the absence of Rabex-5.
Fluorescence anisotropy
RABEX-5 constructs were labeled on a single naturally occurring cysteine residue (Cys404 for RABEX-5401–491 and RABEX-5401–462) or on a cysteine residue incorporated at the N terminus during amplification (RABEX430–491). The thiol labeling reaction was performed according to the manufacturer's instructions (Invitrogen). Briefly, RABEX-5 (100 μM) was combined with 1 mM Alexa Fluor 488 C5-malemide and incubated overnight at 4 °C in 50 mM HEPES (pH 7.4) and 100 mM NaCl. Unlabeled fluorophore was removed using a 10 ml D-Salt column (Pierce) followed by dialysis for 24 h at 4 °C in 50 mM HEPES (pH 7.4) and 100 mM NaCl. Labeled RABEX-5 constructs at 100 nM were incubated at room temperature with varying concentrations of RABAPTIN-5551–862 in 50 mM HEPES (pH 7.4) and 100 mM NaCl. Fluorescence anisotropy was monitored using an ISS spectrofluorometer. Samples were excited at 493 nm and the emission monitored at 516 nm. Anisotropy was calculated as (I‖ − I⊥)/(I‖+ 2I⊥), where I‖ and I⊥ are the emission intensities parallel and perpendicular to the electric field vector of the linearly polarized exciting light. The anisotropy data were analyzed by fitting with a 1:1 binding isotherm.
Crystallization and structure determination
Nucleotide-free complexes of RABEX-5 (residues 132–397) and RAB21 (residues 16–183) were isolated over Superdex-75 following incubation of stoichiometric quantities of each protein in 50 mM Tris (pH 8.0), 100 mM NaCl, 1 mM DTT, 5 mM EDTA and 1 μg ml–1 activated charcoal at 4 °C. The complex was exchanged into 5 mM Tris (pH 8.0) and 50 mM NaCl and concentrated to 15 mg ml−1. Crystals were grown at 4 °C in hanging drops containing 10 mg ml−1 protein in 10% (w/v) PEG 6,000, 200 mM MgCl2 and 50 mM sodium-MES buffer (pH 6.0). Crystals appeared in 1–2 d and grew to maximum dimensions of 0.05 mm × 0.2 mm × 0.3 mm in 1 week. The crystals are in the space group P21212 with unit cell dimensions a = 71.9 Å, b = 113.4 Å and c = 60.5 Å, one molecule each of RABEX-5 and RAB21 in the asymmetric unit and a solvent content of 52%. Crystals were transferred to a cryostabilizer (30% (w/v) PEG 8,000, 50 mM Tris (pH 6.0), 200 mM MgCl2, 10% (v/v) glycerol), flash-frozen in liquid propane and maintained at 100 °K in a nitrogen cryostream. Data were collected at the X25 beam line at the National Synchrotron Light Source and the structure solved by molecular replacement using Phaser53, with search models derived from the RABEX-5 HB-VPS9 tandem (PDB 1TXU) and RAB21 (PDB 1Z0I, residues 26–95 omitted). After simulated annealing in CNS55, a σA-weighted 2Fo − Fc omit map revealed continuous electron density for the omitted regions, with the exception of RAB21 residues 43–46. The structure was completed by iterative cycles of interactive model building with O54 and positional refinement with REFMAC5 (ref. 53). The final model contains residues 139–391 of RABEX-5, residues 17–42 and 47–182 of RAB21 and 462 water molecules. Data collection and refinement statistics are tabulated in Table 1. Structural figures were rendered with PyMOL (http://pymol.sourceforge.net).
Table 1. Data collection and refinement statistics.
| Native crystal | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 72.0, 113.4, 60.5 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 2.1 (20) |
| Rsym | 7.5 (38.5) |
| I / σI | 22.0 (4.0) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 6.0 |
| Refinement | |
| Resolution (Å) | 20–2.1 |
| No. reflections | 28,201 |
| Rwork / Rfree | 0.188 / 0.242 |
| No. atoms | |
| Protein | 3,108 |
| Water | 462 |
| B-factors | |
| Rab21 | 29 |
| Rabex-5 | 31 |
| Water | 55 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.2 |
Data were collected from a single crystal. Values in parentheses are for highest-resolution shell.
Accession codes
Protein Data Bank: Coordinates and structure factors have been deposited with accession code 2OT3.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank the National Synchrotron Light Source X25 beamline staff for assistance with X-ray data collection and J. Goldberg (Memorial Sloan-Kettering Cancer Center) for the coordinates of the nucleotide-free ARF1–Gea2 complex. This work was supported by US National Institutes of Health grants GM56324 and DK60564 (to D.G.L.) and by a Charles A. King Postdoctoral Fellowship (to A.D.).
Footnotes
Competing Interests Statement: The authors declare no competing financial interests.
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol. 1996;16:2369–2377. doi: 10.1128/mcb.16.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem. 1999;274:15284–15291. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- 5.Davies BA, et al. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J Biol Chem. 2003;278:19826–19833. doi: 10.1074/jbc.M301059200. [DOI] [PubMed] [Google Scholar]
- 6.Shih SC, et al. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 2003;22:1273–1281. doi: 10.1093/emboj/cdg140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiuchi H, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 8.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 9.Saito K, et al. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J Biol Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 10.Kajiho H, et al. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J Cell Sci. 2003;116:4159–4168. doi: 10.1242/jcs.00718. [DOI] [PubMed] [Google Scholar]
- 11.Otomo A, et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, et al. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol. 2005;7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, He X, Fu XY, Chang Z. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J Cell Sci. 2006;119:1053–1062. doi: 10.1242/jcs.02810. [DOI] [PubMed] [Google Scholar]
- 14.Gournier H, Stenmark H, Rybin V, Lippe R, Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride HM, et al. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 17.Tam SY, et al. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- 18.Kalesnikoff J, et al. RabGEF1 regulates stem cell factor/c-Kit-mediated signaling events and biological responses in mast cells. Proc Natl Acad Sci USA. 2006;103:2659–2664. doi: 10.1073/pnas.0511191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, et al. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 21.Penengo L, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Vitale G, et al. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 2003;22:78–88. doi: 10.1093/emboj/cdg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu G, et al. Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5. EMBO J. 2004;23:3909–3917. doi: 10.1038/sj.emboj.7600411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 27.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 28.Kauppi M, et al. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- 29.Simpson JC, et al. A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- 30.Magadan JG, Barbieri MA, Mesa R, Stahl PD, Mayorga LS. Rab22a regulates the sorting of transferrin to recycling endosomes. Mol Cell Biol. 2006;26:2595–2614. doi: 10.1128/MCB.26.7.2595-2614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khurana T, Brzostowski JA, Kimmel ARA. Rab21/LIM-only/CH-LIM complex regulates phagocytosis via both activating and inhibitory mechanisms. EMBO J. 2005;24:2254–2264. doi: 10.1038/sj.emboj.7600716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellinen T, et al. Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J Cell Biol. 2006;173:767–780. doi: 10.1083/jcb.200509019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.al-Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure. 1996;4:555–565. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- 34.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 35.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 37.Esters H, et al. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol. 2001;310:141–156. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- 38.Guo Z, Ahmadian MR, Goody RS. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry. 2005;44:15423–15429. doi: 10.1021/bi0518601. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 40.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 41.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding–structure of Rab8 in complex with MSS4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton J, Roberts D, Montaldi M, Novick P, De Camilli P. A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature. 1993;361:464–467. doi: 10.1038/361464a0. [DOI] [PubMed] [Google Scholar]
- 43.Burton JL, Burns ME, Gatti E, Augustine GJ, De Camilli P. Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J. 1994;13:5547–5558. doi: 10.1002/j.1460-2075.1994.tb06892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuoffer C, Wu SK, Dascher C, Balch WE. Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell. 1997;8:1305–1316. doi: 10.1091/mbc.8.7.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mossessova E, Corpina RA, Goldberg J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell. 2003;12:1403–1411. doi: 10.1016/s1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- 46.Weigert R, Yeung AC, Li J, Donaldson JG. Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol Biol Cell. 2004;15:3758–3770. doi: 10.1091/mbc.E04-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aghazadeh B, Lowry WE, Huang XY, Rosen MK. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 48.Margarit SM, et al. Structural evidence for feedback activation by Ras. GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 49.Sondermann H, et al. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 51.Chardin P, et al. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 52.Zhu Z, Dumas JJ, Lietzke SE, Lambright DG. A helical turn motif in Mss4 is a critical determinant of Rab binding and nucleotide release. Biochemistry. 2001;40:3027–3036. doi: 10.1021/bi002680o. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Compuational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 55.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.