Abstract
Chicken endogenous viruses, ALVE (Avian Leukosis Virus subgroup E), are inherited as LTR (long terminal repeat) retrotransposons, which are negatively correlated with disease resistance, and any changes in DNA methylation may contribute to the susceptibility to neoplastic disease. The relationship between ALVE methylation status and neoplastic disease in the chicken is undefined. White Leghorn inbred lines 72 and 63 at the ADOL have been respectively selected for resistance and susceptibility to tumors that are induced by avian viruses. In this study, the DNA methylation patterns of 3∼6 CpG sites of four conserved regions in ALVE, including one unique region in ALVE1, the promoter region in the TVB (tumor virus receptor of ALV subgroup B, D and E) locus, were analyzed in the two lines using pyrosequencing methods in four tissues, i.e., liver, spleen, blood and hypothalamus. A significant CpG hypermethylation level was seen in line 72 in all four tissues, e.g., 91.86±1.63% for ALVE region2 in blood, whereas the same region was hemimethylated (46.16±2.56%) in line 63. CpG methylation contents of the ALVE regions were significantly lower in line 63 than in line 72 in all tissues (P<0.01) except the ALVE region 3/4 in liver. RNA expressions of ALVE regions 2 and 3 (PPT-U3) were significantly higher in line 63 than in line 72 (P<0.01). The methylation levels of six recombinant congenic strains (RCSs) closely resembled to the background line 63 in ALVE-region 2, which imply the methylation pattern of ALVE-region 2 may be a biomarker in resistant disease breeding. The methylation level of the promoter region in the TVB was significantly different in blood (P<0.05) and hypothalamus (P<0.0001), respectively. Our data disclosed a hypermethylation pattern of ALVE that may be relevant for resistance against ALV induced tumors in chickens.
Introduction
Epigenetic information is heritable but not encoded in the DNA sequence [1]. DNA methylation is an important epigenetic mechanism for pretranscriptional control, and plays an essential role in regulating genes' expression and maintaining cellular function [2]. DNA methylation is restricted to CpG dinucleotides in mammals, which commonly locate on transposable elements and promoters [3]. Numerous studies have shown that changes in DNA methylation patterns may contribute to neoplastic disease or cancer susceptibility [4], [5], [6], [7], [8], [9], [10], [11]. The nonrandom occurrence and observed patterns of CpG-rich methylation events further suggest that gene-specific methylation provides potentially useful markers for molecular diagnostics and detection of neoplastic disease risks [12].
Neoplastic diseases are defined as any malignant growth or tumor resulted from abnormal or uncontrolled cell division. It may spread to other parts of the body through the lymphatic system or the blood stream. Neoplastic diseases are a serious concern to the poultry industry due to the cost of routine vaccination against an herpesvirus that induces Marek's disease lymphomas [13] and the eradication of avian leukosis viruses from breeders [14]. Avian leukosis viruses (ALVs) are associated with a variety of tumors including lymphoid leukosis, myeloblastosis, erythroblastosis, osteopetrosis, myxosarcomas and fibrosarcomas. The ALVs are retroviruses and have six subgroups [14]. Among the avian retroviruses, ALVE is the unique endogenous virus (ev). The other five ALVs, including ALVA, ALVB, ALVC, ALVD and ALVJ, are exogenous viruses [15], [16]. ALVE is inherited as LTR (long terminal repeat) retrotransposons [17], [18], [19], [20].
Chicken ALVE loci are present in the genome of most chickens and can be inherited as normal cellular genes [21]. Since ALVE were first identified in chicken cells as ALV group-specific antigens [22], more than 22 ALVE loci have been defined in the genome of White Leghorn chickens (Crittenden, 1991). Some ALVE are actively transcribed from their inherited chromosomal locations, whereas others (e.g., ALVE1) are silent [23]. ALVEs relevant to this study are ALVE1, ALVE2 and ALVE3. ALVE1 is located on chromosome 1, ALVE2 is located on chromosome 2, and ALVE3 is found on one of the many microchromosomes. Inbred line 63 has one copy of ALVE1 and ALVE3 whereas line 72 has one copy of ALVE1 and ALVE2 [24]. The cellular receptor of each ALV subgroup is encoded by a tumor virus locus (TV), which mediates viral entry. TVA and TVC encode the cellular receptors for ALV of subgroup A and C, respectively, TVB is the cellular receptor gene for ALV of subgroup B, D and E [25], [26].
The highly inbred line 63 at the Avian Disease and Oncology Laboratory (ADOL) are resistant to Marek's disease (MD) tumors but susceptible to both Marek's disease virus (MDV) and avian leukosis viruses (ALV), whereas the highly inbred line 72 chickens are resistant to ALV (subgroup A, B, D, and E) but susceptible to both MDV and MD tumors [27]. Therefore, these inbred lines constitute unique models for epigenetic research by making it possible to explore the mechanisms of resistance and susceptibility to neoplastic diseases [27], [28] .
In this research, we hypothesize that the methylation status of ALVE and TVB genes are associated with resistance and susceptibility to neoplastic disease. To examine the hypothesis, we did DNA methylation analysis by pyrosequencing, which was recently developed as a quantitative technique to detect changes in methylation patterns [29], [30]. This technique is advantageous for analyzing and quantifying the degree of methylation of multiple CpG sites in one reaction. In order to advance our understanding of genetic mechanisms and to develop a better strategy for disease prevention, we have investigated epigenetic differences in methylation patterns between lines 63 and 72, as well as six recombinant congenic strains (RCSs) that developed from background inbred line 63 and donor line 72. This unique model system provides a way to elucidate mechanisms that may induce susceptibility or enhance resistance to viral induced tumors.
We first analyzed variations of DNA methylation patterns in four consensus regions among ALVE1, 2 and 3, including one region unique for ALVE1 as well as the promoter region of the TVB gene in ALV-susceptible line 63 and ALV-resistant line 72 using pyrosequencing. Next, we investigated line-specific gene expression for ALVEs and TVB with real-time RT-PCR. Subsequently, we characterized potential differences in ALVEs PPT (polypurine tract) region and U3 region of 3′LTR between the lines by DNA sequencing. The effects of ALVEs and TVB methylation status on the tumor susceptible and resistant lines and six recombinant congenic strains are discussed. Our results suggest that variation in methylation patterns of ALVE, TVB and variations in the PPT region may be factors that contribute to viral induced tumors in chicken.
Results
Profiling DNA methylation patterns of ALVEs in two inbred chicken lines
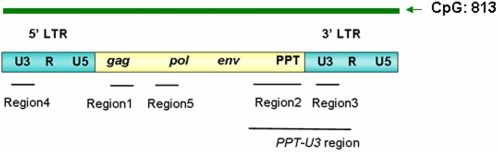
The whole ALVE1 is a CpG island that including 813 CpG sites within 7.5 kp based on the BLAT with UCSC Genomic Browser (Figure 1). To determine the DNA methylation levels of ALVEs, we selected four conserved regions among ALVE1, 2 and 3 that cover the gag gene, PPT region and U3 regions of 5′ and 3′ LTR, then quantitative evaluated the methylation differences between inbred chicken line 63 and line 72 using pyrosequencing method in four different tissues (blood, liver, spleen and hypothalamus) (Figure 1). Figure 2A, 2B and 2C show the typical methylation pyrograms for the six CpG sites in ALVE region 1 (p19 matrix protein in gag gene), the five CpG sites in ALVE region 2 (Direction Repeat Sequence, DRS, adjacent to 3′ LTR of ALVE), and the four CpG sites in ALVE region 3/4 (U3 region in 5′ and 3′ LTR of ALVE) in blood of line 72 and line 63, respectively. The averages from five individuals showed that hypermethylated CpG sites were identified in blood of line 72 , 85.56±2.66% for ALVE region 1 and 91.86±1.63% for ALVE region 2, whereas the same regions are hypomethylated (10.05±2.56%) and hemimethylated (46.16±3.41%) in the line 63 (Figure 2). The measurements using pyrosequencing technique for each conserved region of ALVE were proved to be highly reproducible between biological replicates. Moreover, the methylation percentages of region 2 in ALVE from bisulfite cloning and sequencing methods were very comparable to that from pyrosequencing as shown in Figure S2, which validated the pyrosequencing data.
Figure 1. Four conserved regions (region1∼4) on ALVEs and one unique region (region5) of ALVE1 for pyrosequencing and real-time RT-PCR.
5′LTR: 5′ long terminal repeat; U3: U3 region in LTR; R: R region in LTR; U5: U5 region in LTR; gag: gag gene; pol: pol gene; env: env gene; PPT: polypurine tract; 3′LTR: 3′ long terminal repeat of ALVE. PPT-U3 region: combined PPT and U3 regions for real-time RT-PCR. Green arrow shows the CpG islands that including 813 CpG sites within 7525 bp of ALVE1 (Blat result from UCSC Genome Browser website).
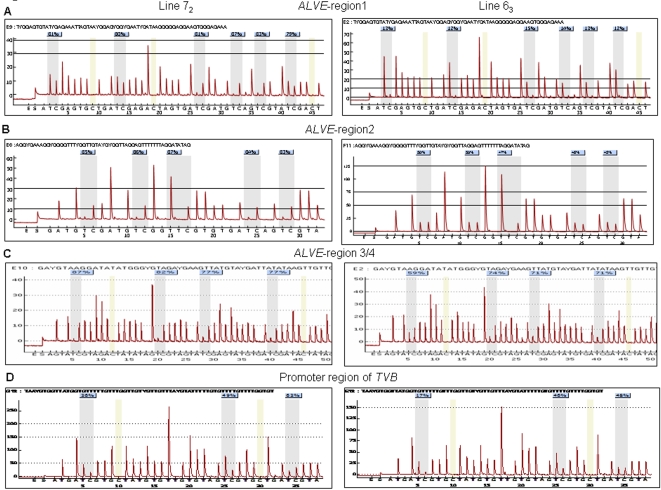
Figure 2. Representative pyrograms for each gene region in line 72 and 63.
The percentage on each CpG site is the methylation percentage of mC/(mC+C) on this site. mC: methylated cytosine, C: unmethylated cytosine. The sequence above the peak is the sequence to assay. The x axes are the dispensation nucleotides to the sequencing reaction based on the assayed sequences. The y axes show light emission obtained as relative light units. A: ALVE region1 in blood (the mean value of methylation percentage is 81.8±2.9 in line 72 and 13.2±1.2 in 63); B: ALVE region2 in blood (the mean value of methylation percentage is 85±1.6 in line 72 and 47.6±2.3 in 63); C: ALVE region3/4 in blood (the mean value of methylation percentage is 80.8±4.8 in line 72 and 68.8±6.7 in 63); D: Promoter region of TVB in Hypothalamus (the mean value of methylation percentage is 45.7±14.8 in line 72 and 37.0±14.1 in 63).
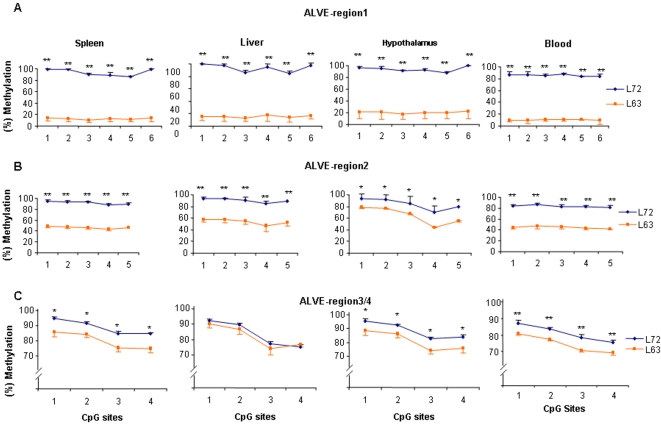
In order to compare the DNA methylation differences of the conserved ALVE regions between line 72 and line 63 in spleen, liver, hypothalamus and blood, point-wise comparison method was carried out at each CpG site. As depicted in Figure 3A and 3B and Table S1 and S2, we found that the methylation contents for each CpG site in ALVE region 1 and ALVE region 2 were very significantly higher in the four tissues in line 72 than in line 63 (P<0.0001), except for ALVE region 2 in hypothalamus (P<0.01). Hypermethylated patterns (methylation level >60%) in ALVEs region 1 and 2 existed in line 72, while line 63 showed a hypomethylated pattern (methylation level <30%) in ALVEs region 1 and a hemimethylated pattern (30%< methylation level <60%) in ALVEs region 2 except for a hypermethylated pattern in the hypothalamus. As for ALVEs region 3/4, although line 72 and line 63 had a hypermethylated status in the four tissues, the DNA methylation level was significantly higher in line 72 than in line 63 in spleen (P<0.01), hypothalamus (P<0.01) and blood (P<0.0001), but not in the liver (P>0.5) (Figure 3C and Table S3).
Figure 3. Results of quantitative DNA methylation analysis on conserved regions of ALVEs in spleen, liver, hypothalamus and blood.
**P<0.0001. *P<0.01. A: Methylation levels for each CpG site in the ALVE region1. B: Methylation levels for each CpG site in the ALVE region2. C: Methylation levels for each CpG site in the ALVE region3 and 4. n = 5 for each line.
mRNA expression level of ALVEs
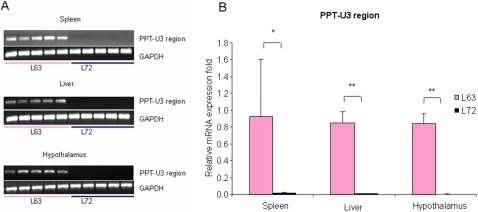
There is a typical retroviral polypurine tract (PPT) adjacent to the U3 region of the 3′ LTR in ALVEs (Figure 1). The PPT is a short RNA sequence that generally serves as a primer for plus-strand DNA synthesis during reverse transcription and initiation of DNA synthesis, then, the PPT primer is accurately removed from nascent DNA to create a double-stranded, integration-competent DNA provirus. Due to the importance of PPT-U3 region, real-time quantitative RT-PCR on the PPT-U3 region was used to examine the transcriptional activation level of ALVEs. As shown in Figure 4A and 4B, RNA expression levels of the PPT-U3 region were significantly higher in line 63 than in line 72 in the spleen (P<0.01), liver and hypothalamus (P<0.0001). There was no significant difference among tissues within either line (P>0.05).
Figure 4. mRNA expression of PPT- U3 region on 3′ ALVE in line 63 and line 72.
A: RT-PCR results. B: Real-time quantitative RT-PCR analysis. n = 5 for each line. *P = 0.01; ** P<0.0001.
Variation of Polypurine Tract (PPT) Site among ALVEs
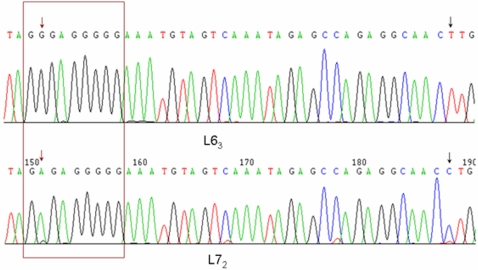
To further ascertain the genetic background of the two inbred lines and explore the potential mechanisms of methylation content influencing the mRNA expression levels, we did sequencing analysis for the PPT-U3 region in ALVE as shown in Figure 5. We found two variation sites in the two lines. One variation located in the PPT site changed from guanine (GGGAGGGGG) in line 63 to adenine (GAGAGGGGG) in line 72, and another variation mutated from thymidine (T) in line 63 to cytosine (C) in line 72. Based on the distinct feature for ALVE in the two lines, it shows that the sequences of the PPT site for ALVE1 and ALVE3 are GGGAGGGGG (upper panel in Figure 5), and that of the PPT for ALVE2 is GAGAGGGGG (lower panel in Figure 5).
Figure 5. DNA sequencing of PPT region and partial U3 region of 3′ LTR in ALVE between line 63 and line 72.
Brown box shows the PPT region. Brown arrow shows one variation located on PPT region changed from G in line 63 to A in line 72. Black arrow shows another variation located in U3 region of 3′ LTR changed from T in line 63 to C in line 72. n = 11 for each line.
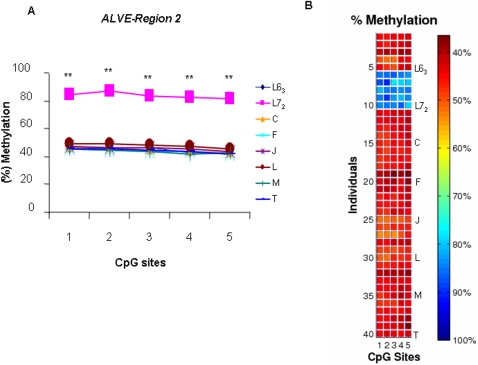
DNA methylation analysis of ALVEs region 2 in inbred lines and recombinant congenic strains
Due to the importance of PPT site in ALVEs region 2, the quantitative measurements of methylation level in the region 2 enable monitoring of epigenetic inheritance or non-inheritance of parental methylation patterns of ALVEs and help us to uncover the inner mechanisms between ALVEs and susceptibility or resistance of neoplastic diseases in the unique population. There are 19 recombinant congenic strains (RCSs) established by a cross between the two inbred line 72 and line 63. The F1 was then consecutively backcrossed to the background line 63 twice. After more than 13 generations of sib-matings within strain, each RCS is expected to contain random 7/8 background line 63 and 1/8 donor line 72 genome. We measured the methylation status of the ALVE-region 2 in blood in the two parental lines and 6 randomly selected RCS C, F, J, L, M and T. We found that the methylation patterns can easily be categorized into two groups, as exemplified in Figure 6A and 6B. One of the categories is extremely similar within the six RCSs and is also similar to line 63 than to line 72 (P<0.0001). Specifically, the methylation patterns of six RCSs can be acknowledged and were inherited from the background line 63, but not from the donor line 72. The similarity of the methylation patterns of region 2 between RCSs and background line 63 is mainly a result of the two backcrosses to line 63, and it could be a potential biomarker to predict susceptibility of neoplastic disease in chickens.
Figure 6. Methylation profiles of ALVE-region 2 in chicken inbred lines 63 and 72 and six recombinant congenic strains C, F, J, L, M and T.
A. Statistical analysis results. ** P<0.0001. B. The methylation levels are displayed in the form of a matrix. The matrix contains the data obtained form all the samples. n = 5 for each line or RCS.
DNA methylation patterns of the TVB promoter region
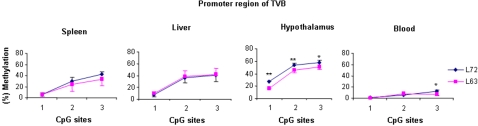
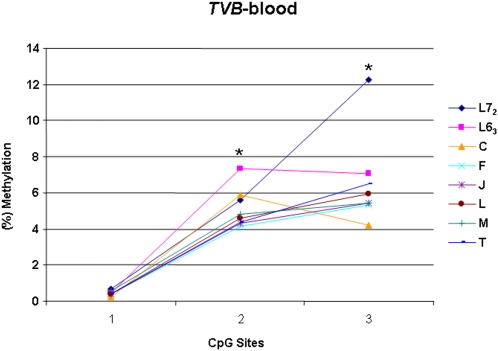
The tumor virus B (TVB) locus is an important gene encoding the cellular receptor of avian leukosis virus, different TVB alleles can encode different receptors permitting infection by exogenous ALVB, or ALVD and endogenous ALVE. To uncover epigenetics factors contributing to susceptibility or resistance of neoplastic disease in chickens, we examined the DNA methylation status of the TVB gene. Figure 2D shows the presentative pyrograms for three CpG sites in the promoter region of the TVB gene in the hypothalamus tissue of both lines. The results show that the methylation level of TVB promoter was significantly higher in line 72 than that in line 63 (P<0.0001, Figure 7). Unlike the consensus methylation trends of the ALVEs regions 1 and 2 (Figure 3A and 3B), the promoter region of TVB shows tissue-specific methylation patterns (Figure 7) similar to the promoter region 3/4 in ALVEs (Figure 3C). As shown in Figure 7, CpG site 3 in the promoter region of TVB showed a hemimethylated pattern in the spleen, liver and hypothalamus in both lines, whereas a hypomethylated pattern was disclosed in the blood. In addition, CpG site 3 of TVB in blood was significantly higher in line 72 than in line 63 (P<0.05). We found some variations regarding methylation contents among the 6 RCSs, especially for CpG sites 2 and 3, although the methylation patterns of RCSs were similar to background line 63 (Figure 8).
Figure 7. Results of quantitative DNA methylation analysis of inbred lines 63 and 72 in promoter region of TVB in spleen, liver, hypothalamus and blood.
n = 5 for each line. **P<0.0001, * P<0.05.
Figure 8. Results of quantitative DNA methylation analysis in promoter region of TVB in blood.
Line 63 and 72 are two parental lines. C, F, J, L, M and T are six recombinant congenic strains (RCS) chicken. n = 5 for each line and RCS. *P<0.05.
mRNA expression level of TVB
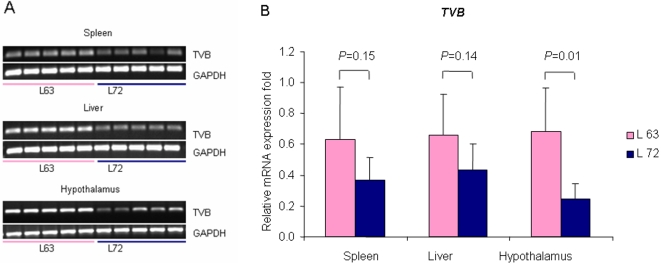
The mRNA expression level of the TVB gene in different tissues between the line 63 and line 72 was further examined with real-time quantitative PCR. The results were shown in Figure 9A and 9B. We found that, in general, the mRNA expression levels of the TVB gene were higher in the liver, spleen and hypothalamus in line 63 than in line 72. However, this difference between the two lines was only statistically significant in the hypothalamus (P<0.05), differences were not significant in the spleen (P = 0.15) or the liver (P = 0.14) (Figure 9B).
Figure 9. mRNA expression difference of TVB in line 63 and line 72.
P values indicates the statistical significance for the differences of relative TVB mRNA expression levels between lines 63 and 72 in spleen, liver, and hypothalamus, respectively. A: RT-PCR results. B: Real-time quantitative RT-PCR analysis. n = 5 for each line.
Association analysis between mRNA expression levels and DNA methylation contents
To further explore variation resources and clarify the association between mRNA expression and DNA methylation levels, two generalized linear models were used to quantitatively evaluate the effects of lines, tissues, and the methylation contents of ALVEs on the expression levels of ALVEs PPT-U3 region in ALVE and the methylation contents of TVB on its expression levels. Statistical analysis revealed that the effect of chicken inbred lines on the mRNA expression levels of the PPT-U3 region in 3′ ALVE was statistically significant (P<0.0001) (the first model). However, the effect of tissue on the mRNA expression of the PPT-U3 region in ALVE was not statistically significant (P>0.05). A further analysis of the line-specific results indicated that the mRNA expression level of PPT-U3 region in ALVE was significantly higher in line 63 than in line 72. Subsequently, we analyzed the association between mRNA expression level of the PPT-U3 region in ALVE and DNA methylation level of the ALVE region 2 using the second model. It shows that the effect of DNA methylation level on the mRNA expression level of PPT-U3 region in ALVE was statistically significant (P<0.0001).
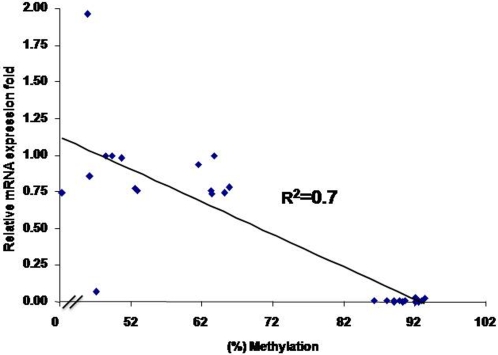
Thereafter, we explored the relationship between DNA methylation level of TVB and mRNA expression level. There was a negative effect of methylation levels of the TVB promoter region on its mRNA expression from hypothalamus (P<0.05). Finally, a regression analysis was done for exploring the relationship between DNA methylation level of the region 2 in ALVE and RNA expression level of the PPT-U3 region in ALVE. As shown in Figure 10, there was a higher negative relationship (R2 = 0.7) between them. Figure S3A and S3B also showed a negative relationship between mRNA expression level of ALVE PPT-U3 region and methylation level of ALVE-region 1 (R2 = 0.7) or region 3/4 (R2 = 0.55). Taken together, we found that the higher the methylation levels, the lower the mRNA expression level for PPT-U3 region of ALVE in all tissues (P<0.0001) and for the TVB gene in only the hypothalamus (P<0.05).
Figure 10. Regression analysis of mRNA expression level of PPT-U3 region of ALVE and DNA methylation contents of ALVE region2.
Discussion
In this study, we observed distinct DNA methylation patterns of avian endogenous viruses (ALVEs) between the two inbred chicken lines, and also analyzed the DNA methylation pattern variations of ALVE and TVB genes. To the best of our knowledge, this is the first study in chickens to elucidate the variation of DNA methylation patterns variations in ALVEs.
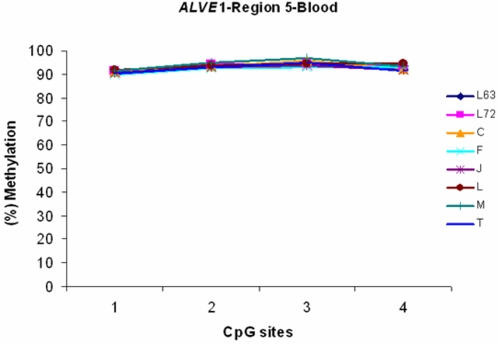
The previous studies have reported that the line 63 contains ALVE1 and ALVE3, while line 72 possesses ALVE1 and ALVE2 in the genome respectively [16], [31], [32], [33]. The reports were confirmed in our study as shown in Figure S1. Notably, we identified a unique region in ALVE1 (named as region5, Figure 1) with four CpG sites and examined methylation status of the region in blood among the inbred line 63 and 72 and 6 RCSs as shown in Figure 11. We found that both inbred lines and six RCSs have nearly the same hypermethylation patterns. Therefore, our results not only provide further evidence and strong support for the previous finding that the complete ALVE1 exists in both lines, but also indicate that ALVE1 in the two lines may not contribute to the unique features of resistant and susceptibility of neoplastic diseases and possess inhibitory methylation [32], [33]. Since ALVE1 did not have methylation differences between the both lines, any DNA methylation differences in conserved ALVE regions between line 72 and line 63 must be attributed to ALVE2 and ALVE3, respectively.
Figure 11. The results of DNA methylation analysis in a unique region of ALVE1 from blood in line 72, line 63 and RCS C, F, J, L, M and T blood.
n = 5 for each line or strain. P>0.05.
In vitro and in vivo studies in mammals have shown that retrotransposons are generally hypermethylated in normal tissues, but are hypomethylated in malignant tissues [11], [34], [35], [36], [37], [38], [39]. ALVEs belong to LTR-retrotransposons in chicken genome. In this study, the methylation level of ALVE conserved regions in line 72 was nearly twice to six times as high as that observed in line 63, which is consistent with the hypermethylated retrotransposon frequently found in normal tissues of mammals [11], [40], [41]. The methylation variations detected in this study were large in size and probably represent a strong link between DNA hypermethylation pattern of ALVE and resistance to ALV in the chicken. Based on association analysis, an up-regulating effect of hypomethylation in ALVEs region 1 and 2 was observed on ALVE PPT-U3 expression in line 63, which was in contrast to a down-regulating effect of hypermethylation in line 72. However, a directly transcriptional repression effect of ALVEs DNA methylation on gene expression level remains to be characterized.
Notably, the methylation pattern of ALVE-region 2, which possesses the primer PPT, could be an epigenetic biomarker and has an important potential value in prevention of chicken neoplastic disease. In our study, the similarity of the methylation pattern in ALVE region 2 between the RCSs and background line 63 is mainly because of the two continuative backcrosses to line 63. Based on sequencing analysis, we found that the 6 RCSs and background line 63 have the exactly same sequences on this region without any alterations (Unpublished data). On the other hand, although the methylation patterns of 6 RCSs in this region are similar to their background line 63, there are some variations in the methylation patterns among the 6 RCSs. Furthermore, we found some phenotypic variations in susceptible ALV infections among 6 RCSs (Private communication, unpublished data). The epigenetic profiles of this region in RCSs were likely to be transmitted from the line 63, and they retained the parental methylation patterns after over 13 generations of sib-mating. These results suggest us to further build the association between the methylation patterns and the phenotypic variations, and explore the possibility of the region as a biomarker in resistant breeding.
The polypurine tract (PPT) of the long terminal repeat-retrotransposons and retroviruses is a short RNA sequence from which the second or plus-strand DNA synthesis is initiated [42]. The PPT sequence required for plus-strand initiation was the key step for the replication of retrotransposon and retroviruses [43]. Previous studies demonstrated that the mutation in both the 5′ and 3′ halves of the human immunodeficiency virus type 1 (HIV-1) PPT has an effect on virus replication and titer [44], [45]. One mutation discovered in the PPT of 3′ ALVEs in our study was involved a change from a guanine (GGG) in line 63 to an adenine (GAG) in line 72 (Figure 5). Further study in vitro is to examine the function of the mutation in the PPT of chicken ALVEs on neoplastic disease.
The TVB locus, a single copy located on chromosome 22, is very complex and has three alleles encoding different receptors to accommodate the viral entry of different subgroups [46]. The receptors encoded by two susceptible alleles TVB*S1 and TVB*S3 support viral entries of ALVB, ALVD and ALVE subgroups. TVB*R is the resistant allele and encodes no functional receptors. Previous researches found that the ALVE susceptibility between line 63 and line 72 should be dependent on allelic differences at the receptor TVB locus (line 63 is susceptibility SNP TVB-S and line 72 is resistant SNP TVB-R) [33], [47]. Our study showed that the methylation contents of TVB were moderately higher in line 72 than in line 63, and the mRNA expression levels were reversely lower in line 72 than in line 63. It is worth noting that we found a variation appeared coalescence with of resistance to ALVA and ALVB in the 6 RCSs, although they have the same susceptibility SNP TVB-S as their background line 63 (Unpublished data). In this study, the methylation profiles of TVB promoter in six RCSs were much close to background line 63 than to line 72. However, some differences in the CpG site 2 and 3 among the six RCSs chickens were identified (Figure 8) and they may contribute to the varied ALVA or ALVB induced tumor incidences among the six RCSs in addition to the TVA genotype variations.
In conclusion, our data define the consensus CpG sites methylation patterns of the conserved ALVE regions in the ALV-resistant line 72 and -susceptible line 63 are attributed to ALVE2 and ALVE3, respectively. The results disclose that the mRNA expression levels of PPT-U3 in ALVEs and TVB gene are negatively associated with the CpG methylation status in the primer region of ALVEs and the promoter region of TVB, which suggest that the hypermethylaion profiles may contribute to ALV resistance in the chickens. The ALVE-region2, the PPT located region, could be considered as an epigenetic biomarker for resistant breeding against neoplastic disease in chicken.
Materials and Methods
Animal Samples
All samples were collected from highly inbred chickens of lines 63, 72, and recombinant congenic strains (RCS). Five 15 month old females from chicken line 63 and line 72, and five 12 month old females from line 63, 72 and six RCSs (C, F, J, L M and T) were tested. Heparinized blood was collected from each chicken prior to euthanasia. Then tissue samples from 15 month-olds chickens were obtained from three organs: hypothalamus, liver and spleen. Tissues were frozen in liquid nitrogen prior to storage at −80°C.
DNA extraction and bisulfite treatment
DNA was extracted from 20 µl blood or 3 mm3 tissue samples using a phenol-chloroform method. DNA concentration was measured by a spectrophotometer (Bio-Rad). 1 µg DNA of each sample was treated with bisulfite with EZ DNA Methylation Golden Kit (ZYMO Research) as the manufacture's protocols. Bisulfite converted DNA was eluted in 20 µl elution buffer (ZYMO Research).
PCR and pyrosequencing primers
PCR and pyrosequencing primers were designed to amplify 3∼6 CpG dinucleotides sites in each gene region, including four conserved regions among ALVE1, ALVE2 and ALVE3, a unique region in ALVE1 [48], and one promoter region of TVB gene (Figure 1 and Table 1). Forward and reverse primers used in PCR and the sequencing primers used in pyrosequencing methylation assays were designed with PSQ Assay Design software (Biotage, Swedan). To save time and cost, a biotin labeled universal primer (5′-GGGACACCGCTGATCGTTTA-3′) was used in the PCR assays [29]. The 5′ end of each reverse primer was tailed with the same sequence as the universal primer (Table 1).
Table 1. PCR and pyrosequencing primers and assays for each gene.
| GeneA | Assay | CpG sitesB | Primers | SequenceC |
| 6: 882 | 892 | Forward | 5′-TTAGGGGGAGGGAGGGTTT-3′ | |
| ALVE-region1 | 905 | 911 | Reverse | 5′-GGGACACCGCTGATCGTTTA CCATCTTCACATCTCGCTACACAA-3′ |
| 914 | 919 | Sequencing | 5′-GAGGGTTTTTTTTTTAGGT-3′ | |
| Assay | 5′-TYGGAGTGTA TYGAGAAATT AGTAAYGGAG YGGYGAATYG-3′ | |||
| 5: 7171 | 7178 | Forward | 5′-GTATATGGGTGGTGGTATGAAATTTG-3′ | |
| ALVE-region2 | 7186 | 7194 | Reverse | 5′-GGGACACCGCTGATCGTTTA TTCCCCCTCCCTATACAAAAAC-3′ |
| 7196 | Sequencing | 5′-GAGGGGATTATAGTATGTAT-3′ | ||
| Assay | 5′-AGGYGAAAGG YGGGGTTTYG GTTGTAYGYG GTTAGGAGTT-3′ | |||
| 3′ LTR | 5′LTR | Forward | 5′-TGGYGATTAGATAAGGAAGGAATG-3′ | |
| ALVE -region3/4 | 4: 7359 | 4: 108 | Reverse | 5′-GGGACACCGCTGATCGTTTA TATCCATCTACCCAAATACACACCA-3′ |
| 7375 | 124 | Sequencing | 5′-YGATTAGATAAGGAAGGAAT-3′ | |
| 7381 | 130 | Assay | 5′-GAYGTAAGGA TATATGGGYG TAGAYGAAGT TATGTAYGATTATATAAGTT-3′ | |
| 7393 | 142 | |||
| 4: 3793 | Forward | 5′-AGGCGTTTATTGTTTGGTTAGAAG-3′ | ||
| ALVE -region5 | 3795 | Reverse | 5′-GGGACACCGCTGATCGTTTA CAAAAAAATATCAACCTCCTTACC-3′ | |
| 3800 | Sequencing | 5′-TTATTTTTTTGATTATTAAG-3′ | ||
| 3808 | Assay | 5′-TTAYGYGTTT YGGTAGTGYG AATTTTTGGT AAGGAGGTTG ATAT-3′ | ||
| Forward | 5′-ATGTGTAGGTTATGGGAAGGGTAT-3′ | |||
| TVB | 3: 1282407 | Reverse | 5′-GGGACACCGCTGATCGTTTA AAAACTAAACTACTCCCACCATTT-3′ | |
| 1282435 | Sequencing | 5′-GGTTATGGGAAGGGTA-3′ | ||
| 1282444 | Assay | 5′-TAAYGTGGTT ATGGTGTTTT TGTTTGGTTG TYGTTGTTTA YGTA-3′ | ||
| Universal | 5′-/Biotin labeled/GGGACACCGCTGATCGTTTA-3′ | |||
ALVE-region1, 2, 3/4: based on the conserved DNA sequence between ALVE1 (AY013303) and ALVE3 (AY013304). The relative product of ALVE region1 is p19 matrix protein of gag gene. ALVE region2 is the consensus sequence of oncogene Direction Repeat Sequence (DRS). ALVE region3/4 is the conserved DNA sequence-U3 region, which located in 5′LTR and 3′LTR of ALVEs. ALVE region5: based on the unique DNA sequence (reverse transcriptase alpha subunit) in ALVE1 (AY013303,[48]. TVB: based on the UCSC DNA sequence (May 2006, Chr22) that BLAT from TVB cDNA sequence (AF161712), it is on the promoter region of TVB.
The CpG site numbers and positions.
Y and R stand for C/T and G/A, respectively. Bold Y is the CpG sites assayed in each region.
Hot start PCR amplification
The hot start PCR was carried out in 30 µl solution for ALVE and TVB genes: 1.5 µl bisulfite treated DNA (1∶5 dilution), 1×PCR buffer, 0.2 mM dNTPs, 0.5 µM forward primer, 0.05 µM reverse primer with universal tail, 0.45 µM biotin labeled universal primer, and 0.75 U Qiagen's Hotstar Taq DNA polymerase. PCR cycling conditions were 95°C for 15 min, followed by 50 cycles at 94°C for 30 sec, 50∼60°C for 45 sec, and 72°C for 45 sec, and a final incubation at 72°C for 10 min. PCR product quality verification was defined using 1.5% agarose gels with ethidium bromide.
Pyrosequencing methylation analysis
Based on the concentration of the PCR product, 10∼25 µl PCR product was used for each pyrosequencing reaction. Pyrosequencing methylation analysis was carried out using the Pyro Q-CpG system (PyroMark ID, Biotage, Sweden) according to the manufacture's protocol. In brief, the PCR product was bound to Streptavidin coated Sepharose beads (GE Healthcare Bio-sciences AB, Sweden). The Sepharose beads containing the immobilized PCR product were purified in 70% ethanol for 5 sec, denatured in Denature buffer (Biotage) for 5 sec, and washed with Washing buffer (Biotage) for 10 sec using the pyrosequencing Vacuum Prep Tool (Biotage). Then, 0.5 µM sequence primer was annealed to the purified single-stranded PCR product and pyrosequencing was carried out using the Pyro Q-CpG system. The level of methylation was expressed for each cytosine locus on CpG sites as the percentage of mC/(mC+C) (Figure 2, mC is methylated cytosine, C is unmethylated cytosine). Non-CpG cytosine residues were used as controls to verify bisulfite conversion.
Real-time RT-PCR
Total RNA of 5 individuals from each line was extracted from liver, spleen and hypothalamus using an RNeasy Midi kit (Qiagen). The first strand cDNA was synthesized from total RNA using SuperScript™ III Reverse Transcriptase (Invitrogen). Samples were then analyzed by real time RT-PCR using an iCycler iQ PCR system (Bio-Rad). The real time RT-PCR reactions were performed in a final volume of 20 µl with a QuantiTect SYBR Green PCR Kit (Qiagen) according to the manufacture's instructions. The mRNA expression of PPT (Polypurine tract) with U3 region (termed as PPT-U3) on 3′ LTR of ALVE and TVB was normalized against the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA in the corresponding samples.
Statistical analysis
Statistical analyses were conducted with the SAS 9.1.3 package. Point-wise comparison was carried out to analyze the difference of methylation contents between two lines at different CpG sites. Student's t test was done for analyzing mRNA expression levels of the PPT-U3 region in 3′-ALVE between line 63 and line 72. The GLM (generalized linear model) program was used to analyze the association between the RNA expression levels and lines, tissues or DNA methylation levels. The first model is: y = μ+l+t+e, where y is the RNA expression level of PPT-U3 region in 3′LTR of ALVE, µ is the overall mean, l is the class of line (Line 63 and Line 72), t is the type of tissue (liver, spleen or hypothalamus), and e is residual effect. The second model is: y = μ+m+t+e, where y, µ , t and e have the same meaning as that in the first model, and m is the DNA methylation level of ALVE region 2 or the promoter region in TVB, respectively. We also did the regression analysis of DNA methylation contents of ALVE region 1, 2 and 3/4 on mRNA expression level of the PPT-U3 region in ALVEs.
Supporting Information
The methylation percentage (%) of ALVE-region1 in line 63 and line 72
(0.03 MB TIF)
The methylation percentage (%) of ALVE-region2 in line 63 and line 72
(0.03 MB DOC)
The methylation percentage (%) of ALVE-region3/4 in line 63 and line 72
(0.03 MB DOC)
PCR diagnostics for ALVE1, ALVE2 and ALVE3 in line 63 and line 72. n = 3 for each line. L63: line 63; L72: line 72. M: 100 bp markers. “-” is negative control. A. Left panel of Marker lane shows that line 63 and line 72 are all positive ALVE1 birds. Right panel of Marker lane shows that line 72 is ALVE2 positive birds, however, line 63 is ALVE2 negative birds. B. Line 63 is ALVE3 positive birds, and line 72 is ALVE3 negative birds.
(0.99 MB DOC)
Validation of pyrosequencing results by bisulfite cloning and sequencing methods. TA Cloning Kit (Invitrogen Inc.) was used in cloning. The sequencing was done by ABI 3730. Black dots show methylated CpG sites, while open dots show unmethylated CpG sites. The same bisulfite treated spleen DNA from line 72 (A) and line 63 (B) was tested with cloning and sequencing (right panel) and pyrosequencing (left panel).
(2.12 MB TIF)
Regression analysis of mRNA expression level of PPT-U3 region of ALVE and DNA methylation contents of ALVE region1 (Figure 3A) and ALVE region3/4 (Figure 3B).
(0.87 MB TIF)
Acknowledgments
We would like to thank Naili Liu for DNA cloning technical support, and Apratim Mitra, Laura Ellestad and Dongxia Chen for some drawing services, Mardi Byerly for sampling help.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Maryland State Fund
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Weber AP, Weber KH, I., Stadler MB, Ramos L, Carr KS, et al. SamplingDistribution, silencing potential and evolutionary impact of promoter DNA methylation in the Arabidopsis transcriptome with massively parallel pyrosequencinghuman genome. Plant PhysiolNat Genet. 2007;14439:32-42457-42466. [Google Scholar]
- 3.Jeltsch A, Friedrich T, Roth M. Kinetics of methylation and binding of DNA by the EcoRV adenine-N6 methyltransferase. J Mol Biol. 1998;275:747–758. doi: 10.1006/jmbi.1997.1492. [DOI] [PubMed] [Google Scholar]
- 4.Scholzova E, Malik R, Sevcik J, Kleibl Z. RNA regulation and cancer development. Cancer Lett. 2007;246:12–23. doi: 10.1016/j.canlet.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 6.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, et al. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 7.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 8.Davis TL, Tremblay KD, Bartolomei MS. Imprinted expression and methylation of the mouse H19 gene are conserved in extraembryonic lineages. Dev Genet. 1998;23:111–118. doi: 10.1002/(SICI)1520-6408(1998)23:2<111::AID-DVG3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Davis T, Kirk D, Rinaldi A, Burdon RH, Adams RL. Delayed methylation and the matrix bound DNA methylase. Biochem Biophys Res Commun. 1985;126:678–684. doi: 10.1016/0006-291x(85)90238-4. [DOI] [PubMed] [Google Scholar]
- 10.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2006 doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- 11.Davis CD, Uthus EO. DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 12.Waterland RA, Michels KB. Epigenetic Epidemiology of the Developmental Origins Hypothesis. Annu Rev Nutr. 2007 doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 13.Nair V. Evolution of Marek's disease – a paradigm for incessant race between the pathogen and the host. Vet J. 2005;170:175–183. doi: 10.1016/j.tvjl.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Fadly AM, Payne LN. Leukosis/sarcoma group. In: Saif YM, Barnes HJHJ, Glisson JR, Fadly AM, Mcdougald LR, editors. Disease of Poultry 11th end: 465–516. Ames, Iowa, USA: Iowa State Press, Blackwell Publishing Company; 2003. [Google Scholar]
- 15.Crittenden LB, Fadly AM. Responses of chickens lacking or expressing endogenous avian leukosis virus genes to infection with exogenous virus. Poult Sci. 1985;64:454–463. doi: 10.3382/ps.0640454. [DOI] [PubMed] [Google Scholar]
- 16.Crittenden LB, Smith EJ, Fadly AM. Influence of endogenous viral (ev) gene expression and strain of exogenous avian leukosis virus (ALV) on mortality and ALV infection and shedding in chickens. Avian Dis. 1984;28:1037–1056. [PubMed] [Google Scholar]
- 17.Crittenden LB, Hayward WS, Hanafusa H, Fadly AM. Induction of neoplasms by subgroup E recombinants of exogenous and endogenous avian retroviruses (Rous-associated virus type 60). J Virol. 1980;33:915–919. doi: 10.1128/jvi.33.2.915-919.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen BR, Skalka AM, Ju G. Endogenous avian retroviruses contain deficient promoter and leader sequences. Proc Natl Acad Sci U S A. 1983;80:2946–2950. doi: 10.1073/pnas.80.10.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crittenden LB, Astrin SM, Smith EJ. Independent segregation of ev 10 and ev 11, genetic loci for spontaneous production of endogenous avian retroviruses. Virology. 1983;129:514–516. doi: 10.1016/0042-6822(83)90192-7. [DOI] [PubMed] [Google Scholar]
- 20.Borisenko L. Avian endogenous retroviruses. Folia Biol (Praha) 2003;49:177–182. [PubMed] [Google Scholar]
- 21.Conklin KF. Activation of an endogenous retrovirus enhancer by insertion into a heterologous context. J Virol. 1991;65:2525–2532. doi: 10.1128/jvi.65.5.2525-2532.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crittenden LB, Smith EJ, Weiss RA, Sarma PS. Host gene control of endogenous avian leukosis virus production. Virology. 1974;57:128–138. doi: 10.1016/0042-6822(74)90114-7. [DOI] [PubMed] [Google Scholar]
- 23.Baker B, Robison H, Varmus HE, Bishop JM. Analysis of endogenous avian retrovirus DNA and RNA: viral and cellular determinants of retrovirus gene expression. Virology. 1981;114:8–22. doi: 10.1016/0042-6822(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 24.Crittenden LB. Retroviral Elements in the Genome of the Chickens: Implications for Poultry Genetics and Breeding. Critical Reviews in Poultry Biology. 1991;3:73–109. [Google Scholar]
- 25.Elleder D, Plachy J, Hejnar J, Geryk J, Svoboda J. Close linkage of genes encoding receptors for subgroups A and C of avian sarcoma/leucosis virus on chicken chromosome 28. Anim Genet. 2004;35:176–181. doi: 10.1111/j.1365-2052.2004.01118.x. [DOI] [PubMed] [Google Scholar]
- 26.Bates P, Young JA, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 27.Bacon LD, Hunt HD, Cheng HH. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult Sci. 2000;79:1082–1093. doi: 10.1093/ps/79.8.1082. [DOI] [PubMed] [Google Scholar]
- 28.Bacon LD, Ch'ng LK, Spencer J, Benedict AA, Fadly AM, et al. Tests of association of immunoglobulin allotype genes and viral oncogenesis in chickens. Immunogenetics. 1986;23:213–220. doi: 10.1007/BF00373015. [DOI] [PubMed] [Google Scholar]
- 29.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 30.Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing(r) technology. Methods Mol Biol. 2006;373:89–102. doi: 10.1385/1-59745-377-3:89. [DOI] [PubMed] [Google Scholar]
- 31.Bacon LD, Fulton JE, Kulkarni GB. Methods for evaluating and developing commercial chicken strains free of endogenous subgroup E avian leukosis virus. Avian Pathol. 2004;33:233–243. doi: 10.1080/0307943042000195731. [DOI] [PubMed] [Google Scholar]
- 32.Cooper GM, Silverman L. Linkage of the endogenous avian leukosis virus genome of virus-producing chicken cells to inhibitory cellular DNA sequences. Cell. 1978;15:573–577. doi: 10.1016/0092-8674(78)90025-9. [DOI] [PubMed] [Google Scholar]
- 33.Bacon LD, Hunt HD, Cheng HH. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult Sci. 2000;79:1082–1093. doi: 10.1093/ps/79.8.1082. [DOI] [PubMed] [Google Scholar]
- 34.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 35.Lubbert M, Oster W, Ludwig WD, Ganser A, Mertelsmann R, et al. A switch toward demethylation is associated with the expression of myeloperoxidase in acute myeloblastic and promyelocytic leukemias. Blood. 1992;80:2066–2073. [PubMed] [Google Scholar]
- 36.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conklin KF, Coffin JM, Robinson HL, Groudine M, Eisenman R. Role of methylation in the induced and spontaneous expression of the avian endogenous virus ev-1: DNA structure and gene products. Mol Cell Biol. 1982;2:638–652. doi: 10.1128/mcb.2.6.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 39.Feinberg AP. Cancer epigenetics takes center stage. Proc Natl Acad Sci U S A. 2001;98:392–394. doi: 10.1073/pnas.98.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 41.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 42.Rausch JW, Le Grice SF. Purine analog substitution of the HIV-1 polypurine tract primer defines regions controlling initiation of plus-strand DNA synthesis. Nucleic Acids Res. 2007;35:256–268. doi: 10.1093/nar/gkl909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atwood-Moore A, Ejebe K, Levin HL. Specific recognition and cleavage of the plus-strand primer by reverse transcriptase. J Virol. 2005;79:14863–14875. doi: 10.1128/JVI.79.23.14863-14875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McWilliams MJ, Julias JG, Sarafianos SG, Alvord WG, Arnold E, et al. Combining mutations in HIV-1 reverse transcriptase with mutations in the HIV-1 polypurine tract affects RNase H cleavages involved in PPT utilization. Virology. 2006;348:378–388. doi: 10.1016/j.virol.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 45.Miles LR, Agresta BE, Khan MB, Tang S, Levin JG, et al. Effect of polypurine tract (PPT) mutations on human immunodeficiency virus type 1 replication: a virus with a completely randomized PPT retains low infectivity. J Virol. 2005;79:6859–6867. doi: 10.1128/JVI.79.11.6859-6867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith EJ, Cheng HH. Mapping chicken genes using preferential amplification of specific alleles. Microb Comp Genomics. 1998;3:13–20. doi: 10.1089/omi.1.1998.3.13. [DOI] [PubMed] [Google Scholar]
- 47.Zhang HM, Bacon LD, Cheng HH, Hunt HD. Development and validation of a PCR-RFLP assay to evaluate TVB haplotypes coding receptors for subgroup B and subgroup E avian leukosis viruses in White Leghorns. Avian Pathol. 2005;34:324–331. doi: 10.1080/03079450500179491. [DOI] [PubMed] [Google Scholar]
- 48.Johnson JA, Heneine W. Characterization of endogenous avian leukosis viruses in chicken embryonic fibroblast substrates used in production of measles and mumps vaccines. J Virol. 2001;75:3605–3612. doi: 10.1128/JVI.75.8.3605-3612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The methylation percentage (%) of ALVE-region1 in line 63 and line 72
(0.03 MB TIF)
The methylation percentage (%) of ALVE-region2 in line 63 and line 72
(0.03 MB DOC)
The methylation percentage (%) of ALVE-region3/4 in line 63 and line 72
(0.03 MB DOC)
PCR diagnostics for ALVE1, ALVE2 and ALVE3 in line 63 and line 72. n = 3 for each line. L63: line 63; L72: line 72. M: 100 bp markers. “-” is negative control. A. Left panel of Marker lane shows that line 63 and line 72 are all positive ALVE1 birds. Right panel of Marker lane shows that line 72 is ALVE2 positive birds, however, line 63 is ALVE2 negative birds. B. Line 63 is ALVE3 positive birds, and line 72 is ALVE3 negative birds.
(0.99 MB DOC)
Validation of pyrosequencing results by bisulfite cloning and sequencing methods. TA Cloning Kit (Invitrogen Inc.) was used in cloning. The sequencing was done by ABI 3730. Black dots show methylated CpG sites, while open dots show unmethylated CpG sites. The same bisulfite treated spleen DNA from line 72 (A) and line 63 (B) was tested with cloning and sequencing (right panel) and pyrosequencing (left panel).
(2.12 MB TIF)
Regression analysis of mRNA expression level of PPT-U3 region of ALVE and DNA methylation contents of ALVE region1 (Figure 3A) and ALVE region3/4 (Figure 3B).
(0.87 MB TIF)