Abstract
Here we characterize the expression of the full system of genes which control the segmentation morphogenetic field of Drosophila at the protein level in one dimension. The data used for this characterization are quantitative with cellular resolution in space and about 6 min in time. We present the full quantitative profiles of all 14 segmentation genes which act before the onset of gastrulation. The expression patterns of these genes are first characterized in terms of their average or typical behavior. At this level, the expression of all of the genes has been integrated into a single atlas of gene expression in which the expression levels of all genes in each cell are specified. We show that expression domains do not arise synchronously, but rather each domain has its own specific dynamics of formation. Moreover, we show that the expression domains shift position in the direction of the cephalic furrow, such that domains in the anlage of the segmented germ band shift anteriorly while those in the presumptive head shift posteriorly. The expression atlas of integrated data is very close to the expression profiles of individual embryos during the latter part of the blastoderm stage. At earlier times gap gene domains show considerable variation in amplitude, and significant positional variability. Nevertheless, an average early gap domain is close to that of a median individual. In contrast, we show that there is a diversity of developmental trajectories among pair-rule genes at a variety of levels, including the order of domain formation and positional accuracy. We further show that this variation is dynamically reduced, or canalized, over time. As the first quantitatively characterized morphogenetic field, this system and its behavior constitute an extraordinarily rich set of materials for the study of canalization and embryonic regulation at the molecular level.
Keywords: Drosophila embryo, Segmentation genes, Blastoderm, Gene expression, Quantitative expression data, Positional information
Introduction
During Drosophila embryogenesis the segmented body plan is established through a cascade of maternally and zygotically expressed segmentation genes (reviewed in Akam, 1987; Ingham, 1988). The zygotic genes have been classified according to their mutant phenotypes and expression patterns. ‘Gap’ genes are expressed in one to three broad domains, ‘pair-rule’ genes initially form seven transverse stripes and ‘segment polarity’ genes manifest in patterns of fourteen stripes about one cell wide. Of these genes, maternal, gap, and pair-rule genes act during the blastoderm stage, giving rise to the initial expression of segment polarity genes at the onset of gastrulation, by which time the segmental pattern is determined (Simcox and Sang, 1983). This process is of general interest because it is perhaps the best characterized example of a “morphogenetic field” (Gilbert et al., 1996).
The morphogenetic field is a fundamental object in developmental biology. It was shown in the late 19th century that groups of cells underwent collective determination events (morphallaxis) in which cell fate was stably assigned to individual cells with exquisite spatial precision. Although early workers (Driesch, 1914) were pessimistic regarding the question of whether critical phenomena connected with determination could be understood on a chemical basis, the introduction of high-throughput technologies gives rise to optimism. Success in elucidating genomes, proteomes, and so on suggests the importance of understanding the “morphome”, by which we mean the complete set of determinants of a morphogenetic field. In general, the morphome will consist of a description of the quantities of morphogenetic determinants at a resolution in space and time sufficient to uniquely determine the biological trajectory of the system. Because of the central role of cells and their genomes, the information about the morphome must be of at least cellular resolution in space, must include the expression levels of all the genes encoding cell fate determinants, and must be of a time resolution shorter than the time in which significant changes in the levels of these determinants can take place.
In this paper we present a preliminary description of the morphome of segment determination in the Drosophila blastoderm in terms of the one-dimensional protein expression levels of segmentation genes.
During this period the embryo is syncytial and only a very limited number of zygotic genes are expressed. Saturation mutagenesis (Nüsslein-Volhard and Wieschaus, 1980) has identified most or all of the segmentation genes. Of these, only 14 act in the blastoderm as determinants of the segmentation morphogenetic field. All of these genes code for transcription factors (Rosenberg et al., 1986; Mohler et al., 1989; Nauber et al., 1988; Tautz et al., 1987; Pignoni et al., 1990; Driever and Nüsslein-Volhard, 1988; Macdonald et al., 1986; Kuroiwa et al., 1984; Ish-Horowicz et al., 1985; Gergen and A., 1988; Coulter et al., 1990; Frigerio et al., 1986; Benedyk et al., 1994; Grossniklaus et al., 1992). This fact, together with the syncytial nature of the blastoderm suggests that cell–cell communication by means of signaling pathways does not occur in the segmentation morphogenetic field, but rather that spatial interactions occur through diffusion of these transcription factors. Mechanical forces and cell migration appear to be uncoupled from the segment determination process as well, since mutations in segmentation genes do not affect morphology until after gastrulation. The segmental field stretches from 30% egg length (EL) from the anterior pole to about 93% EL. In this region genes controlling the anterior–posterior (A–P) axis are uncoupled from those controlling the dorso-ventral (D–V) axis, and hence a one-dimensional treatment is a good approximation. The description of the segmentation morphome given here is quantitative, at cellular spatial resolution, and has a temporal resolution of about 6 min. The treatment presented here is nevertheless preliminary as it does not consider the full three-dimensional blastoderm, signaling events in the terminal regions, or certain late acting segmentation genes.
In order to characterize the segmentation morphome, it is necessary to assemble the full time-dependent expression pattern from many individual embryos of different ages which have been stained for different combinations of proteins. We first characterize this typical, or integrated, pattern of gene expression for each of the segmentation genes. We demonstrate that domains form in a characteristic way over time, and in some cases shift position during the course of development. We also consider the question of how close the integrated patterns are to those of actual individuals. Such comparisons as well as comparisons between individuals give insight into the relationship between natural variation and the regulation, or error correction, properties of this morphogenetic field. Classically, regulation was assayed after surgical manipulation, but recent work has demonstrated that regulation can be observed in the variations of the levels of gene products from one individual to another without surgical manipulation (Houchmandzadeh et al., 2002; Spirov and Holloway, 2003b; Holloway et al., 2006).
In this work we show that there is substantial variation between individual expression patterns early, but this variation is canalized into highly uniform patterns by the onset of gastrulation. We further demonstrate that as early broad expression domains transform themselves into an increasing number of subdomains, the new subdomains show variational behavior that at early times is independent of that of other subdomains of the same gene. The data described here are accessible on the Internet database FlyEx (Poustelnikova et al., 2004, http://urchin.spbcas.ru/flyex or http://flyex.ams.sunysb.edu/flyex).
A large number of studies published for the most part in the late 1980s and early 1990s gave a good overall picture of segmentation gene expression (see Supplementary Information, Section 1 for a brief review). In certain cases, diagrams of the relative patterns of expression of several genes were produced for a particular region of blastoderm at a particular time. Nevertheless, three very basic questions remained unanswered. First, it is evident that determination proceeds using the cell (or nucleus) as a fundamental processing unit. Hence the biologically essential information is not the overall shape of an expression pattern, but rather what quantities of gene products are present in each cell. Second, the expression patterns are known to change in time, and hence it does not suffice to know the expression levels at any one time; it is necessary to characterize the entire time course. Finally, the differences between individuals must be categorized. We present partial answers to these questions in what follows.
Materials and methods
Quantitative gene expression data
Approximately 1600 embryos from Oregon-R flies were fixed and incubated with primary antibodies as described (Kosman et al., 1998), using sera raised against bacterially expressed protein products of bcd and cad; the gap genes Kr, kni, gt, hb, and tll; the pair-rule genes eve, ftz, h, run, odd, prd, and slp. All of these serums were raised by us as described (Kosman et al., 1998), except for rabbit anti-Eve, which was a generous gift of Manfred Frasch (Azpiazu and Frasch, 1993). Some embryos were stained with secondary sera conjugated to FITC, Texas Red, and Cy5 (Jackson Labs), and others with sera conjugated to Alexa Fluor 488, 555, 647, and 700 as described (Janssens et al., 2005).
Each embryo was stained for Eve protein and two other segmentation proteins. Approximately half of these were also counterstained with nuclear marker as described (Janssens et al., 2005), while in the remainder nuclei were detected by constructing a pixel maximum image from the three gene expression channels (Kosman et al., 1997). Embryos were imaged at stages ranging from cleavage cycle 10 (when proteins synthesized from maternal transcripts begin to appear) up to the onset of gastrulation. Embryos showing any morphological signs of gastrulation were not scanned. Only laterally oriented embryos were selected for scanning, although it is difficult to judge orientation in early cleavage cycles.
Quantitative confocal microscopy was performed as described (Kosman et al., 1997; Myasnikova et al., 2005; Janssens et al., 2005). Approximately half of the embryos were imaged with the 16× oil immersion plan objective of a Leica TCS4D confocal microscope (Kosman et al., 1997), and about half with a 20× Plan Apo dry objective of a Leica TCS SP2 confocal system (Janssens et al., 2005). The gain of the microscope photomultiplier is set for each channel by selecting an embryo exhibiting the spatial pattern characteristic of maximal expression and adjusting gain so that a few pixels are saturated. Offset for each channel is set by setting pixels away from the embryo equal to zero. This calibration is used for scanning all embryos on one slide and is highly reproducible between slides (Janssens et al., 2005). Quantification is relative with respect to maximum protein levels, so it is possible to make quantitative comparisons of the expression of one gene in different nuclei of the same embryo or between two different embryos stained in the same or different experiments, but it is not possible to quantitatively compare the expression levels of different proteins. Expression data were taken to 8-bit accuracy and hence vary between 0 and 255. Embryos were mounted in such a way that the curved surface of the blastoderm was flattened so that over a third of the embryo can be scanned for gene expression using only three optical sections separated by 1 μm.
Each image, containing three channels of expression data, was computationally transformed (“segmented”) into a text file as described (Janssens et al., 2005). Each embryo yields a single tabular ASCII file containing one line per segmented nucleus with about 2300 segmented nuclei per file for an embryo in cleavage cycle 14A. Each line of the file contains the identity number of the nucleus, the x and y coordinates of its centroid, and the average fluorescence level of each of three channels, and hence of three genes. The x and y axes are chosen such that they are tangent to the anterior and ventral sides respectively of an embryo image and hence the x axis corresponds to the anteroposterior (A–P) axis of the embryo and the y axis to the dorsal–ventral (D–V) axis. In the segmented data files, x and y coordinates are expressed as percent of the maximum size of the embryo in the x and y directions with 0% at the anterior pole and most ventral position respectively. This compensates for size differences from embryo to embryo. When necessary, physical coordinates can be regenerated from stored data.
We remove background from expression patterns using a method based on the observation that in our data the background signal is well fit by a very broad two-dimensional paraboloid. The paraboloid is determined from the area of the embryo in which a given gene is not expressed and then extrapolated throughout the entire embryo. Background is removed by a linear mapping of intensity that transforms fluorescence at or below background level to zero and transforms maximum fluorescence (255) to itself (Myasnikova et al., 2005).
Because we wish to reconstruct the time course of expression from individual fixed embryos, it is necessary to classify the embryos in terms of developmental age. We use a combination of methods to solve this problem. The cleavage cycle can be determined by noting the number of nuclei. This gives reasonable resolution for early cycles, as the duration of interphase during cleavage cycles 10–13 is only 6–14 min. Cleavage cycle 14A is about 50 min long and therefore during this cycle other morphological markers must be used for age detection.
We classified embryos in cycle 14A into 8 temporal equivalence classes on the basis of thorough visual inspection of the expression pattern of eve, which was scanned in all embryos (Myasnikova et al., 1999). The operational definition of a temporal equivalence class is that an experienced observer will always see a difference in expression pattern between two embryos in different temporal classes, but typically cannot distinguish an age difference in two embryos of the same class.
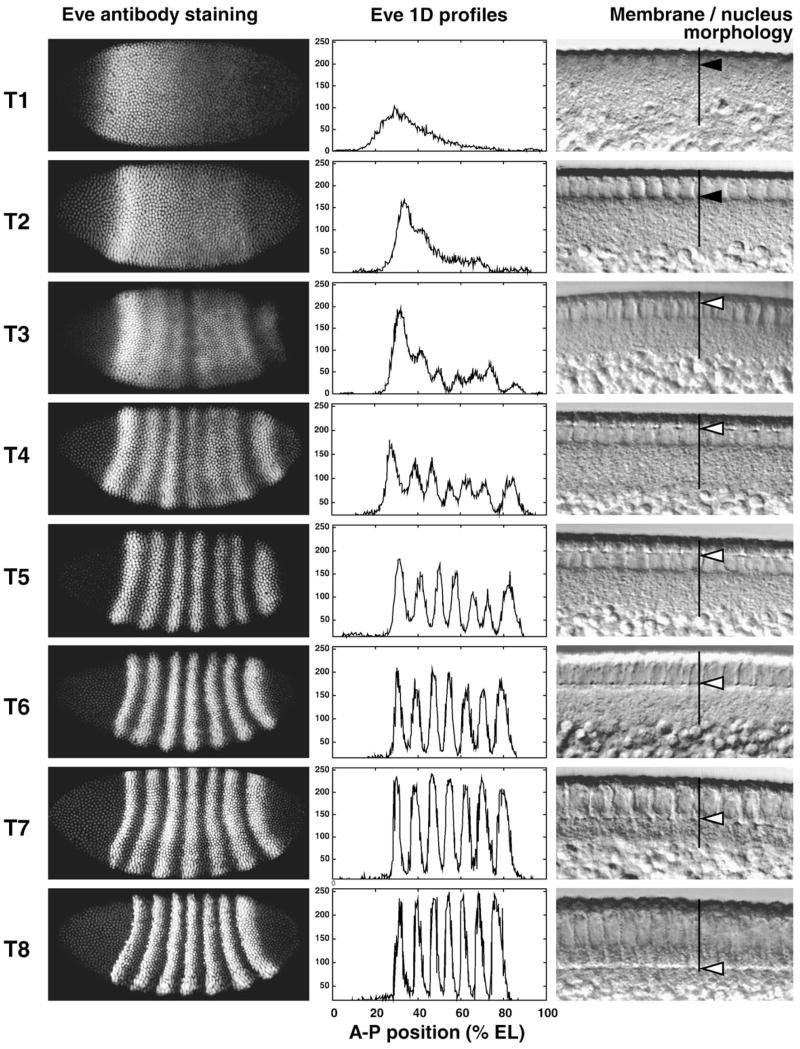
The 8 classes are approximately equally populated, and each class represents an age range of a little over 6 min. To confirm this classification, we made use of morphological features of the embryo that can be observed in both fixed and living tissue. Such features include the extent of membrane invagination and changes in nuclear shape. We confirmed the temporal classification by rephotographing 120 embryos from cycle 14A in Differential Interference Contrast (DIC) optics. We measured the length of membrane as percent cortex length in these photographs and compared this ratio with a published standard curve (Merrill et al., 1988). This showed that the gene expression based classification method was consistent with other methods and that the time classes were uniform in duration, as shown in Fig. 1. The Leica TCS SP2 is equipped with DIC, and we acquired blastoderm morphology images for all embryos scanned on this system.
Fig. 1.
The 8 temporal classes of cycle 14A. A typical embryo of each class is shown in each row. The left-hand panel shows an image of eve expression in that embryo; the middle panel shows the segmented expression pattern from the central 10% strip; the right-hand panel shows a high magnification DIC image of the blastoderm morphology. In the DIC images vertical black lines indicate the cortical cytoplasm, the black arrows in time classes 1 and 2 indicate the elongation of nuclei, and the white arrows in time classes 3–8 show the position of membrane front.
The in vivo studies reported here were conducted as follows. Embryos were dechorionated and mounted between Biofolie 25 semipermeable membranes (In Vitro Systems and Services Gmbh) with halocarbon oil 27 (Sigma). Embryos were imaged in a Leica TCS SP2 Confocal System using transmission mode with a 40× objective, 2.5× digital zoom and DIC optics. Images are 8-bit and 1024×1024-pixel resolution with averaging over 4 scans.
We wish to map the expression patterns of all genes in the morphome but can only image the products of three genes at once. However, each embryo is stained for eve expression, and by small coordinate transformations, it is possible to align expression patterns from multiple embryos so as create aligned patterns which can be averaged. This was usually done by aligning peaks and valleys of the eve pattern as described (Myasnikova et al., 2001). Embryos in the first temporal class of cleavage cycle 14A or younger are not susceptible to this method because of a lack of features and were not registered.
In the central area of the blastoderm that constitutes the presumptive segmented germ band, segmentation and homeotic genes expression is a function of A–P position in the embryonic coordinate system, and D–V gene expression is a function of the embryonic D–V coordinate. Because of the asymmetric curvature of the embryo, this coordinate system is orthogonal but curvilinear. Although the extent of the curvature is exaggerated by mechanical deformation in the embryos considered here, it is clearly seen in non-deformed whole mounts (J. Reinitz, unpublished observations) and in tissue sections hybridized to RNA in situ (Reinitz and Levine, 1990). Although we have developed methods for working with 2D data (Spirov et al., 2000, 2001; Kozlov et al., 2002), the essential biology of the segmentation genes is well represented in one dimension and hence it is sufficient to consider data extracted from the narrow strip of nuclei comprising the central 10% of the embryo in the D–V direction. This strip yields gene expression measurements from about 350 nuclei per embryo.
Finally, we obtain an integrated pattern for the full set of gene products by dividing this central 10% strip into A–P zones of 1% egg length (EL) which is very close to the observed average nuclear width of 0.97% EL. We take the average expression level of each gene in each A–P zone and consider this to be a one-dimensional nuclear model of an expression pattern. We take 0% EL to be at the anterior pole and 100% EL at the posterior pole, contrary to the normal convention, so that A–P position increases from left to right in graphs. We refer to these data as “one-dimensional integrated data” or “integrated data for the 10% strip” (Myasnikova et al., 2001).
Statistical analysis of gene expression data
The most natural way to study the dynamics of formation of segmentation gene expression domains is to describe these domains by a small number of characteristic features of the pattern and to monitor how these features change in time. We consider data in the 10% central strip along the A–P axis of an embryo in order to ensure a sufficient number of samples. As the characteristic features of segmentation gene expression domains we take the A–P positions of expression maxima as well as points where expression is a predefined percentage of maximum, typically 50% (Supplementary Fig. 1).
The characteristic features of expression domains of pair-rule genes were extracted using the fast dyadic wavelet transform (FRDWT) (Kozlov et al., 2000; Myasnikova et al., 2001), or in a small number of specific cases that are noted in the text by singular spectrum analysis (SSA; see below under Classification methods). FRDWT is good enough for accurate detection of the extremal points of the pattern. However the domains of gap genes and maternal gradients occupy larger territories than pair-rule gene expression domains and contain local spikes which do not correspond to domain maxima. To identify the most essential features of each domain and to eliminate noise on the curves, we approximate the gene expression pattern by quadratic splines, which provide a smooth approximation of the domains, and classifies each pattern by a set of features (Myasnikova et al., 1999, 2001). The quality of automatic extraction of features was checked by thorough visual inspection.
To validate the significance of temporal changes in the positions of expression domains (“shifts”), we performed ANOVA (StatSoft Statistica version 6.0) to test the differences between mean positions of characteristic features of expression domains in each time class for statistical significance at a confidence level of P=0.05. The positional variability of expression patterns was estimated by computing the standard deviations of the positions of characteristic features.
Classification methods
Individual eve patterns subjected to automated classification were treated as follows. First, the embryos were subjected to a coordinate transformation that straightens their stripes, as described (Spirov et al., 2000; Spirov and Holloway, 2003a). Further processing was performed on a strip containing the central 50% of D–V values, which captures the expression levels of about 1500 nuclei in cleavage cycle 14A. In order to classify expression variation among individual embryos in terms of the pattern over the whole A–P axis, the D–V coordinate of each nucleus was suppressed, and the resulting noisy pattern was smoothed by means of singular spectrum analysis (SSA) (Broomhead and King, 1986; Fraedrich, 1986; Vautard and Ghil, 1989). In SSA, a single 1D sequence of length N whose ith value is xi is considered as an M-dimensional series of length N−M+1 whose ith element is given by the M-tuple (xi−M+1, … xi−1, xi) (Elsner and Tsonis, 1996). M is called the “window length”. Principal component analysis is performed on this M-dimensional series, resolving the total variance into M components of which the first few terms contain most of the variance. The series can then be reconstructed from these resolved components. Much of the behavior of the series can be reconstructed from the first few terms, and such reconstruction was used here as an adaptive smoothing method (Golyandina et al., 2001). Initial studies (Holloway et al., 2006) were made with M=N/2 to determine a suitable noise model; here M is chosen small in light of that noise model to ensure proper smoothing.
In order to classify the patterns entirely on the basis on their intensity profiles, variations of intensity were eliminated by renormalizing intensity to 100, and patterns were spatially registered by placing the maximum of the SSA smoothed pattern at 35% EL classification was performed by the method of self-organizing maps (SOM) (Kohonen, 1990; Team, 1995; Kohonen, 1997; Tamayo et al., 1999). We chose the number of classes to best visualize the types of patterns seen in intensive visual inspection; classification was performed with SOM pak 3.1 (Team, 1995) with the number of trials set to 5, hexagonal topology, bubble neighborhood type, the training length, rate, and radius were equal to 500, 0.1, and 10 respectively in the first part and 104, 0.01, and 1 respectively in the second part.
Results
In what follows, we first present the results of a characterization of the expression pattern of each gap and maternal gene, together with qualitative aspects of variability in this system. We next consider the pair-rule genes, where the pattern of variability is considerably more complex. For the pair-rule gene eve, we present a full characterization of transitional patterns leading to the formation of stripes. This is followed by a quantitative characterization of domain shifts and variability.
Quantitative atlas of segmentation gene expression
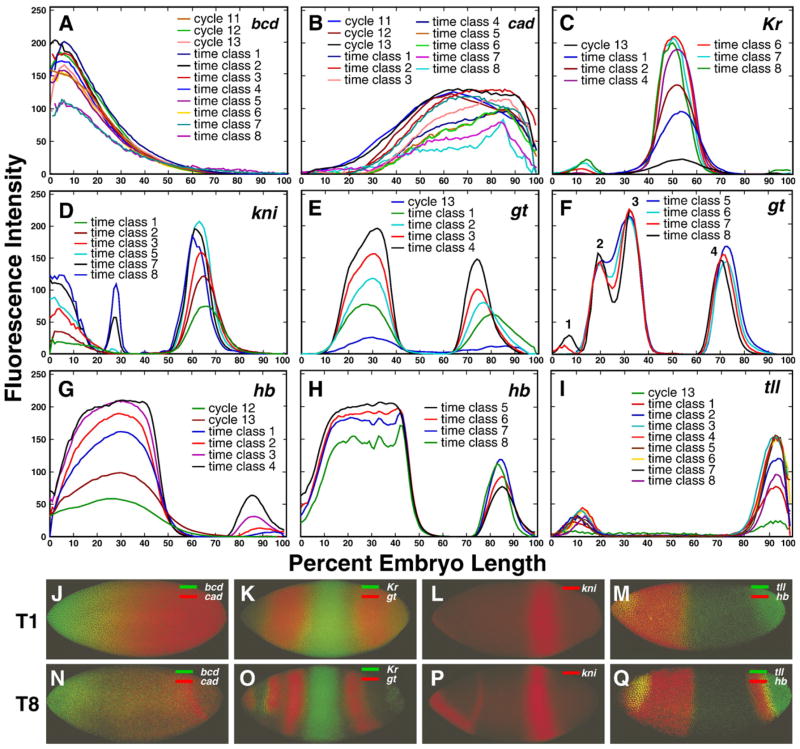
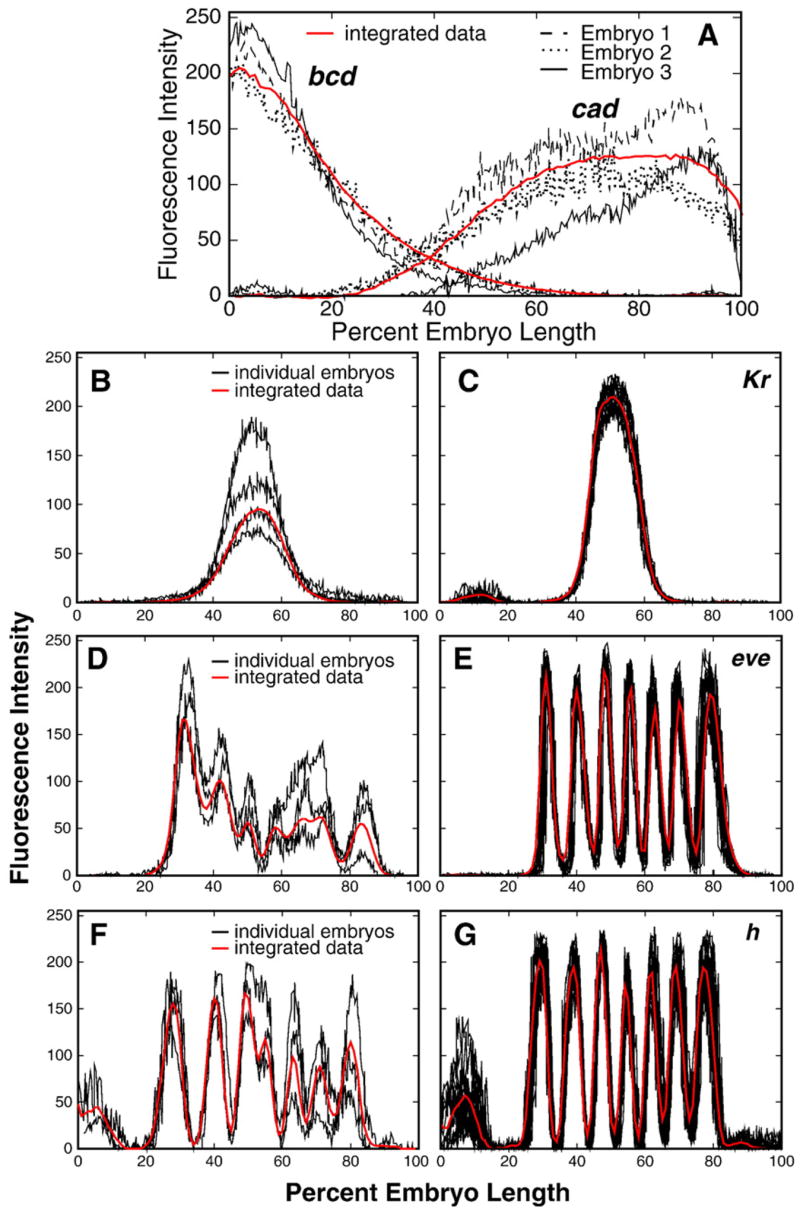
Integrated expression data for the maternal gene bcd and for maternal expression of hb and cad are presented in Figs. 2A, B, and G. Representative images of embryos are shown in panels J and N of this figure, although N exhibits zygotic expression of cad. bcd expression is relatively constant until near the end of the blastoderm stage, while the patterns of hb and cad are considerably more dynamic. Zygotic expression of hb and that from the zygotic gap genes Kr, kni, gt and tll is shown in Figs. 2C–I as integrated data, and in images of representative stained embryos in Figs. 2K–M and O–Q. Zygotic gap gene expression at the protein level is first detected, in localized domains, at cleavage cycle 12. Expression levels in most gap domains tend to increase until mid-cycle 14A and then decline (Supplementary Fig. 7). Note that in the posterior half of the embryo there is a tendency for domains to shift to the anterior over time, a point that we return to below. A full description of maternal and gap gene patterns is presented in Supplementary Information, Sections 2.1 and 2.2.
Fig. 2.
Temporal dynamics of expression of maternal and gap genes. The 1D integrated patterns of bcd (A), cad (B), Kr (C), kni (D), gt (E and F), hb (G and H) and tll (I) expression are shown for time points indicated in each panel. Expression domains within the gt expression pattern on panel F are numbered from anterior to posterior as indicated. Panels J–Q show representative confocal images of the expression of the genes shown in panels A–I in individual embryos belonging to temporal classes 1 (T1) and 8 (T8). Genes expressed in the same individual embryo are shown by different colors as given in the key for each panel.
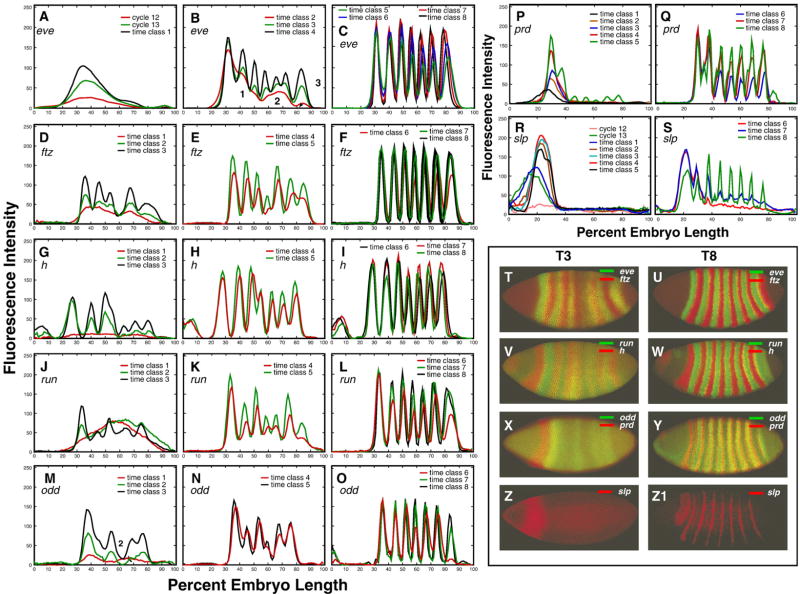
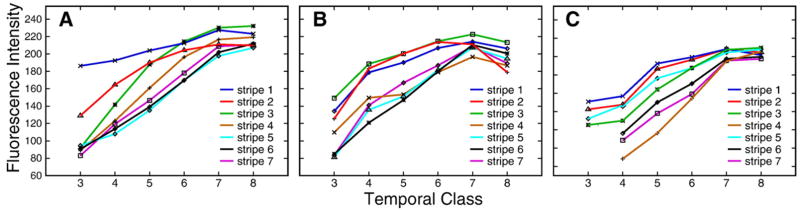
Integrated expression data for the pair-rule genes eve, h, run, ftz, odd, prd, and slp are presented in Figs. 3A–S, together with the expression patterns of these genes in representative individual embryos in time classes 3 (Figs. 3T, V, X and Z) and 8 (Figs. 3U, W, Y, and Z1). Pair-rule expression at the protein level is in general first detected at cycle 12 or 13. This early expression is very broad for eve, h, ftz, run, and odd, but it is restricted to a gap-like domain in the anterior in the case of prd and slp. Each pair-rule gene forms its domains in a characteristic manner, either by the splitting of a preexisting domain, by budding from the boundary of a preexisting domain, or by forming de novo (Supplementary Fig. 2). For eve, h, ftz, run, and odd the formation of new domains begins at time class 2, with stripe 1 tending to form first and stripe 4 last (Supplementary Fig. 30). The pair-rule stripes of prd and slp form significantly later, such that pair-rule stripes 3–7 in prd and 2–7 in slp, are initiated de novo in time classes 5 and 6, respectively (Figs. 3P, S). After pair-rule stripes are formed, most of them change their position in the course of cycle 14A (see below). Levels of pair-rule gene expression change in a characteristic manner for each gene. For example, all ftz stripes start to grow later than stripes of eve and h, while h stripes decrease in amplitude at the end of cycle 14A but ftz stripes do not (Fig. 8). The expression patterns for each pair-rule gene are characterized in detail in the Supplementary Information, Section 2.3.
Fig. 3.
Temporal dynamics of expression of pair-rule genes. The 1D integrated patterns of eve (A–C), ftz (D–F), h (G–I), run (J–L), odd (M–O), prd (P, Q) and slp (R–S) expression are shown for time points indicated in each panel. Numbers in panel B indicate the three transient domains of integrated eve expression during temporal class 2. Representative confocal images showing expression of these genes in individual embryos belonging to temporal classes 3 (T3) and 8 (T8) are shown in panels T–Z1. Genes expressed in the same individual embryo are shown by different colors as given in the key for each panel.
Fig. 8.
Temporal changes in the levels of expression of pair-rule genes. (A) eve, (B) h, (C) ftz; each curve corresponds to a particular stripe as shown in the key.
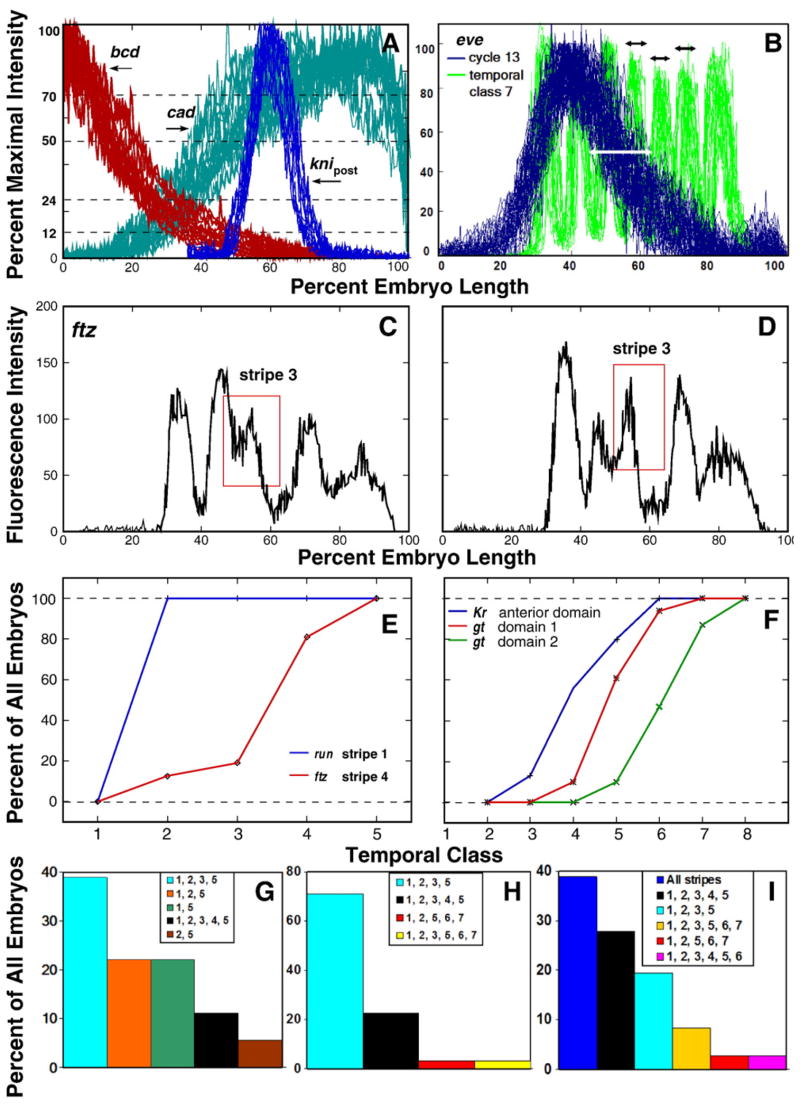
These data revealed certain overall patterns of variability. At a qualitative level, individual variation among maternal genes was large, with cad more variable than bcd (Fig. 4A). Among zygotically expressed genes there is an overall reduction in variability over time, such that individuals in early time classes may vary widely from the integrated pattern (Figs. 4B, D and F), but at late time classes individual patterns are very similar to one another and to the integrated pattern (Figs. 4C, E and G). Qualitatively, variability in gap gene expression tends to involve variations in amplitude, while variability in pair-rule expression appears to involve variability in the formation of new domains which will become stripes.
Fig. 4.
The variability of gene expression in individual embryos. bcd and cad patterns are shown in panel A for three individual embryos belonging to time class 2 and stained for the expression of both genes. Panels B, D and F show several of the most diverse patterns of early expression of Kr, eve and h in individual embryos from time classes 1, 3 and 4, respectively. The expression of Kr in time class 6 (C) eve (E) and h (G) in time class 7 is shown for a randomly chosen set of 13, 12 and 16 individual embryos respectively. For each gene the graph of the corresponding one-dimensional integrated data is shown in red.
These observations introduce an important question concerning the segmentation morphogenetic field. Variability in domain amplitude is compatible with a picture in which the field follows an essentially fixed trajectory over time, but that different individuals traverse this trajectory at different rates at early stages. In this picture, the differing amplitudes of Kr expression in Fig. 4B represent a form of developmental heterochrony, in which embryos at the same clock time are at different places on the same developmental pathway. Alternatively, it is possible that there is no single developmental trajectory early and that embryos reach the low variability state at gastrulation by a variety of pathways. In order to answer this question it is necessary to be able to fully sample the space of possible patterns, which in our dataset can only be performed for eve. We thus complete our characterization of the overall appearance of expression patterns with a comprehensive classification of early individual eve patterns.
Variability of early even-skipped patterns
A characterization of the variability of full expression patterns requires a sufficient number of samples such that each pattern is seen in multiple individuals. The complexity of individual patterns means that computational classification must be used, while the requirement for extensive data restricts the study to eve, since it is the only gene whose expression was monitored in every embryo in our dataset. To separate individual variation in pattern development from nucleus to nucleus variation in expression amplitude, we consider smoothed patterns that have been rescaled and registered (see Materials and methods). These preprocessed patterns were then subjected to classification by self-organizing maps (SOM), the essential feature of which is that the user provides the number of classes and the SOM algorithm adjusts the classes so that each one contains elements which are as similar as possible to one another and as different as possible from members of other classes (Tamayo et al., 1999).
The most diverse eve patterns are found in temporal class 2 (Fig. 5). The seven classes found range from a single domain with a posterior bump (Fig. 5B) to cases where seven peaks are visible (Fig. 5H). Group G, containing 19% of the embryos, was the predominant class. Most groups contain a feature – either a peak or shoulder – corresponding to stripe 2, small but varying expression of region 3, and extremely variable expression of region 2, the areas that will give rise to stripe 7 and stripes 4–6 respectively (Fig. 3B). Patterns with a large number of peaks, such as those shown in Fig. 5H, may belong to the oldest embryos in temporal class 2, and perhaps patterns with only a few features (Figs. 5B and C) belong to the youngest. Overall, however, it is hard to imagine how the patterns shown in Fig. 5 could be sampled from a single temporal sequence. Inspection of unsmoothed data in the central 10% strip supports the machine analysis of smoothed patterns. All temporal class 2 embryos had stripe 1, 54% had some sign of stripes 2 and 7, 34% had stripe 3, 27% had stripe 4, while stripes 5 and 6 were detected in less than 6% of temporal class 2 embryos (Supplementary Fig. 30). Description of the computational classification of eve patterns from cycle 13 and time classes 1 and 3 of cycle 14A is presented in Section 2.4 of the Supplementary Information.
Fig. 5.
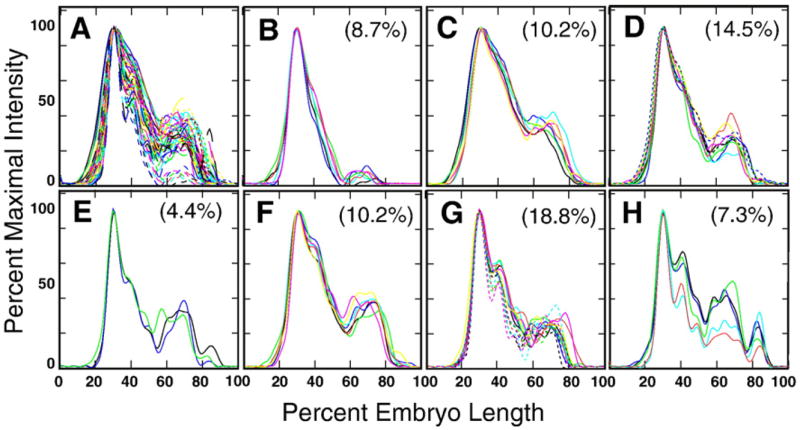
Automated classification of eve profiles for 69 embryos in temporal class 2 by self-organizing maps (SOM). (A) An overlay of all eve profiles from temporal class 2. (B–H) Seven classes of expression patterns found by SOM with the percent of the total number of embryos belonging to that class.
The positional shifts of expression domains
Most expression domains of segmentation genes change their position during cycle 14A (Supplementary Figs. 26 and 27A; Table 1 and Supplementary Table 3). These shifts can be seen in both registered (Jaeger et al., 2004b) and unregistered data. In this section we consider the unregistered data, which enables us to use positional variation within a temporal class to assess the statistical significance of shifts.
Table 1.
Temporal shifts of pair-rule domains
| Gene/Domain | Stripe 1 | Stripe 2 | Stripe 3 | Stripe 4 | Stripe 5 | Stripe 6 | Stripe 7 | |
|---|---|---|---|---|---|---|---|---|
| eve | tc | 3–8 | 3–8 | 3–8 | 3–8 | 4–8 | 4–8 | 3–8 |
| (N=654) | Shift | 0.5 | 1.9 | 2.0 | 3.4 | 3.2 | 2.6 | 5.3 |
| ftz | tc | 3–8 | 3–8 | 3–8 | 4–8 | 3–8 | 4–8 | 5–8 |
| (N=158) | Shift | 0.2* | 1.3 | 1.7 | 1.9 | 2.3 | 3.8 | 1.0* |
| h | tc | 3–8 | 3–8 | 3–8 | 4–8 | 4–8 | 4–8 | 4–8 |
| (N=105) | Shift | −2.2 | 1.5 | 3.6 | 1.1 | 2.6 | 3.3 | 3.8 |
| odd | tc | 3–8 | 4–8 | 3–8 | 5–8 | 3–8 | 3–8 | – |
| (N=85) | Shift | 2.4 | 1.1* | 3.0 | 0.9 | 3.7 | 2.7 | – |
| run | tc | 3–8 | 4–8 | 3–8 | 5–8 | 3–8 | 3–8 | 5–8 |
| (N=65) | Shift | 0.2* | 1.7 | 2.8 | 2.6 | 1.6 | 3.4 | 3.6 |
The shifts are calculated from the stripe’s maxima. Time intervals begin at the time of formation of each stripe and are given in terms of temporal classes (tc). The sample size N is shown for each domain. We do not present shifts for odd stripe 7, as it forms at the very end of cycle 14A. Positive values for shifts indicate motion to the anterior and conversely for negative values. Asterisks indicate shifts that were statistically insignificant at the 0.05 confidence level.
The shifts of the central domain of Kr and posterior domains of kni, gt and hb are statistically significant (Supplementary Table 3) and directed from the posterior to the anterior of an embryo. For example, the posterior boundaries of the Kr, kni and gt expression domains shift by about 5, 6 and 15 nuclei respectively. In each case, posterior borders shift more than anterior ones, resulting in a net contraction of each expression domain as well as the entire pattern over time. This is particularly evident for the Kr, kni, and gt domains, which contract to about two thirds of their original size over the course of cycle 14A (Supplementary Table 4). The reduction in spatial width takes place during early temporal classes while expression levels are rapidly increasing.
The boundaries of the anterior gt domain do not move and this domain does not contract with time (Supplementary Tables 3 and 4). However during temporal class 4 a new domain (domain 2 in Fig. 2F) starts to form at the left boundary of the anterior gt domain. The formation of this new domain is correlated with a shift in the old one (domain 3 in Fig. 2F) by 5 nuclei posteriorly, in the opposite direction of shifts of the posterior gap domains.
From temporal class 1 to temporal class 8 the posterior boundary of the hb anterior domain shifts to the posterior by about 0.9 nuclei as it becomes steeper with time (Supplementary Table 3). The domain as a whole does not move. Although not fully quantified because of D–V dependence, it is clear that the head domains of Kr and gt expression undergo shifts in the posterior direction, by about 4 nuclei from time classes 4 to 8 and by about 2 nuclei from time classes 6 to 8 respectively (Figs. 2C and F).
Most peaks of pair-rule gene expression change their position and shift in the anterior direction (Table 1) in a statistically significant manner. The same is true of interstripes (data not shown). With the exception of h and odd the most anterior peaks of these genes either do not move or move slightly. The first stripe of h shifts to the posterior and that of odd to the anterior; in the case of h this correlates with the formation of the anterodorsal domain 1 (Lardelli and Ish-Horowicz, 1993), and for odd with the formation of stripe 2. The largest shifts were observed for posterior domains of eve, run, and h, with eve stripe 7 shifting by about 5 nuclei. These shifts cause the contraction of expression patterns over time (Supplementary Table 5 and Supplementary Fig. 28). In contrast, the ftz pattern does not shrink with time (Supplementary Fig. 28), as its posterior boundary does not move (Table 1 and Supplementary Table 5). The behavior of the odd pattern is ambiguous in this regard as the posterior domain of odd expression forms only at the end of cycle 14A.
The above analysis was performed by considering shifts at eight discrete time points. In order to demonstrate that the shift takes place continuously, we performed a further analysis by assigning 120 individual embryos a precise developmental age by comparing their degree of membrane invagination to in vivo time lapse data (Supplementary Fig. 27). It is clear that eve stripes move continuously in an anterior direction over time, although the rate of shift for stripes 5 and 6 is less uniform than for stripe 7. The uniform movement of eve stripe 7 is reminiscent of similar smooth shifts seen in the most posterior stripes of h and run (not shown). This smooth motion of these posteriormost stripes is in marked contrast to the nonuniform movement of the other stripes, the shift of which correlates with the formation of new domains within the pattern. For example, odd stripe 1 undergoes a large shift over time classes 3 to 5, when stripe 2 forms by budding. Similarly, the formation of odd stripe 7 during temporal classes 5–7 is associated with a narrowing of odd stripe 6 between temporal classes 6 and 7 (Fig. 3O and Supplementary Fig. 22C), followed by a large shift in the maximum of this stripe between temporal classes 7 and 8 (Fig. 6). The formation of h stripe 4 in time classes 3–5 is accompanied by a shift in position of stripe 3 by 2.5 nuclei during the same period. That constitutes nearly 3/4 of the total shift of stripe 3 during cycle 14A (Table 1).
Fig. 6.
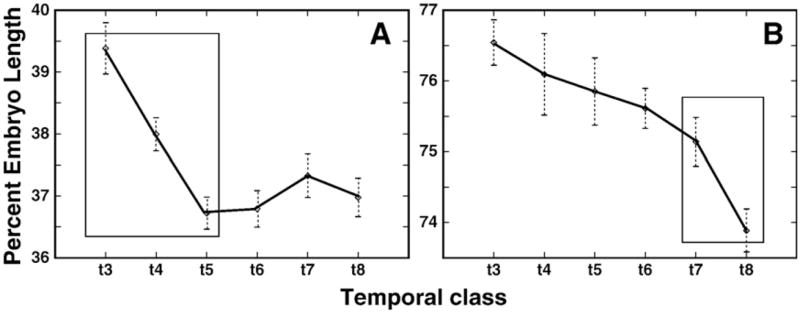
Shifts and stripe formation within the odd pattern. (A) The shifts in position of odd stripes 1 (A) and 6 (B) are shown. The period when the largest shift occurs is marked by a box. Error bars indicate the standard errors of domain positions at each temporal class.
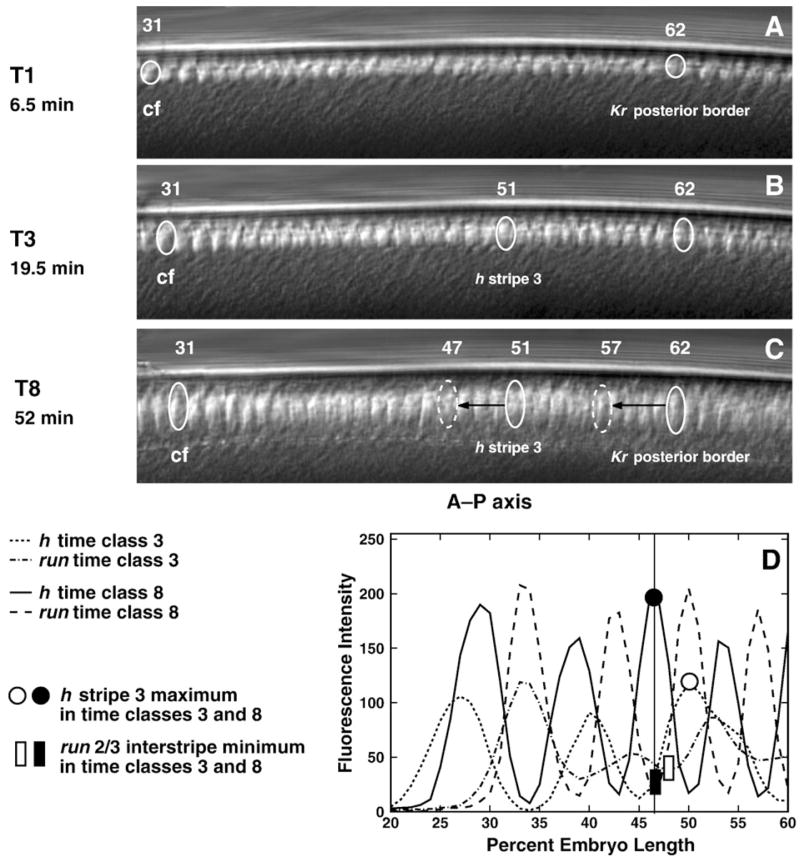
It has recently been shown that nuclei in the pregastrulation blastoderm undergo morphogenetic movements (Keränen et al., 2006), which these authors refer to as “morphological flow.” We investigated the role of this phenomenon by monitoring nuclear movements in living embryos (Figs. 10A–C, Supplementary Table 6) by following the location of three nuclei on the dorsal and ventral surfaces of 4 embryos. These nuclei were located at the position of the presumptive cephalic furrow, the posterior border of the central Kr domain at temporal class 1, and the maximum of h stripe 3 when it first forms during temporal class 3. We find that the nuclei at the positions of the initial Kr border and h stripe 3 shift by about a half nucleus (Figs. 10A–C, Supplementary Table 6), but this movement is nonuniform in time. In early cycle 14A, from about time class 1 to time class 3, the nuclei shift slightly to the posterior, while in the later part of cycle 14A the nuclei move slightly to the anterior. These periods correspond roughly to the slow and fast phase of membrane invagination accompanied by the elongation of nuclei (Loncar and Singer, 1995; Lecuit, 2004).
Fig. 10.
In vivo nuclear motion compared to gene expression. Panels A–C are frames from a movie of the dorsal side of a living embryo, taken at the times indicated. Panel D is a graph of run and h expression patterns at temporal classes 3 and 8. In panels A–C, the identified nucleus 31 is at the approximate position of the presumptive cephalic furrow (cf); the identified nucleus 62 is at the position of the posterior border of Kr at temporal class 1 (Supplementary Table 1); and in panel B, the identified nucleus 51 is at the position of the maximum of h stripe 3 in temporal class 3. We track the positions of these nuclei (solid ovals) during time classes 1–8 for Kr and 3–8 for h. The dashed ovals in panel C indicate the A–P positions of the posterior border of Kr domain and h stripe 3 maximum in temporal class 8. Black arrows show the distance between positions of the tracked nuclei and the real positions of the corresponding expression domains in time class 8. Similar results were obtained from the ventral and dorsal sides of 15 embryos. Panel D shows that the positions of the maximum of h stripe 3 and the minimum of the run 2/3 interstripe, which have the same A–P positions in time class 8, shift position with respect to one another between temporal classes 3 and 8.
Further evidence that shifts in the presumptive germ band are primarily due to shifts in gene expression rather than of nuclei can be adduced by noting that the run 2/3 interstripe and h stripe 3 shift position with respect to one another, a phenomenon that cannot be due to motions of nuclei (Fig. 10D). In the anterior, by contrast, we find that a nucleus at the position of the presumptive cephalic furrow shifts to the posterior by about two nuclei (Figs. 10A–C, Supplementary Table 6), an amount that is as large as the posterior expression shifts which we observed in the anterior part of the embryo (Fig. 9; Table 1 and Supplementary Table 3).
Fig. 9.
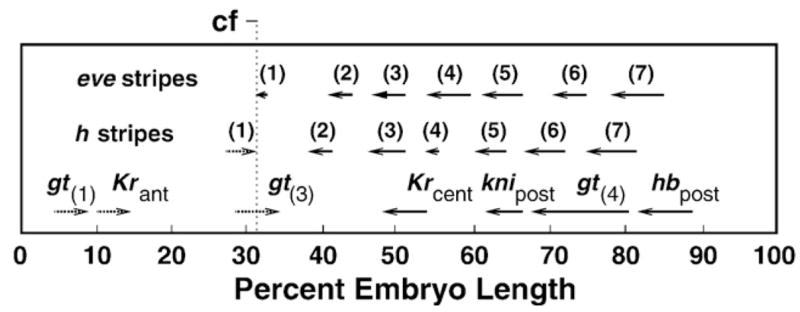
Summary of domain shifts. Direction of shifts is shown by arrows. Digits in brackets denote eve and h stripes, other domains are as indicated. Values of shifts were computed for the domain peaks over the following time intervals: from time classes 1 to 8 for gt(3), gt(4), Krcent and knipost; from time classes 2 to 8 for hbpost; from time class 3 to time class 8 for eve stripes 1–4 and 7 and h stripes 1–3; from time class 4 to time class 8 for Krant, eve stripes 5 and 6 and h stripes 4–7; from time classes 6 to 8 for gt(1). The approximate position of the prospective cephalic furrow (cf) is indicated.
The positional variability of segmentation gene expression patterns
The full characterization of the variability of segmentation gene expression is extremely complex. By examining specific features of the pattern, rather than the pattern as a whole as was done with eve, we can quantitatively assay variation in expression in the full set of segmentation genes. One particular feature which has received considerable attention is the variability in position of particular levels of expression or domain borders with respect to the total length of the embryo. Another measure of variability is with respect to a single domain, where the variability in width or intensity can be assessed. Lastly, it is possible to measure the overall deviation in proportional spacing of multiple domains. This last measure is essentially the variance of domain positions following affine registration and has been presented previously (Myasnikova et al., 2001). In this section we consider the variation of position of a number of domains as well as the variation in the width of eve stripes. We will show that the latter provides a finer scale measure of spatial variability. In this section we consider only unregistered data.
With respect to the variation of border location, it has been shown (Houchmandzadeh et al., 2002) that a particular level of Bcd expression has high embryo-to-embryo positional variability but that this noise in positional information is strongly decreased at the level of expression of the gap gene hb. Here we confirm this result and extend it to other genes. We measured the position in the embryo where the Bcd concentration was respectively 70%, 50%, 24%, and 12% of maximum. In cleavage cycle 14A, the standard deviation of these positions ranges from 3% to 6% EL, with larger standard deviations associated with lower expression levels and more posterior positions (Supplementary Table 7 and Fig. 7A). The standard deviation of the points at which Bcd was at 70% and 50% of maximal expression was the same in cycles 13 and 14A, but there is a distinct rise in the standard deviation of positions of smaller expression levels between cycle 13 and 14A (Table 2 and Supplementary Table 7). This is in distinct contrast to a general fall in positional variation of zygotic gene expression over time.
Fig. 7.
The variability in space, time, sequence and manner of domain formation. (A) The spatial variability of Bcd, Cad, and Kni domains in cycle 14A. The figure shows 26 embryos stained for bcd and cad and 18 embryos stained for kni. We show the posterior part of the kni pattern at 35–100% EL for temporal class 8. The thresholds corresponding to different percents of maximal expression are shown. (B) Spatial variability of the individual eve patterns in cycle 13 and temporal class 7 of cycle 14A. The variability in position of the posterior border of eve pattern is marked by white arrow. Black arrows mark the variability of eve stripes formed at the territory of posterior boundary of early eve. (C, D) Patterns of ftz expression in two embryos from time class 3 showing the variable mode of formation of stripe 3, which is marked with a red rectangle. (E, F) Temporal variability in formation of the pair-rule (E) and gap (F) domains indicated in the key. For each temporal class we show the percent of embryos in which the indicated domain has appeared. (G–I) Variability in the sequence of formation of stripes within the ftz pattern in temporal classes 2 (G), 3 (H), and 4 (I). Each bar shows the percent of embryos in which the indicated stripes are formed.
Table 2.
Spatial variability of the bcd, hb, Kr and eve patterns at cycle 13 and early cycle 14A
| Gene/Developmental time | bcd Cycle 13 (N=80) | hb Cycle 13 (N=25) | Kr Cycle 13 (N=20) | |||||
|---|---|---|---|---|---|---|---|---|
| A–P position | 70% | 50% | 24% | 12% | P | Peak | ||
| SD | 3.1 | 4.4 | 4.0 | 3.8 | 2.3 | 2.5 | ||
| Gene/Dev. time | eve Cycle 13 (N=75) | eve Time class 1 (N=78) | eve Time class 2 (N=63) | |||||
|
| ||||||||
| A–P position | A | Peak | P | A | Peak | P | A | Peak |
| SD | 2.4 | 2.4 | 3.7 | 2.0 | 2.2 | 3.4 | 1.4 | 1.6 |
“A–P position” gives the positions of characteristic features for each gene. The characteristic features of bcd are the positions of points at four indicated thresholds with respect to maximal fluorescence intensity. For the domains of other genes, “peak” indicates the position of maximum expression, and “A” and “P” respectively indicate the positions of 50% maximum expression at the anterior and posterior. The sample sizes N are shown. SD stands for standard deviation.
The variability of cad expression shows significant contrasts to that of bcd. In the posterior half of the embryo, where expression is high, cad expression cannot be characterized as a gradient (Fig. 7A). Although the positional error of lower levels of Cad is of the same general magnitude as that of Bcd, the trend is towards smaller positional variability at lower expression levels, in contrast to bcd (Supplementary Table 7).
We measured the standard deviation of the position of the peaks of Kr and eve expression during cleavage cycle 13, as well as the position of the posterior boundaries at half maximum of hb and eve expression (Table 2). The standard deviations fell into two classes. On the posterior slope of the cycle 13 eve domain, the standard deviation was about that seen for bcd in the same portion of the embryo (at about 12% EL), while all other features measured in cycle 13 had noticeably smaller spatial standard deviations of 2.3 to 2.5% EL (Table 2, Fig. 7B).
The positional error of zygotic expression domains continues to decrease until temporal class 2 of cycle 14A. For example, the dense coverage of eve in our dataset enables us to assess this gene’s positional variability in every temporal class. Once stripes form, there is no statistically significant change in their positional variation over time (Supplementary Table 10). This appears to indicate two very distinct temporal regimes of spatial eve variability. During the early phase, the posterior gradient of eve expression behaves like bcd, but later takes on an extremely high level of precision (Fig. 7B). Supplementary Table 10 also indicates that errors in the specification of eve stripes increase from the center of an embryo towards its ends, and F-statistics computed for minimal and maximal values of errors shows that their difference is statistically significant at a 0.05 confidence level. A similar effect can be seen with other pair-rule genes (Supplementary Table 9). We believe that this phenomenon is a consequence of variability in orientation.
We summarize the positional variation of other genes in temporal class 5. At this time, the positional standard deviation of spatial expression of the four nonterminal gap genes ranged from 0.97 to 1.5% EL (Supplementary Table 8). Because the posterior boundaries of kni, gt and the hb posterior domains shift significantly even within the limits of one time class, part of the positional variation of these borders stems from limited temporal resolution. For eve the increase in spatial precision is quite marked. At temporal class 1 the positional error in the specification of eve expression domains is almost the same as it was in cycle 13, but it declines by temporal class 2 (Table 2). The spatial variability of expression domains of pair-rule genes in the nonterminal regions of the embryo approaches an accuracy of one nucleus (Supplementary Tables 9 and 10).
While the positional variation with respect to the entire embryo appears to become essentially static by temporal class 2, variation within individual expression domains continues to decrease. For example, the variation in width of eve stripes can be estimated by evaluating the standard deviation of the distance between the stripe peak and an adjacent minimum. This measure of variation is shown in Supplementary Table 11, which extends from temporal class 3, when nearly all stripes are visible, to temporal class 8. The table shows that by this measure, standard deviation decreases until temporal class 5 or 6, with a small rise in temporal class 8. A similar result was found for ftz (data not shown).
The temporal variability of segmentation gene expression patterns
The detailed characterization of patterns which we have generated permits us to estimate the temporal variability of the formation of a single domain with a resolution of one temporal class, that is about 6 min. The natural way to estimate the temporal variability of domain formation is to monitor how quickly each domain appears in 100% of embryos. The pair-rule genes provide a particularly rich set of features with which to do this. The results of a stripe by stripe analysis for each gene are shown in Supplementary Fig. 30. These data give us the raw material with which to estimate the temporal precision with which each pair-rule stripe is formed. For example, the temporal precision of the formation of run stripe 1 is less than 6 min, since this stripe begins to form at temporal class 1 and is formed by temporal class 2 in 100% of all embryos observed (Fig. 7E). In sharp contrast, the temporal variability of formation of ftz stripe 4 is about 24 min. This stripe starts to form at time class 1 and is not observed in all embryos until temporal class 5. These two stripes are the most and least variable, while other cases lie in between (Supplementary Fig. 30).
Each pair-rule stripe forms with a distinctive temporal precision, so that the temporal precision of stripe formation is different for each stripe of one pair-rule gene. Moreover, it is clear that for many pair-rule genes stripes do not form in a fixed order. ftz provides an excellent example of such behavior. Although stripe 5 always appears first, it can be followed by the appearance of either stripe 1 or 2. Stripe 3 can appear before or after stripe 6, and the last stripe to form can be either 4 or 7 (Figs. 7G–I). At our current level of temporal resolution, these possibilities are in no way exhaustive. Not only can the order of stripe formation vary, but the same stripe can form by more than one mode. For example, ftz stripe 3 can form by splitting or posterior budding (Figs. 7C, D).
Temporal variability in the formation of gap domains differs significantly from that of pair-rule stripes. During the middle part of cycle 14A, there is considerable variation in the time of appearance of the anteriormost domains of Kr and gt. We detected the Kr anterior domain (Fig. 2C) in 13%, 56% and 80% of embryos from time classes 3, 4 and 5 respectively; domain 1 of gt (Fig. 2F) in 10%, 47%, and 87% of embryos in time classes 5, 6 and 7 respectively; and gt domain 2 (Fig. 2F) in 10%, 61% and 94% of embryos at temporal classes 4, 5 and 6 respectively. Thus the variability in the time of formation of these gap domains spans about 24 min in all cases, although the start of this period varies from domain to domain (Fig. 7F). Moreover, the rate of formation of these domains is more uniform over time compared to ftz stripe 4, which is also formed during about 24 min of cycle 14A (Fig. 7E).
Discussion
Here we summarize the major biological conclusions arising from this work. First, we have achieved a comprehensive characterization of the segmentation morphome at the systems level. Secondly, we have shown that the morphome exhibits canalizing behavior. This couples our work to classics studies of embryonic regulation, but while these studies were performed using unnatural methods of microsurgery, we have characterized regulation in its natural context. Thirdly, we show that pattern formation in the Drosophila takes place by a rich but highly varied set of trajectories; this has serious implications for efforts to model and understand the system. Finally, we have demonstrated that patterns do not form “in place”, but rather involve a set of precise spatial movements which differ in characteristic ways for each gene. We now discuss each of these points in turn.
The segmentation morphome
In this paper we have performed a detailed characterization of the expression patterns of 14 segmentation genes during the blastoderm stage. In most previous studies, the expression of segmentation genes was characterized by visual inspection of patterns, typically of the products of one or at most two genes. The vast majority of these studies were focused on the measurement of dynamics of mRNA accumulation. Much less attention has been paid to the assessment of segmentation gene expression at the protein level. This work contains a full characterization of the protein patterns of kni, odd, run, and slp, which have not been published except in a method paper and two theoretical analysis based on the data which is fully presented in this work (Kosman et al., 1998; Jaeger et al., 2004a,b). In the case of ftz we have reported expression in cleavage cycle 13, although previous studies did not see expression prior to cycle 14A (Carroll and Scott, 1985; Karr and Kornberg, 1989).
We believe that the characterization of the entire morphome is essential for understanding its biological function. Indeed, we have made use of such data previously in semiquantitative (Reinitz et al., 1995; Reinitz and Sharp, 1995; Reinitz et al., 1998) and fully quantitative forms (Jaeger et al., 2004a,b) to answer specific questions about the function of the segmentation system. More recently, there has been an increased interest in obtaining quantitative spatially resolved expression data from the blastoderm at both the protein and RNA levels. The first of these studies provided the first description of canalizing phenomena at the level of Hb response to the Bcd gradient (Houchmandzadeh et al., 2002, 2005). A large-scale study of the morphome at the RNA level is also in progress (Luengo-Hendriks et al., 2006; Keränen et al., 2006).
The work reported here provides the first detailed characterization of protein expression levels throughout the entire period of blastoderm expression at high time resolution. A drawback of our approach is that it is limited to one dimension, although this limitation is of little importance in the presumptive segmented germ band, where the separation between the A–P and D–V morphogenetic systems is rather strict. Although there is a weak coupling to the D–V system which causes the “splaying” effect of stripes (Spirov et al., 2000; Keränen et al., 2006), this effect does not influence one-dimensional data because images are obtained in a standard lateral orientation, and any remaining error is compensated for during the registration process. In the presumptive head, this decoupling is lost, and one-dimensional data must be regarded with caution. Nevertheless, our ability to reconstruct a lateral hemisphere of the embryo in 2D enables good visualization of many head patterns (see Supplementary Fig. 13).
These data are of very high resolution in intensity, space, and time. Because all measurements of expression levels are calibrated against embryos at maximum expression levels for their respective proteins, it is possible to compare expression levels of one gene at different stages (see for example Fig. 8). The spatial resolution of the data has enabled us to discover shifts in the pattern (Jaeger et al., 2004a,b and this work) that were overlooked in qualitative studies. The temporal resolution of 6.5 min has permitted us to describe transient patterns in considerable detail, and to characterize the systems level variation of gene expression precisely.
We have described the expression of the segmentation morphome both at the level of individual embryos and in terms of averaged reference data, which we refer to as one-dimensional integrated data. A full discussion of the detailed findings from this data is presented in Section 3 of the Supplementary Information.
The motion of expression domains
In this work we present an analysis of dynamic changes in positions of expression domains during cleavage cycle 14A by means of statistical tests on high-resolution data. Nevertheless, the particularly large shift in the posterior domain of gt was noted from qualitative observations (Eldon and Pirrotta, 1991). Here we demonstrate that this phenomenon is inherent to most domains of segmentation genes; certain aspects of these results have been reported previously by us in a modeling study (Jaeger et al., 2004b) and other observations of shifts have recently been made at the RNA level (Keränen et al., 2006).
At cycle 14A central and posterior domains of gap genes and most expression domains of pair-rule genes shift anteriorly (Table 1 and Supplementary Table 3), while the anterodorsal domain of Kr, the first and third domains of gt and h stripe 1 move in the opposite direction (Fig. 9). Thus, all domains moving in the posterior direction are located to the anterior of the future cephalic furrow, which arises at the juxtaposition of the head and trunk patterning systems and is one of the first morphological manifestations of the patterning process (Vincent et al., 1997; Blankenship and Wieschaus, 2001). Given that, we conclude that shifts to the posterior are inherent to domains formed in the future head region. Thus, the cephalic furrow serves as a watershed which separates regions of anterior and posterior domain shifts.
An important difference exists between the posterior shifts in the head and anterior shifts in the presumptive germ band, however. The shifts in the head appear to arise from shifts of nuclei (Keränen et al., 2006), rather than from gene regulation (Fig. 10). We show here that in the presumptive germ band nuclear movements are much smaller than expression shifts and that expression patterns of different segmentation genes shift with respect to one another (Figs. 9 and 10, Supplementary Table 6). That, together with the asymmetric distribution of mRNA with respect to protein in gap domains (Jaeger et al., 2004b), shows that in the presumptive germ band shifts occur as a consequence of gene regulation.
We have found that the Tll protein has different dynamics of accumulation in comparison to the “classical” gap genes. In cycle 13 and early cycle 14A tll expression sharply increases and reaches maximum (Fig. 2I). It remains constant during about 30 min of development and declines before gastrulation. Furthermore, in contrast to the posterior domains of the other gap genes, the tll posterior domain does not shift position with time (Fig. 2I).
Of particular significance are the shifts of the posterior boundaries of the Kr central domain and the posterior domains of kni and gt, which range from 5 to 15 nuclei. The shifts of domains inside the expression pattern of pair-rule genes usually occur at the time when a new stripe arises in the vicinity and range from about 1 to 4 nuclei, while the posterior stripes of eve, run and h move by about 3–5 nuclei. It is possible that the large shift of run stripe 7 is partly caused by a dynamic change of its shape because this stripe becomes asymmetric by temporal class 8. Given that the width of pair-rule stripes ranges from 3 to 5 nuclei at the end of cleavage cycle 14A, it is clear that the shifts of gap and pair-rule domains are on the order of the size of a pair-rule stripe. Hence these shifts are critical for the positioning of domains of downstream genes and are biologically essential to the pattern formation process.
Central and posterior gap domains contract with time due to larger shifts of the posterior boundaries than the anterior ones (Supplementary Tables 1 and 4). Large shifts of posterior stripes result in the contraction of the expression patterns of eve, h and run (Supplementary Table 5, Supplementary Fig. 28). In contrast, the ftz pattern does not shrink with time, as its posterior boundary does not move (Table 1). It is well known that the complementary patterns of eve and ftz expression set the positions of en stripes (Hughes and Krause, 2001). Our results demonstrate that the complementary phasing of eve and ftz stripes comes about by the shift of eve pattern in relation to ftz and is not simply the result of refinement of these striped patterns in one place (Supplementary Fig. 29). Currently the subdivision of pair-rule genes into primary and secondary ones is a matter of controversy. It has been pointed out that ftz is not a secondary pair-rule gene (Yu and Pick, 1995) in the initial sense of this term (Ingham and Martinez-Arias, 1986; Howard and Ingham, 1986). Nevertheless there are some features which set this gene apart from other pair-rule genes. It is well known from the literature that ftz has different regulatory inputs and a different organization of its regulatory region than other pair-rule genes (Hiromi et al., 1985; Klingler et al., 1996). In this study we have found that ftz stripes form and refine in a different way than stripes of other pair-rule genes: stripe 5 is the first stripe to form and stripe 7 does not move with time (Supplementary Fig. 30, Table 1).
With respect to the anterior, it has been previously noted that the region of the blastoderm anterior to the presumptive cephalic furrow is a domain of low nuclear density which forms by cleavage cycle 11 and persists until gastrulation (Blankenship and Wieschaus, 2001). During cycle 14A a region with a greater depth of cellularization forms at the location of the future cephalic furrow, a fact that may be connected with the observed nuclear movements in the anterior part of the embryo. These processes were shown to be under control of the genes bcd and prd, which thus makes it the earliest manifestation of morphological control by the segmentation system (Blankenship and Wieschaus, 2001).
Variation and canalization of blastoderm expression patterns
Our analysis shows that by the onset of gastrulation some gap genes and all pair-rule genes form patterns which are highly uniform from embryo to embryo. However, there is large embryo-to-embryo variability of these patterns in cycle 13 and early temporal classes of cycle 14A. This variability manifests itself in variation in gene expression levels, variation in the time of formation of individual domains, variation in the manner in which a domain forms, variation in the sequence of formation of individual domains, and variation in the position of expression domains. These types of variability place limitations on what we can conclude about the morphome, and at the same time are of fundamental biological interest in their own right. We discuss each form of variability in turn.
Variability in gene expression levels and the limitations of the integrated data
The integrated data on the expression of 14 genes presented here were assembled from many individual isogenic embryos, each stained for the products of three genes. Even in an isogenic population, there are differences between individuals. The fundamental criterion for the validity of our integrated data is that it should represent the possible actual dynamics of one individual in the isogenic population. Without the ability to simultaneously monitor the expression of all 14 segmentation genes in vivo, it is impossible to check directly whether or not this criterion is satisfied. Nevertheless, inspection of the figures which compare individual patterns to integrated patterns permits a number of conclusions to be drawn.
First of all, late in cleavage cycle 14 the integrated data and individual data are very close, and the integrated data likely well represent an individual. Variation between individuals involves position and amplitude, the latter of which also subsumes the order of appearance of domains. Here we confine our attention to variations in amplitude, which in any case is much larger than variation in position (see the following section). In the first half of cleavage cycle 14 there is considerable variation in amplitude among individuals, which has a markedly different character for gap and pair-rule genes. Gap gene domains (see Fig. 4B and Supplementary Fig. 10) which vary in amplitude have very similar profiles, and it is evident from the superimposed mean expression patterns that the means are very close to a median individual pattern.
In the case of early pair-rule patterns, the variation in the order in which stripes arise (see Figs. 4D and F) causes the early integrated patterns to be flatter than actual individuals because there is a tendency for peaks and valleys in different individuals to cancel out. We discuss the individual variations in the order of stripe appearance below. We believe that this variation in early pair-rule stripes is driven by the variation in gap gene amplitudes, since there have been numerous studies indicating that the first pair-rule interstripes are driven by repressive inputs from gap genes (Small et al., 1992; Hartmann et al., 1994; Reinitz and Sharp, 1995; Klingler et al., 1996). Unfortunately, it is not feasible to generate integrated data from median individuals without being able to select all median patterns simultaneously from one individual.
Finally, we note that because the integrated data are in one dimension, there will be some variation in the expression along the D–V axis that will not be captured. In the present study, this problem may manifest itself in the increased variation in positioning of pair-rule stripes near the ends of the embryo (Supplementary Tables 9 and 10). Otherwise, inspection of our images and a related study in 3 dimensions indicate that this error is small (Keränen et al., 2006).
Variability in time of domain formation
Our detailed characterization of patterns allowed us estimate the variability of the time of formation of each stripe. Currently this variability can be estimated with a precision of about 6 min, as this is the temporal resolution of our dataset. We have found that the temporal precision of stripe formation can vary in a wide range from less than 6 to as much as 24 min of development (Figs. 7E and Supplementary Fig. 30). This is a very high level of temporal variability, especially if one recalls that the whole pair-rule pattern forms within about 50 min. Each pair-rule stripe forms with its own characteristic temporal precision. Examination of the time of formation of three anterior gap domains showed that the temporal precision of their formation is about 24 min of development independent of when each domain first appears (Fig. 7F).
Variation in the manner of stripe formation
We have found that the stripes of pair-rule genes can arise de novo, by splitting, or by budding (Supplementary Fig. 2), and there is variation in the manner in which specific stripes of individual genes form. For example, ftz stripe 3 can form in two different ways, either by the splitting of one large domain or by posterior budding from stripe 2 (Figs. 7C, D). The classification of modes of domain formation can be extended to the formation of gap domains, but here there is little indication of variation. Most domains of gap genes form de novo, however, the second domain of gt arises at the boundary of the third domain by budding (Fig. 2F). Different modes of pair-rule stripe formation may reflect different regulatory mechanisms governing the appearance of these stripes.
Variability in the sequence of domain formation
We can assess variability in the order of domain formation from the patterns of ftz and eve. At temporal class 2, all embryos have ftz stripe 5. However, among these there are embryos in which stripe 1 is formed, embryos with stripe 2, and embryos with both these stripes. This means that after formation of stripe 5 the next stripe to appear may be stripe 1 or stripe 2 (Fig. 7G). In the case of eve, the large number of available embryos analyzed by machine classification procedures demonstrates an extremely diverse set of possible developmental trajectories (Fig. 5, Supplementary Figs. 16 and 17). These trajectories are indeed too diverse to permit a classification in terms of the order of appearance of particular stripes and provide a striking contrast to the precise and stereotyped arrangement of eve stripes that exists by temporal class 6. It is likely that such diversity is characteristic of all pair-rule stripes at early stages of formation. As such it is a dramatic illustration of the phenomenon of canalization proposed by Waddington (1940). On a fundamental level, it demonstrates that there is no fixed developmental pathway in the segmentation system, but rather a range of developmental trajectories compatible with viability, which converge as gastrulation approaches. These findings concerning canalization are strongly reinforced by studies of the reduction of positional error, which we consider in the next section.
Variability in the position of expression domains
The analysis of spatial variability of segmentation gene expression patterns is directly connected to the idea of positional information (Wolpert, 1969; Jaeger and Reinitz, 2006) and hence has received considerable attention. It has been shown that the variability of positional information is strongly reduced in the transfer of this information from the Bcd gradient to hb (Houchmandzadeh et al., 2002), a process which these authors and ourselves refer to as the filtration of positional error. We have confirmed these results and extended them to other maternal gradients, as well as gap and pair-rule genes (Figs. 7A, B; Table 2; and Supplementary Tables 7–10). The canalization described in the previous paragraph manifests itself here as filtration of noise inherent in maternal gradients over time (Fig. 11). It is evident that at the gap gene level partial filtration of bcd error has already happened at cycle 13. The filtration of this error continues at temporal class 1 and is completed by temporal class 2. In comparison with gap genes, the filtration of bcd positional error at the pair-rule gene level begins later and happens faster: at temporal class one, eve positional error is comparable with that of bcd, however by temporal class 3 eve variability is comparable with that of gap genes.
Fig. 11.
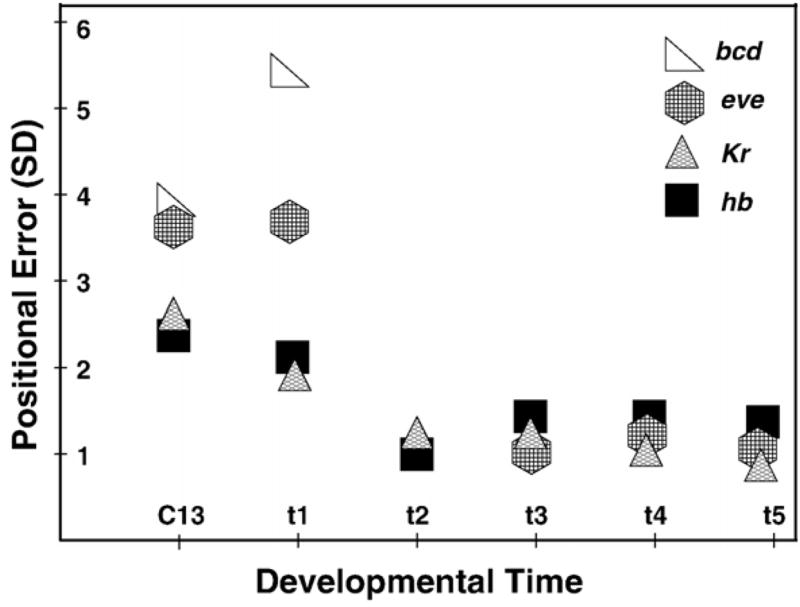
Dynamic filtration of the positional error at the level of zygotic gene expression. We consider the standard deviations (SD) in the positions of the posterior border of the anterior hb domain, maximum of the central Kr domain, and the posterior border of the early eve domain, which later corresponds to the position of eve stripe 3. Changes in the positional error of these domains are compared to the level of variability in position of the 12% concentration threshold of the Bcd protein gradient. All these features have approximately the same positions along the A–P axis of an embryo (45–55% EL). In cycle 14A we show the Bcd variability only for time class 1, as it remains at approximately the same level thereafter.
Supplementary Material
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2007.10.037.
Acknowledgments
This work was supported by grant RR07801 from the US NIH, by GM072022 jointly from the US NIH and NSF, by CRDF GAP Awards RBO-1286 and RUB1-1578, by contract 02.467.11.1005 from the FASI of the RF, and by grant 047.011.2004.013 from the NWO-RFBR. We thank N. Golyandina and T. Alexandrov for the SSA methods and software; A. Samsonov, H. Janssens, and J. Jaeger for their valuable discussions; and D. Holloway for the helpful comments about the manuscript.
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Wieschaus E. Two new roles for the Drosophila AP patterning system in early morphogenesis. Development. 2001;128:5129–5138. doi: 10.1242/dev.128.24.5129. [DOI] [PubMed] [Google Scholar]
- Broomhead D, King G. Extracting qualitative dynamics from experimental data. Physica D. 1986;20:217–236. [Google Scholar]
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Swaycus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990;8:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesch H. In: The History and Theory of Vitalism. Ogden CK, translator. MacMillan and Company; London: 1914. [Google Scholar]
- Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Eldon ED, Pirrotta V. Interactions of the Drosophila gap gene giant with maternal and zygotic pattern-forming genes. Development. 1991;111:367–378. doi: 10.1242/dev.111.2.367. [DOI] [PubMed] [Google Scholar]
- Elsner J, Tsonis A. A New Tool in Time Series Analysis. Plenum Press; New York: 1996. Singular Spectrum Analysis. [Google Scholar]
- Fraedrich K. Estimating the dimensions of weather and climate attractors. J Atmos Sci. 1986;43:419–432. [Google Scholar]
- Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Gergen JP, Butler BA. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988;2:1179–1193. doi: 10.1101/gad.2.9.1179. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Opitz JM, Raff RA. Resynthesizing evolutionary and developmental biology. Dev Biol. 1996;173:357–372. doi: 10.1006/dbio.1996.0032. [DOI] [PubMed] [Google Scholar]
- Golyandina N, Nekrutkin V, Zhigljavsky A. Analysis of Time Series Structure: SSA and Related Techniques. Chapman and Hall/CRC; Boca Raton: 2001. [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Taubert H, Jäckle H, Pankratz MJ. A two-step mode of stripe formation in the Drosophila blastoderm requires interactions among primary pair rule genes. Mech Dev. 1994;45:3–13. doi: 10.1016/0925-4773(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Holloway DM, Harrison LG, Kosman D, Vanario-Alonso CE, Spirov AV. Analysis of pattern precision shows that Drosophila segmentation develops substantial independence from gradients of maternal gene products. Dev Dyn. 2006;235:2949–2960. doi: 10.1002/dvdy.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Precise domain specification in the developing Drosophila embryo. Phys Rev, E Stat Nonlinear Soft Matter Phys. 2005;72:061920. doi: 10.1103/PhysRevE.72.061920. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham PW. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Hughes SC, Krause HM. Establishment and maintenance of parasegmental compartments. Development. 2001;128:1109–1118. doi: 10.1242/dev.128.7.1109. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Martinez-Arias A. The correct activation of antennapedia and bithorax complex genes requires the fushi tarazu gene. Nature. 1986;324:592–597. doi: 10.1038/324592a0. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Howard K, Pinchin SM, Ingham PW. Molecular and genetic analysis of the hairy locus in Drosophila. Cold Spring Harbor Symp Quant Biol. 1985;50:135–144. doi: 10.1101/sqb.1985.050.01.019. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Reinitz J. On the dynamic nature of positional information. BioEssays. 2006;28:1102–1111. doi: 10.1002/bies.20494. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Blagov M, Kosman D, Kozlov KN, Manu, Myasnikova E, Surkova S, Vanario-Alonso CE, Samsonova M, Sharp DH, Reinitz J. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics. 2004a;167:1721–1737. doi: 10.1534/genetics.104.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, Manu, Myasnikova E, Vanario-Alonso CE, Samsonova M, Sharp DH, Reinitz J. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004b;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- Janssens H, Kosman D, Vanario-Alonso CE, Jaeger J, Samsonova M, Reinitz J. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Dev Genes Evol. 2005;215:374–381. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- Karr TL, Kornberg TB. Fushi tarazu protein expression in the cellular blastoderm of Drosophila detected using a novel imaging technique. Development. 1989;106:95–103. doi: 10.1242/dev.106.1.95. [DOI] [PubMed] [Google Scholar]
- Keränen SVE, Fowlkes C, Luengo C, Sudar D, Knowles DK, Malik J, Biggin MD. Quantitation of temporal changes in morphology and gene expression in the Drosophila cellular blastoderm. Genome Biol. 2006;7:R124. doi: 10.1186/gb-2006-7-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler M, Soong J, Butler B, Gergen JP. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev Biol. 1996;177:73–84. doi: 10.1006/dbio.1996.0146. [DOI] [PubMed] [Google Scholar]
- Kohonen T. The self-organizing map. Proc IEEE. 1990;78:1464–1480. [Google Scholar]
- Kohonen T. Self-Organizing Maps. 2. Springer; Heidelberg: 1997. [Google Scholar]
- Kosman D, Reinitz J, Sharp DH. Automated assay of gene expression at cellular resolution. In: Altman R, Dunker K, Hunter L, Klein T, editors. Proceedings of the 1998 Pacific Symposium on Biocomputing, PSB Proceedings. World Scientific Press; Singapore: 1997. pp. 6–17. http://www.smi.stanford.edu/projects/helix/psb98/kosman.pdf. [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal anti-bodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Kozlov K, Myasnikova E, Samsonova M, Reinitz J, Kosman D. Method for spatial registration of the expression patterns of Drosophila segmentation genes using wavelets. Comput Tech. 2000;5:112–119. [Google Scholar]
- Kozlov K, Myasnikova E, Pisarev A, Samsonova M, Reinitz J. A method for two-dimensional registration and construction of the two-dimensional atlas of gene expression patterns in situ. In Silico Biol. 2002;2:125–141. [PubMed] [Google Scholar]
- Kuroiwa A, Hafen E, Gehring WJ. Cloning and transcriptional analysis of the segmentation gene fushi-tarazu of Drosophila. Cell. 1984;37:825–831. doi: 10.1016/0092-8674(84)90417-3. [DOI] [PubMed] [Google Scholar]
- Lardelli M, Ish-Horowicz D. Drosophila hairy pair-rule gene regulates embryonic patterning outside its apparent stripe domains. Genes Dev. 1993;2:1021–1036. doi: 10.1242/dev.118.1.255. [DOI] [PubMed] [Google Scholar]
- Lecuit T. Junctions and vesicular trafficking during Drosophila cellularization. J Cell Sci. 2004;117:3427–3433. doi: 10.1242/jcs.01312. [DOI] [PubMed] [Google Scholar]
- Loncar D, Singer SJ. Cell membrane formation during the cellularization of the syncytial blastoderm of Drosophila. Proc Natl Acad Sci U S A. 1995;92:2199–2203. doi: 10.1073/pnas.92.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo-Hendriks CL, Keranen SV, Fowlkes CC, Simirenko L, Weber GH, Henriquez C, Kaszuba D, Hamann B, Eisen M, Malik J, Sudar D, Biggin MD, Knowles DW. 3D measurement of morphology and gene expression in the Drosophila blastoderm at cellular resolution. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Mohler J, Eldon ED, Pirrotta V. A novel spatial transcription pattern associated with the segmentation gene, giant, of Drosophila. EMBO J. 1989;8:1539–1548. doi: 10.1002/j.1460-2075.1989.tb03538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikova E, Kosman D, Reinitz J, Samsonova M. Spatio-temporal registration of the expression patterns of Drosophila segmentation genes. In: Lengauer T, Schneider R, Bork P, Brutlag D, Glasgow J, Mewes H-W, Zimmer R, editors. Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology; AAAI Press, Menlo Park, CA. 1999. pp. 195–201. [PubMed] [Google Scholar]
- Myasnikova E, Samsonova A, Kozlov K, Samsonova M, Reinitz J. Registration of the expression patterns of Drosophila segmentation genes by two independent methods. Bioinformatics. 2001;17:3–12. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- Myasnikova E, Samsonova M, Kosman D, Reinitz J. Removal of back-ground signal from in situ data on the expression of segmentation genes in Drosophila. Dev Genes Evol. 2005;215:320–326. doi: 10.1007/s00427-005-0472-2. [DOI] [PubMed] [Google Scholar]
- Nauber U, Pankratz MJ, Kienlin A, Seifert E, Klemm U, Jäckle H. Abdominal segmentation of the Drosophila embryo requires a hormone receptor-like protein encoded by the gap gene knirps. Nature. 1988;336:489–492. doi: 10.1038/336489a0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Poustelnikova E, Pisarev A, Blagov M, Samsonova M, Reinitz J. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Levine M. Control of the initiation of homeotic gene expression by the gap genes giant and tailless in Drosophila. Dev Biol. 1990;140:57–72. doi: 10.1016/0012-1606(90)90053-l. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Sharp DH. Mechanism of eve stripe formation. Mech Dev. 1995;49:133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Mjolsness E, Sharp DH. Cooperative control of positional in-formation in Drosophila by bicoid and maternal hunchback. J Exp Zool. 1995;271:47–56. doi: 10.1002/jez.1402710106. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Kosman D, Vanario-Alonso CE, Sharp DH. Stripe forming architecture of the gap gene system. Dev Genet. 1998;23:11–27. doi: 10.1002/(SICI)1520-6408(1998)23:1<11::AID-DVG2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Rosenberg UB, Schröder C, Preiss A, Kienlin A, Cóté S, Riede I, Jäckle H. Structural homology of the product of the Drosophila krüppel gene with Xenopus transcription factor IIIA. Nature. 1986;319:336–339. [Google Scholar]
- Simcox AA, Sang JH. When does determination occur in Drosophila embryos? Dev Biol. 1983;97:212–221. doi: 10.1016/0012-1606(83)90078-7. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirov AV, Holloway D. Evolutionary techniques for image processing to construct integrated dataset of early Drosophila embryo genes activity. EURASIP Journal on Applied Signal Processing, Special Issue on Genetic and Evolutionary Computation for Signal Processing and Image Analysis 2003a [Google Scholar]
- Spirov AV, Holloway D. Making the body plan: precision in the genetic hierarchy of Drosophila embryo segmentation. Silico Biology. 2003b;3:0009. http://www.bioinfo.de/isb/2003/03/0009/ [PubMed]
- Spirov AV, Timakin DL, Reinitz J, Kosman D. Experimental determination of Drosophila embryonic coordinates by genetic algorithms, the simplex method, and their hybrid. In: Cagnoni S, Poli R, editors. Proceedings of the Second European Workshop on Evolutionary Computation in Image Analysis and Signal Processing, Springer Verlag Lecture Notes in Computer Science, Number 1803; Springer. 2000. pp. 97–106. [Google Scholar]
- Spirov AV, Timakin DL, Reinitz J, Kosman D. Use of evolutionary computations in image processing for quantitative atlas of Drosophila gene expression. Appl Evol Comput Proc Lect Notes Comput Sci. 2001;2037:374–383. [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Lander EDES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Lehmann R, Schnürch H, Schuh R, Seifert E, Kienlin A, Jones K, Jäckle H. Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature. 1987;327:383–389. [Google Scholar]
- Team SP. Helsinki University of Technology; Helsinki, Finland: 1995. SOM PAK: The Self-Organizing Map Program Package. Available at ftp://cochlea.hut.fi. [Google Scholar]
- Vautard R, Ghil M. Singular spectrum analysis in nonlinear dynamics, with applications to paleoclimatic time-series. Physica D. 1989;35:395–424. [Google Scholar]
- Vincent A, Blankenship JT, Wieschaus E. Integration of the head and trunk segmentation systems controls cephalic furrow formation in Drosophila. Development. 1997;124:3747–3754. doi: 10.1242/dev.124.19.3747. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Organisers and Genes. Cambridge Univ. Press; Cambridge, UK: 1940. [Google Scholar]
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Pick L. Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mech Dev. 1995;50:163–175. doi: 10.1016/0925-4773(94)00333-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2007.10.037.