Abstract
Histone deactylase enzymes are responsible for the deacetylation of histone tails, and consequently influence gene regulation through their ability to modify chromatin structure surrounding promoter regions. We analyzed the microarray collection of the National Brain Databank to investigate differential expression of these enzymes in the prefrontal cortices of control, schizophrenia and bipolar subjects. HDAC1 expression levels were significantly higher in schizophrenia versus normal subjects. The mRNA expression level of an epigenetically regulated gene GAD67 was strongly and negatively correlated with the mRNA expression levels of HDAC1, HDAC3 and HDAC4 levels. These findings provide additional support for the proposal that epigenetic factors are operative in the brain pathology of patients with schizophrenia.
Keywords: HDAC enzymes, GAD67 expression, schizophrenia, postmortem brain, microarray
INTRODUCTION
Chromatin, a DNA–protein complex, commonly conceptualized as the efficient “packaging” of several billion bases of genomic DNA, functions as an interactive platform for the regulation of gene transcription. Chromatin participation in gene regulation is based on physical and chemical adaptations in the vicinity of regulatory DNA sequences, the mechanics of which are determined by linear patterns of covalent modifications of cytosine bases in DNA and amino acid residues in histone protein tails. These covalent modifications, along with their attendant enzymes and cognate regulatory proteins, are broadly classified under the general term “epigenetic mechanisms.” Acetylation of site specific lysine residues along the N-terminal tail of histone 3 and histone 4 proteins is one such modification that putatively releases the histone tail from its position around the DNA strand, exposing regulatory regions and facilitating DNA-protein interactions with transcriptional regulators. Acetyl groups are covalently attached to these residues by histone acetyltransferases (HATs) and removed by histone deaceytlases (HDACs). The HDAC family of enzymes (HDAC 1–11) is ubiquitously expressed in most tissues including the brain (de Ruijter et al 2003), and is widely recognized as potential therapeutic targets for brain disorders (Sharma 2005a, Tsankova et al 2006, Simonini et al 2006,).
Epigenetic gene regulation in the brain is instrumental for neuronal viability and survival (Fan et al 2001), membrane depolarization (Chen et al 2003, Martinowich et al 2003, Sharma et al 2007), synaptic plasticity (Levenson et al 2006), and long-term potentiation, cognition and memory-consolidation (Miller and Sweatt 2007; Alarcon et al 2004, Korzus et al 2004). Abnormalities in epigenetic gene regulation as a cause of complex disorders has emerged as a powerful heuristic method towards the study of molecular mechanisms in psychiatric disorders (for review Costa et al 2004, Petronis 1999, Sharma 2005a). Since that time, a mounting body of evidence has developed, to support abnormalities in epigenetic gene regulation of prototypic schizophrenia candidate genes such as reelin and GAD67, from cell studies (Chen et al 2002, Noh et al 2005, Kundakovic et al 2007), animal investigations (Tremolizzo et al 2005, Dong et al 2005) and from DNA methylation and expression/protein studies in the postmortem brains of schizophrenia subjects (Akbarian et al 2005, Grayson et al 2005, Abdolmaleky et al 2005, 2006, Veldic et al 2004, 2005, 2007, Ruzicka et al 2007, Guidotti et al 2007, Benes et al 2007). Extending these concepts to clinical populations, we have recently reported that schizophrenia patients may have a comparatively restrictive chromatin structure measured in their peripheral blood lymphocytes (Sharma et al 2006), and that GAD67 is directly regulated by an inhibitor of histone deactylases in primary lymphocyte cultures (Gavin et al 2007 submitted). These findings have motivated an investigation of diagnostic differences in the expression of histone modifying enzymes in postmortem brains that would suggest the presence of an environment conducive to the development of restrictive chromatin. Further, because of the connection between levels of HDAC activity and schizophrenia candidate genes such as GAD67 (extensively replicated down-regulation in the postmortem brains of schizophrenia patients as reviewed by Akbarian and Huang 2006), we looked for a functional association between the mRNA expression levels of HDAC enzymes and GAD67. These questions are examined using a restricted and targeted analysis of the National Brain Databank (NBD) microarray collection.
METHOD
The National Brain Databank is made available by the Harvard Brain Tissue Resource Center as a publicly accessible data repository for neuroscience investigators. The databank provides microarray expression results from postmortem brain tissue samples obtained from the prefrontal cortices of subjects with psychiatric and neurological illness. In particular, the collection includes samples from 27 control subjects, 16 schizophrenia subjects and 18 bipolar subjects as well as 3 subjects with schizoaffective disorder. Further, the collection includes 44 males and 20 females (Table 1). Thirty out of 37 patients were being treated with a psychotropic; of these all but 5 subjects (83%) were treated with multiple medications. Medications included typical and atypical antipsychotics, mood stabilizers (including valproic acid), antidepressants, stimulants and sedatives.
Table 1. Description of the National Brain Databank Microarray Collection.
RNA ratios represent the ratio 28S:18S of ribosomal RNA as a measure of underlying mRNA stability. 3’/5’ expression ratios for GAPDH and β-actin reflect differential degradation at the 5’ end of the gene versus 3’ end where the polyadenylation sequence is used for hybridization to the primer oligonucleotides. ‘Percent probe sets present’ is the percent of microarray wide probes providing a signal.
| Normal Subjects | Schizophrenia | Bipolar Disorder | Schizoaffective Disorder | ||
|---|---|---|---|---|---|
| Age range | 20–31 | 1 | 1 | 1 | |
| 31–40 | 7 | 2 | 3 | ||
| 41–50 | 3 | 6 | 2 | 1 | |
| 51–60 | 2 | 1 | |||
| 61–70 | 7 | 3 | 1 | ||
| 71–80 | 5 | 3 | 8 | 1 | |
| 80+ | 2 | 1 | 2 | 1 | |
|
| |||||
| Gender | |||||
| Male | 19 | 13 | 12 | ||
| Female | 8 | 3 | 6 | 3 | |
|
| |||||
| Post Mortem Interval | 20.34 (5.74) | 20.27 (4.35) | 21.12 (9.73) | 24.31 (10.27) | |
| Brain PH | 6.43 (0.31 | 6.41 (0.25) | 6.45 (0.24) | 6.52 (0.35) | |
|
| |||||
| 3′/5′ G3PDH ratio | 1.43 (0.40) | 1.58 (0.67) | 1.53 (0.48) | 1.45 (0.24) | |
| 3′/5′ β-Actin Ratio | 2.32 (0.86) | 2.45 (0.83) | 2.68 (1.03) | 2.19 (0.68) | |
| RNA Ratio | 1.07 (0.34) | 1.12 (0.50) | 1.01 (0.32) | 0.99 (0.43) | |
| Percent Probe Sets Present | 45.94 (3.98) | 46.1 (4.23) | 45.17 (4.76) | 44.26 (4.07) | |
The microarray platform utilizes the Affymetrix HG-U133A gene chip. This platform represents each interrogated sequence as a set of oligonucleotides synthesized onto the chip. A collection of 22 such oligos for a given genomic sequence is called a probe set. Each probe set is comprised of 11 perfectly matched oligos and 11 mismatched oligos (by one base) – the latter serve as a type of internal standard. The abundance of a given transcript, also referred to as the ‘signal’ is computed by the Gene Chip Operating System (GCOS) as a one-step biweighted estimate of the combined differences of all the probe pairs in the probe set.
A. Default controls at the level of RNA quality and microarray integrity
Four independent parameters of total RNA quality and cellular integrity (18S/28S ribosomal ratio, 3’/5’ GAPDH ratio and 3’/5’ β-actin ratio) and Genechip validity (percent probe sets present) are provided by the NBD website.
B. Scaling at the level of the total array
For single-array analysis, GCOS software executes a mathematical scaling procedure to allow for comparisons between single array results from different experiments (subjects). This minimizes the discrepancy due to variables such as sample preparation, hybridization conditions, staining or probe array lot. The analytic software surveys signal intensity from all probes on the microarray and computes a scaling factor that scales the ‘trimmed’ mean signal to the user defined ‘target signal’. Single arrays that are scaled to the same ‘target signal’ can be compared.
C. Effectiveness of individual probe design
The HG-U133A chip can technically interrogate the expression of HDACs 1–6, 7, 9, 11). However, a given tissue such as the prefrontal cortex may not express all HDAC transcripts at an abundance level that is reliably detected by the chip. This is compounded by the additional possibility that the default probe may hybridize poorly to a tissue specific gene isoform. Among the listed HDACs, we included only those that demonstrated reliable and consistent expression in prefrontal cortex across the whole collection. In the absence of additional information, the p-value threshold for the Detection Call (α2) was set at 0.06 (the default value suggested by the manufacturer). If a given probe (HDAC1 or 2 etc) yielded a greater than 10% ‘absent’ detection call rate, this probe was not included in the analysis. This resulted in exclusion of HDAC5, HDAC7 and HDAC11. Subsequently, all ‘signal’ values for the remaining HDAC genes were analyzed. GAD67 was interrogated using probe ID 205278. Signal levels for HDACs (1–4, 6, 9) as well as for GAD67 were then entered into an SPSS data base and analyzed for diagnostic differences and associations. Diagnostic analyses were conducted only in data from controls, schizophrenia and bipolar subjects. Parametric tests for independent groups (one-way ANOVA and independent t-tests) were used for group comparisons. Pearson correlations were used to establish bivariate associations. All p-values are two-tailed.
RESULTS
There were no significant differences between diagnostic groups in the postmortem brain interval, brain pH or in values of the quality control parameters (18S/28S ribosomal ratio, 3’/5’ GAPDH ratio and 3’/5’ β-actin ratio).
In the total sample, females subjects across diagnostic groups had a significantly higher level of HDAC1 signal (312.23 vs 247.09; df=62; p<0.02). There was no significant difference in the distribution of gender among the four diagnostic groups. Age was significantly related to levels of HDAC4 in a bimodal pattern, with marginally higher levels between 20–40 years of age, decreasing between 40–60 years and significantly higher (p<0.02) in patients above 70 years of age. All HDACs examined were equivalently expressed in either hemisphere (29 left and 35 right).
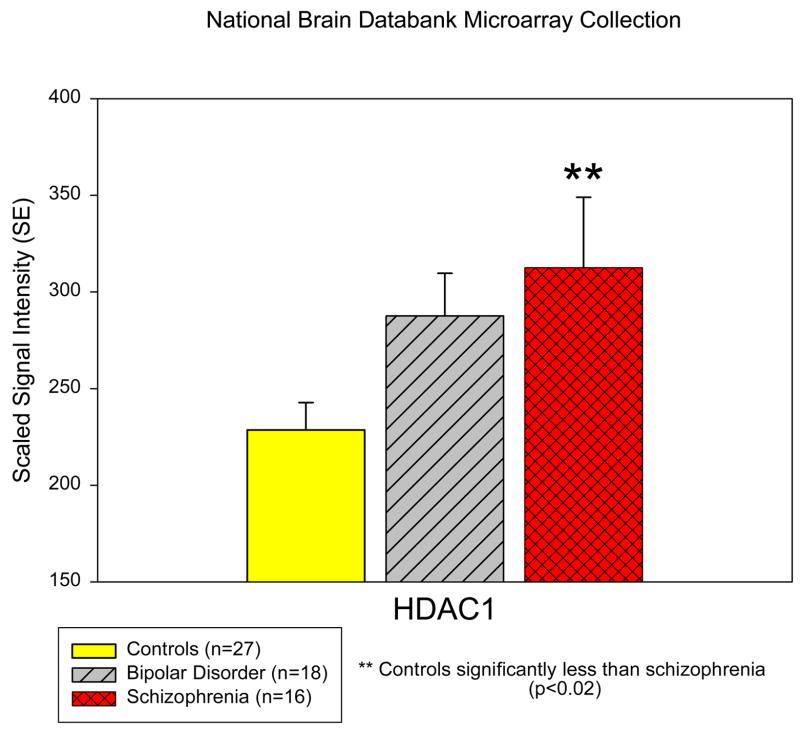
Schizophrenia patient samples had a significant increase in the mRNA expression of HDAC1 in the prefrontal cortex when compared to control samples (313.5 vs. 228.6; p<0.02; Figure 1). No significant diagnostic differences were found for the expression of HDACs 2, 3, 4, 6, and 9.
Figure 1. Histone Deactylase-1 is increased in the prefrontal cortex of schizophrenia subjects.
A restricted analysis (on the identified genes in the text) was conducted using the microarray collection of the National Brain Databank. Expression of HDAC enzymes in samples of postmortem prefrontal cortical tissue was compared between the diagnostic groups as presented. There is a significant increase in HDAC1 expression levels in samples obtained from schizophrenia subjects compared to controls.
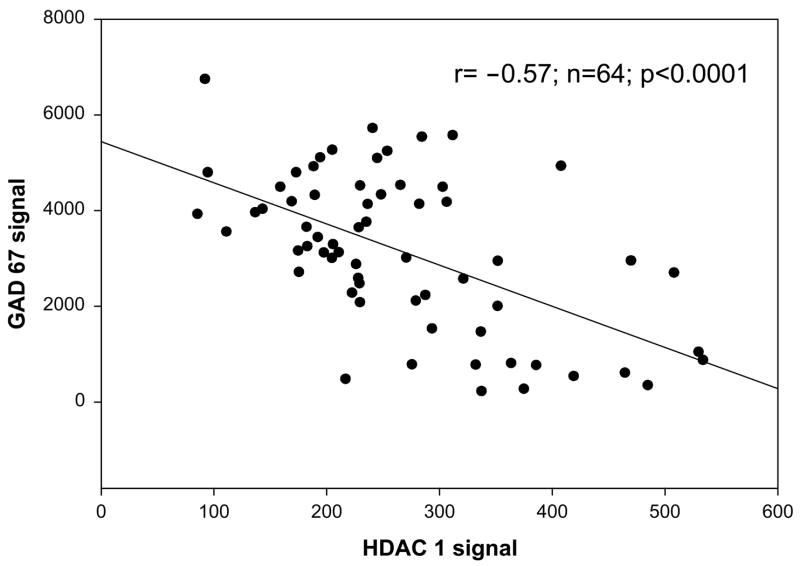
GAD67 expression was significantly and negatively correlated with the expression levels of HDAC1 (r= −0.57; n=64; p<0.0001; Figure 2) and HDAC3 (r= −0.35; n=64; p<0.004) and HDAC4 (r= −0.41; n=64; p<0.001) suggesting an overlapping regulation of this gene by several HDACs in the prefrontal cortices of patients with schizophrenia. Higher HDAC levels were associated with lower levels of GAD67 expression. Based on the findings of the bivariate correlations, a multiple regression with HDAC 1, 3, and 4 as independent variables and GAD67 as the dependent measure revealed a highly significant effect for the complete model (t=8.8; p<9.9E -14; and of HDAC1 by itself (t=−4.27; p<0.00006). The part correlation between HDAC1 and GAD67 was −0.41 controlling for the other independent variables (i.e., HDAC3 and HDAC4) in the model.
Figure 2. HDAC1 and GAD 67 mRNA expression levels are correlated in the prefrontal cortex.
GAD 67 is an epigenetically controlled gene whose regulation is influenced by the level of acetylated (increased expression) or deacetylated chromatin (decreased expression) that surrounds its promoter sequence. There was a significant correlation between HDAC1 and GAD67 expression signals in 64 subjects (Controls=27, Bipolar=16, Schizophrenia=18, Schizoaffective=3)
Because of the HDAC inhibiting properties of valproic acid, we analyzed for medication effects of valproic acid on HDAC enzyme mRNA expression in the prefrontal cortex. Patients being treated with valproic acid (8 bipolars, 1 schizophrenia and 2 schizoaffective subjects) had significantly elevated levels of HDAC 9 when compared to all other subjects (including controls and patients; t=2.13; df=62; p<0.036) but not when compared only to other patients being treated with psychotropic medications (only patients p=ns). All other HDAC enzymes were comparably expressed in the valproic acid treated group. No other systematic analyses were attempted with the other medications given the number of subjects across diagnostic groups with multiple prescribed medications of currently unknown effects on epigenetic enzyme systems.
DISCUSSION
The availability of a well characterized microarray database allows for the preliminary test of a hypothesis in-silico. Given the validity of the technology, the caveat is to avoid analyzing a broad array of targets thereby creating an over-determined statistic as we have previously reported (Gibbons et al 2005). We surveyed expression levels in nine HDAC genes, and accepted six for statistical analysis. This effort is a direct extension of our ongoing examination of chromatin structure in schizophrenia and bipolar subjects (Sharma et al 2005a,2005b, Sharma et al 2006, Gavin et al 2007).
During the preparation of this manuscript, Benes et al. (2007) reported elevated HDAC1 levels from an independent sample of postmortem hippocampal tissue (seven normal, seven schizophrenia and seven bipolar subjects) using laser microdissected samples of GABA neurons obtained from the stratum oriens of CA2/3 layers. They reported elevated levels of HDAC1 by microarray analysis, which was validated using quantitative RT-PCR. Overall a 2 fold increase of HDAC1 in the GABA neurons of the stratum oriens of the CA2/3 region of the hippocampus from schizophrenia subjects was found.
The increase in HDAC1 in the prefrontal cortices of schizophrenia subjects (as well as the specific region of the hippocampus as described by Benes et al 2007) is noteworthy because enhanced expression of these enzymes is predictive of chromatin that is deactylated at site specific residues and consequently repressive to transcriptional activity. Akbarian et al (2005) have examined histone modifications in protein extracts from the prefrontal cortices of schizophrenia patients and normal subjects using Western blot. Although not finding overall differences in levels of modified histones, they report reduced levels of targeted gene expression in subsets of patients with higher levels of specific modifications including acetylation at H3 Lysine 14. We have earlier reported that histone protein obtained from the peripheral lymphocytes of clinical schizophrenia patients is less acetylated than that obtained from bipolar patients, and remains unyielding when subjects are treated with an HDAC inhibitor (Sharma et al 2006). Other details of our analysis that should be considered preliminary gender differences particularly in the schizophrenia subsample, where female schizophrenia subjects (n=3) had almost twice the amount of HDAC1 in their prefrontal cortices than did their male counterparts (n=13) (516.86 vs. 265.33; p<0.003). Also, the confounding effects of psychotropic medication cannot be entirely resolved in this clinical sample because of multiple medications and lengths of treatment. Preliminary observations from our lab indicates that HDAC inhibitors such as Trichostatin-A and Valproic acid do not reliably change HDAC1 mRNA expression in either primary lymphocyte cultures from human subjects (Gavin and Sharma unpublished) or in NT2 dividing cells (Kundakovic and Grayson unpublished). However, MS-275, a new research HDAC inhibitor, can downregulate protein levels of HDAC1 in NT2 cells (Kundakovic and Grayson personal communication). Furthermore, the effects of the typical and atypical antipsychotics as well as antidepressant medications could not be systematically explored. The effects of valproic acid on HDAC9 are interesting and should be considered preliminary. Nonetheless, the effects of psychotropic medications on chromatin structure and its attendant enzymes will be a fruitful area for future investigation.
Prior basic work in cell lines (Kundakovic et al., 2006 ), primary neuron cultures (Noh et al., 2005; Sharma unpublished data), rodent models (Tremolizzo et al., 2002, 2005, Simonini et al., 2006) and peripheral blood lymphocytes from human subjects (Gavin et al., 2007) indicate that the GAD67 promoter is regulated by epigenetic mechanisms. These studies show that GAD67 expression is profoundly impacted by the acetylation of histone proteins surrounding its promoter as well as the intensity of DNA methylation in this region. Our observation of a negative correlation between the expression of GAD67 and several HDAC enzymes is encouraging in that it provides independent validity to the relevance of these findings, and is consistent with previous published reports
HDAC1 is a nuclear protein of the Class I type (de Ruijter et al 2003). It is ubiquitously distributed (brain as well as lymphoid tissue for example), is activated by phosphorylation and associates with larger multiprotein repressor complexes (such as Co-REST in neurons) to stabilize its enzymatic activity (Saha and Pahan 2006). HDAC1 is also inhibited by small molecule pharmacological agents such as the widely prescribed mood stabilizer valproic acid (mM range) and trichostatin A (nM range). Deactylase activity occurs in partnership with DNA binding proteins that not only recruit the enzyme to the targeted promoter but also enhance catalytic activity. HDAC1 has two phosphorylation sites (Serine421 and Serine423) which regulate catalytic activity as well as protein-protein complex formation.
Enhanced deactylase activity and restrictive chromatin in the prefrontal cortices of schizophrenia patients is a heuristic explanation for a variety of clinical findings, from reduced metabolism, cognitive disturbances, negative symptoms and low motivation. Alternatively, restrictive chromatin may be a consequence of the impoverished environmental, emotional and cognitive stimulation that is the daily experience of these subjects or even perhaps the cognitive blunting imposed by ongoing antipsychotic treatment. In either case, a catalytic enzyme serves as an ideal target for pharmacology and a realistic approach to regulating gene expression in a targeted brain region.
5. AUTHOR DISCLOSURE
RPS is supported by MH 69839, DRG by MH62682, and DPG by MH067631. The NIMH had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. RPS conceptualized the study and analyzed the microarrays. DPG and DRG performed the literature searches, and assisted with the analyses in addition to working on numerous drafts of the manuscript. All authors contributed to and have approved the final manuscript. The authors have no conflicts of interest to disclose.
Acknowledgments
The authors acknowledge the support and critical suggestions of Dr Francine Benes and the National Brain Databank funded through an NIH grant (R24 MH/NS31862) awarded to the Harvard Brain Tissue Resource Center at McLean Hospital.
Footnotes
This work was presented at the Society of Biological Psychiatry meeting in San Diego, 2007.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134(1):60–6. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–45. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Ruehl MG, Bliven E, Luiz LA, Peranelli AC, Baker SP, Roberts RC, Bunney WE, Jr, Conley RC, Jones EG, Tamminga CA, Guo Y. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2005;62(8):829–40. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Brain Res Rev. 2006;52(2):293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–59. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104(24):10164–9. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR. On the epigenetic regulation of human reelin promoter. Nucleic Acids Res. 2002;30(13):2930–9. doi: 10.1093/nar/gkf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Grayson DR, Veldic M, Guidotti A. Neurochemical basis for an epigenetic vision of synaptic organization. Int Rev Neurobiol. 2004;59:73–91. doi: 10.1016/S0074-7742(04)59004-9. [DOI] [PubMed] [Google Scholar]
- Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci U S A. 2005;102(35):12578–83. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, Trumpp A, Poon C, Wilson CB, Jaenisch R. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21(3):788–97. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Rosen C, Jayaraman S, Grayson D, Sharma RP. HDAC inhibitors, valproic acid and trichostatin A increase glutamic acid decarboxylase (GAD67) expression in primary lymphocyte cultures. Biol Psychiatry. 2007;61(8S):210S. [Google Scholar]
- Gibbons RD, Bhaumik DK, Cox DR, Grayson DR, Davis JM, Sharma RP. Sequential prediction bounds for identifying differentially expressed genes in replicated microarray experiments. Journal of Statistical Planning and Inference. 2005;129:19–37. [Google Scholar]
- Guidotti A, Ruzicka W, Grayson DR, Veldic M, Pinna G, Davis JM, Costa E. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. Neuroreport. 2007;18(1):57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(26):9341–6. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–72. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA Methyltransferase Inhibitors Coordinately Induce Expression of the Human Reelin and GAD67. Genes Mol Pharmacol. 2006;71(3):644–53. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(23):15763–73. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–3. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, Costa E, Guidotti A, Grayson DR. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proc Natl Acad Sci U S A. 2005;102(5):1749–54. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A, Paterson AD, Kennedy JL. Schizophrenia: an epigenetic puzzle? Schizophr Bull. 1999;25(4):639–55. doi: 10.1093/oxfordjournals.schbul.a033408. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12(4):385–97. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13(4):539–50. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP. Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr Res. 2005a;72(2–3):79–90. doi: 10.1016/j.schres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Grayson DR, Guidotti A, Costa E. Chromatin, DNA methylation, and neuron gene regulation - the purpose of the package. J Psychiatry Neurosci. 2005b;30(4):257–63. [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Rosen CR, Kartan S, Guidotti A, Costa E, Grayson D, Chase K. Valproic acid and chromatin remodeling in schizophrenia and bipolar disorder: preliminary results from a clinical population. Schizophr Res. 2006;88(1–3):227–31. doi: 10.1016/j.schres.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Kartan S, Grayson DR. DNA Methyltransferase Activity In Primary Cortical Cultures. Submitted, Society of Neuroscience meeting, manuscript in preparation 2007 [Google Scholar]
- Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, Guidotti A. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci U S A. 2006;103(5):1587–92. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, Sharma R, Grayson DR, Costa E, Guidotti A. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci U S A. 2002;99(26):17095–100. doi: 10.1073/pnas.262658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Doueiri MS, Dong E, Grayson DR, Davis J, Pinna G, Tueting P, Rodriguez-Menendez V, Costa E, Guidotti A. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol Psychiatry. 2005;57(5):500–9. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, Costa E. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A. 2004;101(1):348–53. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci U S A. 2005;102(6):2152–7. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, Costa E. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91(1–3):51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]