Abstract
The packaging of genomic DNA into chromatin, often viewed as an impediment to the transcription process, plays a fundamental role in the regulation of gene expression. Chromatin remodeling proteins have been shown to alter local chromatin structure and facilitate recruitment of essential factors required for transcription. Brahma-related gene-1 (BRG1), the central catalytic subunit of numerous chromatin-modifying enzymatic complexes, uses the energy derived from ATP-hydrolysis to disrupt the chromatin architecture of target promoters. In this review, we examine BRG1 as a major coregulator of transcription. BRG1 has been implicated in the activation and repression of gene expression through the modulation of chromatin in various tissues and physiological conditions. Outstanding examples are studies demonstrating that BRG1 is a necessary component for nuclear receptor-mediated transcriptional activation. The remodeling protein is also associated with transcriptional corepressor complexes which recruit remodeling activity to target promoters for gene silencing. Taken together, BRG1 appears to be a critical modulator of transcriptional regulation in cellular processes including transcriptional regulation, replication, DNA repair and recombination.
Introduction
The packaging of genomic DNA into nucleosomes, the fundamental unit of chromatin, creates a barrier to nuclear processes, such as transcription, by obstructing the association of essential factors and gene-specific regulators to recognition sequences within target promoters. The regulation of genetic information from a highly organized chromatin structure is essential for normal cellular growth, development, differentiation and genomic stability [Chen et al., 2006; Hansen, 2002; Workman and Kingston, 1998]. The basic unit of chromatin structure is the nucleosome, which consists of 146 bp of genomic DNA wrapped around two copies each of core histone proteins H2A, H2B, H3 and H4 [Wolffe, 2001]. Regions of DNA between adjacent nucleosomes are occupied with a molecule of linker histone protein, H1 [Aoyagi et al., 2005; Hayes and Hansen, 2001]. The association of H1 facilitates condensation of the chromatin structure into 30-nm fibers which in turn self-assembles and organizes into a higher ordered architecture resulting in the formation of the chromosome [Tremethick, 2007; Wolffe, 1998].
Mammalian genes, assembled into chromatin using a combination of histone and non-histone proteins, can adopt a multilayered chromatin structure that can be spatially divided into transcriptionally “active” or “inducible” euchromatic, or into mostly “silent” or “inactive” heterochromatic conformations, according to different cellular circumstances [Horn and Peterson, 2006]. These modulations of chromatin structure, that often accompany transcriptional regulation, frequently require the enzymatic activity of multi-protein complexes to alter the nucleosomal arrangement [Felsenfeld and Groudine, 2003; Kinyamu and Archer, 2004]. At least two highly conserved chromosome-modifying enzymatic activities have been described that alter chromatin structure through disruption of histone-DNA contacts by ATP-dependent chromatin remodeling complexes, or by covalent modification of histone tails by means of acetylation, methylation, phosphorylation, ubiquitination, sumoylation and/or ADP ribosylation [Kouzarides, 2007; Trotter and Archer, 2007].
Alteration of the chromatin architecture by ATP-dependent remodeling complexes is considered a significant step in transcriptional regulation of many eukaryotic genes. These chromatin-modifying enzymes use energy derived from ATP hydrolysis to actively alter the nucleosomal structure [Johnson et al., 2005]. A number of chromatin remodeling complexes have been identified that modulate the arrangement and stability of nucleosomes in a non-covalent manner. Generally, these ATP-dependent remodeling machines are divided into four major subfamilies, characterized by the identity of their central catalytic subunit, which include BRG1 (or hBrm), ISWI, Mi-2 and Ino80 of their respective complexes SWI/SNF, ISWI, NuRD and INO80 [Eberharter and Becker, 2004; Sif, 2004].
Human SWI/SNF contains either BRG1 or hBRM (Brahma) as the central ATPase subunit. The paralogous catalytic subunits share a high degree of sequence identity (74%) and display similar biochemical activities in vitro [Khavari et al., 1993; Phelan et al., 1999; Randazzo et al., 1994]. Despite their similarities, the two ATPase subunits can play different roles in various cellular processes including proliferation and differentiation [Bultman et al., 2000; Kadam and Emerson, 2003; Reyes et al., 1998]. This review will focus on BRG1, the proteins with which it associates, its recruitment and dynamic interactions with chromatin and the physiological pathways which it participates in as a transcriptional coregulator.
History
The BRG1 (or hBRM) protein is the central catalytic ATPase of the SWI/SNF chromatin-remodeling complex. The BRG1 homologue, Swi2, was first identified in yeast by genetic screens searching for proteins important for mating-type switching (SWI) and sucrose non-fermenting (SNF) [Neigeborn and Carlson, 1984; Stern et al., 1984; Winston and Carlson, 1992]. Further analysis identified suppressors of these SWI and SNF mutations which included genes encoding histones and other chromatin-associated proteins, suggesting these products may be involved in transcriptional regulation through modulation of chromatin structure [Sif, 2004; Winston and Carlson, 1992]. These yeast proteins were later found to assemble into a multi-subunit complex, designated SWI/SNF, which was shown in vitro to alter nucleosomal structure, in an ATP-dependent manner, resulting in an open and accessible chromatin conformation conducive for transcription factor binding [Cairns et al., 1994; Peterson et al., 1994]. The catalytic ATPase activity of the yeast SWI/SNF chromatin remodeling complex is provided by the Swi2 or Snf2 protein [Pazin and Kadonaga, 1997; Peterson and Tamkun, 1995]. In S. cerevisiae, a second, more abundant SWI/SNF-like complex exists, designated RSC (Rsc1 and Rsc2) [Cairns et al., 1996]. Although RSC complexes contain Swi2/Snf2-like ATPase activity, derived from the Sth1 subunit, and demonstrate similar biological activities on chromatin, biochemical studies suggest the SWI/SNF and RSC complexes regulate expression of distinct sets of genes [Lorch et al., 2001; Mohrmann and Verrijzer, 2005]. Homologues of yeast SWI2/SNF2 have been identified in humans, Drosophila and mice [Dingwall et al., 1995; Khavari et al., 1993; Kwon et al., 1994; Laurent et al., 1993; Muchardt and Yaniv, 1993; Randazzo et al., 1994; Tamkun et al., 1992; Wang et al., 1996a].
Protein domains
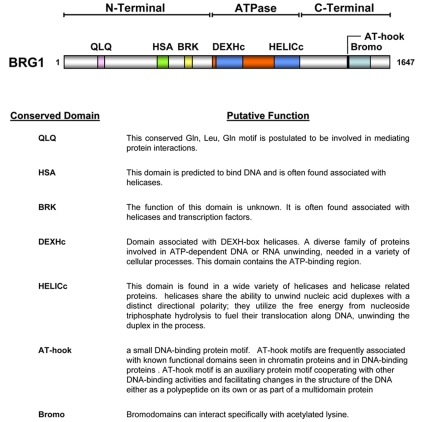
The BRG1 and hBrm proteins share a high degree of sequence identity (74%) and display similar biochemical activities in vitro [Khavari et al., 1993; Phelan et al., 1999; Randazzo et al., 1994]. Despite their similarities, the two ATPase subunits of SWI/SNF play different roles in various cellular processes including proliferation and differentiation using mechanisms of specificity that are currently undefined [Bultman et al., 2000; Kadam and Emerson, 2003; Reyes et al., 1998]. BRG1 is composed of multiple domains, most of which have been identified by primary sequence analysis and include an evolutionarily conserved catalytic ATPase domain, as well as a conserved C-terminal bromodomain, AT-hook motif and the less characterized N-terminal region housing QLQ, HSA and BRK domains [Fan et al., 2005; Khavari et al., 1993] (Figure 1).
Figure 1. Domain architecture of BRG1.
The BRG1 chromatin remodeling protein contains an evolutionarily conserved ATPase region, as well as domains found within the N- and C-terminus. The conserved domain identification and predictions were performed using the NCBI specialized BLAST conserved domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
The bromodomain of BRG1 has been implicated in the recognition of acetylated lysines within histone H3 and H4 tails [Chandrasekaran and Thompson, 2007; Shen et al., 2007]. Such modifications within target promoters may serve as an interaction surface for the assembly and/or recruitment of bromodomain-containing coregulator complexes, including SWI/SNF. In conjunction with the C-terminal bromodomain, BRG1 also contains an AT-hook sequence motif which may aid in DNA binding or recruitment of SWI/SNF to acetylated lysines within histone tails [Singh et al., 2006].
The N-terminus of BRG1 encompasses several regions of interest which could prove essential for BRG1 function. A glutamine-leucine-glutamine motif (QLQ) was identified within amino acids 172-208. Although the functional relevance of this motif remains unclear, QLQ domains are often implicated in protein-protein interactions or may contribute to the conformational structure of the protein [Kim et al., 2003; Williamson, 1994]. Also identified were HSA and BRK domains located between amino acid 475-532 and 612-656, respectively. HSA domains are regions of unknown function found in helicases and other DNA-binding proteins of eukaryotes [Doerks et al., 2002]. BRK domains, also referred to as the TCH domain, are small protein modules of unknown function associated with transcription and CHROMO domain helicases and in DEAD/DEAH box helicase domains [Allen et al., 2007; Doerks et al., 2002].
Taken together, BRG1 is composed of numerous putative domains and motifs, many of which appear to be evolutionarily conserved [Khavari et al., 1993]. Found outside the catalytic ATPase domain, within both the N- and C-terminus, are regions which may serve as potential protein interaction modules. These domains may be utilized in the recognition of modified histone proteins and/or recruit the chromatin remodeling activity of BRG1 to genomic targets.
BRG1 complexes and interacting proteins
In cells, BRG1 is found within the context of various multi-protein complexes, usually as the central enzymatic subunit, with roles in transcriptional regulation, DNA replication, repair and recombination. Among the various BRG1-containing complexes, the SWI/SNF family has been best characterized regarding structure, function and enzymatic activity. Human SWI/SNF contains one of two mutually exclusive ATPase-containing subunits, BRG1 or hBrm, and numerous BAFs (BRG1-associated factors), most of which are orthologous to those found in yeast SWI/SNF and RSC [Nie et al., 2000; Wang et al., 1996b; Xue et al., 2000]. Mammalian BRG1 is usually associated with approximately 10-12 BAF subunits or other proteins involved in regulation of gene expression, as well as nucleosome assembly, genomic stability and maintenance. Human SWI/SNF includes a heterogeneous mixture of proteins, where most purified complexes contain core subunits BRG1 (or hBrm), BAF170, BAF155 and BAF47/INI1, as well as BAF60, BAF57, BAF53 and actin [Wang et al., 1996a]. Human SWI/SNF can be further subdivided into the BAF and PBAF (Polybromo-associated BAF) complexes. These complexes are distinguished only by the presence of specific subunits, BAF250a/b or BAF180, found in BAF or PBAF, respectively [Lemon et al., 2001; Nie et al., 2000; Yan et al., 2005]. This differential makeup of the various BRG1/Brm-based complexes suggests the BAF subunits may play important roles in promoter-specific targeting or to convey a stabilized nucleosomal conformation favorable for SWI/SNF activity. Interestingly, BRG1 protein alone is capable of inducing a remodeled nucleosomal state in vitro; however, addition of the core SWI/SNF subunits, BAF170, BAF155 and BAF47 reconstitutes chromatin remodeling to near optimal levels [Phelan et al., 1999].
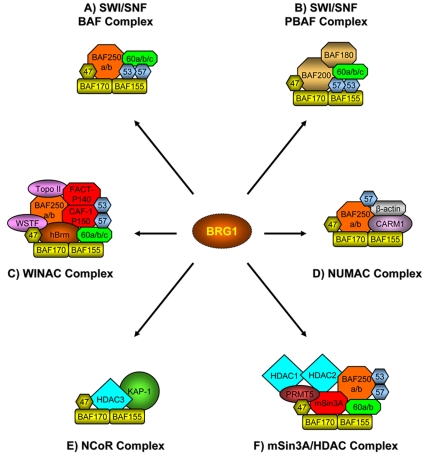
The BRG1 protein can also be found assembled with transcription factor and histone-modifying enzyme complexes to activate or repress nuclear processes including transcription, elongation and DNA replication (Figure 2). For example, the WINAC (WSTF including nucleosome assembly complex), which shares subunits with both SWI/SNF- and ISWI-based chromatin remodeling complexes, consists of at least 13 components, BRG1 and hBrm, as well as member subunits of the SWI/SWI-BAF including BAF250, BAF170, BAF155, BAF60a, BAF57, BAF53 and BAF47, and WSTF (Williams syndrome transcription factor), a reported member of the SNF2h-based remodeling complex [Aoyagi et al., 2005; Kitagawa et al., 2003]. Interestingly, WINAC includes proteins associated with DNA replication, TopoIIβ and CAF-1p150, and transcription elongation factor FACTp140, which are not present in other SWI/SNF-based complexes [Belandia and Parker, 2003]. WINAC promotes both the assembly and disruption of nucleosomal structure, in an ATP-dependent manner, via BRG1 and hBrm ATPase and contributes to the assembly of replicated DNA into chromatin. The WSTF component of WINAC is able to interact with vitamin-D receptor, in a ligand-independent manner, to both activate and repress gene transcription by inducing a remodeled chromatin state and allowing transcription or condensing the nucleosomal architecture to repress gene expression [Kitagawa et al., 2003].
Figure 2. BRG1-containing chromatin-modifying complexes.
The BRG1 chromatin remodeling protein can associate with numerous chromatin-modifying complexes including transcription coactivators and corepressors. BRG1 (or hBrm) is the central catalytic subunit of SWI/SNF-BAF or -PBAF chromatin remodeling complexes, which have been implicated in the transcriptional activation or repression of a variety of genes. Nuclear receptors can associate with many of these complexes through direct interaction with BAF subunits such as BAF250, BAF60a and BAF57. BRG1 can be found in complexes with transcription coactivators and histone modifying enzymes such as WINAC and NUMAC. Conversely, BRG1 can be assembled in complexes known to repress transcription and induce gene silencing including NCoR and mSin3A/HDAC complexes.
Histone-modifying enzymes such as coactivator-associated arginine methyltransferase-1 (CARM1) associate with BRG1 in the NUMAC complex (nucleosomal methylation activation complex) [Xu et al., 2004]. This complex harbors multiple SWI/SNF-associated proteins including BRG1, BAF250, BAF170, BAF155, BAF57, BAF47 and β-actin, and was found to display increased activity for histone methylation via CARM1 activity. In the NUMAC complex, BRG1 and CARM1 directly associate and assemble on estrogen receptor (ER)-responsive target promoters to cooperatively activate transcription [Xu et al., 2004].
BRG1 can also be found associated with other histone-modifying enzymes such as histone deacetylases-3 (HDAC3) and the transcriptional corepressors KAP-1 (Krab associated protein 1) within the NCoR-1 (Nuclear receptor corepressors-1) complex. Four core SWI/SNF proteins, BRG1, BAF170, BAF155 and BAF47, were identified by mass spectrometry or immunoblotting as members of the NCoR-1 complex [Underhill et al., 2000]. Also associated with this complex is KAP-1, which interacts with proteins containing KRAB domains, a common motif found in DNA-binding transcriptional repressors. KAP-1 has also been observed to interact in vivo with HP-1 (heterochromatin protein-1) proteins which are important for gene silencing [Nielsen et al., 1999; Ryan et al., 1999]. Taken together, the presence of BRG1 in NCoR-1 strongly suggests the complex possesses chromatin modifying function which may be required for transcriptional repression. An exciting possibility is that chromatin remodeling activity within NCoR-1 may be necessary to facilitate the binding of specific corepressors with nucleosomal targets to induce gene silencing.
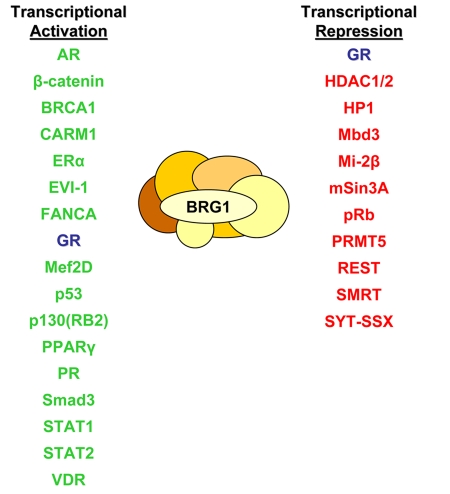
Various studies have identified BRG1-interacting proteins using methods such as immunoaffinity selection, chromatographic purifications and mass spectrometric peptide sequencing, as well as protein-protein interaction assays including yeast two-hybrid cDNA library screens, GST-pull down, immunoblotting and immunoprecipitations. Searching the Myriad ProNet protein-protein interaction database, we identified proteins which have been reported to interact either directly of indirectly with BRG1 [Wixon, 2001]. Figure 3 summarizes these BRG1-associating proteins and classifies the putative interaction according to the transcriptional outcome. Interestingly, BRG1 interacts with a diverse group of nuclear proteins involved in a wide range of processes and include nuclear receptors, members of the transcriptional machinery, chromatin-modifying enzymes and tumor suppressors, as well as core SWI/SNF subunits and proteins critical for genomic stability and maintenance [Chen et al., 2006; Trotter and Archer, 2007]. The number of putative biochemical associations involving BRG1 suggests this evolutionarily conserved DNA-dependent ATPase containing protein may be critical for numerous biological activities. Although these studies demonstrate BRG1 has the potential to associate with vast numbers and types of proteins, the mechanistic consequences in vivo of many of these interactions remain to be determined.
Figure 3. Reported BRG1-interacting proteins.
Numerous studies have been conducted using various techniques which have identified BRG1-interacting proteins. For the purpose of this review, these BRG1-associating proteins have been grouped into two categories according to their transcriptional consequence: activation or repression. BRG1 has been reported to associate with numerous proteins implicated in transcriptional activation including various NRs such as AR (Marshall et al., 2003), ERα (Ichinose et al., 1997), GR (Fryer and Archer, 1998), PPARγ (Debril et al., 2004), PR (Vicent et al., 2006) and VDR (Kitagawa et al., 2003). Tumor suppressor proteins have also been found associated with BRG1, including BRCA1 (Bochar et al., 2000), p53 (Lee et al., 2002), and FANCA (Otsuki et al., 2001). Other proteins which are reported to interact with BRG1 include β-catenin (Barker et al., 2001), CARM1 (Xu et al., 2004), EVI-1 (Chi et al., 2003), Mef2D (Ohkawa et al., 2006), p130(RB2) (Giacinti and Giordano, 2006), Smad3 (Xi et al., 2007, 2008) and STAT proteins (Ni and Bremner, 2007; Pattenden et al., 2002). Proteins involved in transcriptional repression also interact with BRG1 and include GR (Bilodeau et al., 2006), HDACs (Underhill et al., 2000), HP-1 (Nielsen et al., 1999), Mbd3 (Datta et al., 2005), Mi-2β (Shimono et al., 2003), mSin3A (Sif et al., 2001), Rb (Giacinti and Giordano, 2006), PRMT5 (Pal et al., 2003), REST (Ooi et al., 2006), SMRT (Jung et al., 2001) and SYT-SSX (Ito et al., 2004; Perani et al., 2003). This list highlights a number of BRG1-interacting proteins which are considered transcription coactivators, corepressors or tumor suppressor proteins.
Role of BRG1 in transcription control
Recruitment to target genes
The SWI/SNF enzymatic complexes are large multimeric assemblies which are thought to be recruited to specific gene targets through association with DNA-binding transcription factors, coregulators, or by members of the general transcriptional machinery. These BRG1 assemblies contain a multitude of distinct DNA-binding motifs which are thought to play vital roles in targeting SWI/SNF activity to gene-specific promoters. It has been suggested that these DNA-binding motifs do not direct the complex to specific genomic sequences, but rather work in concert with gene-specific activation domains within transcriptional or histone-binding factors to allow efficient binding and chromatin remodeling [Peterson and Workman, 2000].
Multiple interactions are probably involved in the recruiting and stabilization of BRG1-containing SWI/SNF to hormone responsive promoters. Depending on the stage of transcription, nucleosomal composition, and promoter architecture, one or more subunits may play leading roles in SWI/SNF binding. Several SWI/SNF components including BRG1, BAF250, BAF60a/c and BAF57 have been shown to mediate critical interactions between several Type-I NRs, including glucocorticoid receptor (GR), estrogen receptor (ER), progesterone receptor [Roopra et al., 2004] and androgen receptor (AR) [Link et al., 2005], as well as Type-II NRs, retinoic acid receptor (RAR), vitamin D3 receptor (VDR) and the peroxisome proliferator-activated receptor γ (PPARγ) [Aoyagi et al., 2005; Chen et al., 2006; Debril et al., 2004; Dilworth et al., 2000; Ichinose et al., 1997; Lemon et al., 2001; Link et al., 2005; Trotter and Archer, 2004; Trotter and Archer, 2007].
Components of SWI/SNF with bromodomains such as BRG1, hBrm and BAF180 are thought to target acetylated histone tails [Singh et al., 2007]. In addition, SWI/SNF subunits, including BRG1, BAF250, BAF60a and BAF57, have been reported to mediate critical interactions with NRs. These associations are thought to be important mediators for recruiting the remodeling complex to genomic targets [Belandia et al., 2002; Garcia-Pedrero et al., 2006; Hsiao et al., 2003; Inoue et al., 2002]. Consequently, it is likely that multiple interactions mediate recruitment and stabilization of the SWI/SNF remodeling complex to gene-specific promoters through direct or indirect interactions involving one or more BAF proteins. The interaction between GR and BRG1 can occur through BAF250 and/or BAF60a. The BAF250 subunit, which contains intrinsic DNA-binding capabilities through an ARID (AT-rich interaction domain) motif, directs SWI/SNF activity to GR-responsive promoters using both DNA-binding and protein-protein interactions in combination to confer specificity of action [Trotter and Archer, 2007]. Interestingly, BAF60a utilizes distinct protein-binding surfaces, one each for interaction with GR and BRG1, to specifically regulate GR-responsive promoters. Glucocorticoid receptor binding of BAF60a has been shown to take place in a ligand-independent manner, but is required for efficient GR-mediated transcriptional activation and remodeling of the mouse mammary tumor virus (MMTV) promoter or endogenous promoters in vivo [Hsiao et al., 2003]. By contrast, the interaction between GR and BAF250a is dependent on the presence of ligand [Nie et al., 2000].
The BAF57 subunit has been shown to be a critical subunit by which SWI/SNF interfaces with various NRs including GR, ER and AR [Garcia-Pedrero et al., 2006; Hsiao et al., 2003; Link et al., 2005]. BAF57 interacts with both ER and p160 coactivators to recruit SWI/SNF, in a ligand-dependent manner, to estrogen-responsive promoters for transcriptional activation [Belandia et al., 2002]. Additionally, BAF57 has been shown to associate directly with AR to recruit BRG1-based complexes to AR-responsive promoters to alter transcriptional activity [Link et al., 2005; Marshall et al., 2003].
Recruitment of BRG1-containing complexes is also thought to occur through association with zinc finger proteins (ZFP), which represent a family of eukaryotic regulatory factors that have been shown to control numerous cellular processes such as differentiation, proliferation, signaling and apoptosis. The association between SWI/SNF and ZFPs were shown to take place exclusively through specific DNA-binding domains (DBD) and BRG1 to initiate ZFP-dependent transcription [Kadam and Emerson, 2003]. This enables BRG1-based chromatin modifying complexes to selectively target distinct sets of promoters. Taken together, BRG1 complexes can be recruited to target promoters through association with BAF subunits or transcription factors which display DNA-binding capabilities.
Transcriptional activation
A role for BRG1 chromatin remodeling complexes in transcriptional regulation by NRs was initially proposed for GR in yeast studies and substantiated in vivo using mammalian cell models [Fryer and Archer, 1998; Muchardt and Yaniv, 1993; Trotter and Archer, 2004; Yoshinaga et al., 1992]. Numerous studies suggest ATP-dependent chromatin remodeling by BRG1 influences ligand-induced transcriptional activation by a variety of NRs. Integrated hormone-responsive promoter systems such as MMTV and the estrogen-dependent pS2 have been employed to investigate the molecular mechanisms involved in NR-mediated chromatin remodeling and transcriptional activation by BRG1 [Chen et al., 2006].
The steroid hormone responsive MMTV promoter acquires a phased array structure of six positioned nucleosomes when stably integrated as chromatin. These nucleosomes are highly organized over the long terminal repeat (LTR), which encompasses several regulatory elements including hormone response elements (HREs) and binding sites for transcription factors, as well as for TATA binding protein (TBP) [Archer et al., 1991; Trotter and Archer, 2004]. These transcriptional elements found within MMTV allow for strong promoter activation by glucocorticoids through mechanisms requiring chromatin modifying proteins such as BRG1. Chromatin remodeling proteins are thought to increase DNA accessibility, thus permitting recruitment and binding of transcription factors to site-specific sequences and allowing promoter activation.
Using techniques such as ultrafast UV-laser crosslinking have provided insight into the progression and possible mechanism of GR-mediated chromatin remodeling by SWI/SNF at the MMTV promoter. Using this assay, the highly dynamic interactions between GR and MMTV were found to be dependent on the GR-mediated activity of SWI/SNF, suggesting the transient association of GR occurs in concert with chromatin remodeling [McNally et al., 2000; Nagaich et al., 2004]. Collectively, these studies demonstrate rapid binding of GR to the chromatin followed by active displacement during the remodeling process. In the absence of GR, the BRG1 remodeling complex is associated with random positions on the chromatin template, but is recruited to specific regions in the presence of ligand-bound receptor [Fletcher et al., 2002; Georgel et al., 2003; Nagaich et al., 2004]. Studies using a stably integrated MMTV reporter, in the BRG1/hBrm null SW-13 cell line, also further establish the requirement for BRG1 in GR-dependent promoter activation. Interestingly, non-BRG1-based ATP-dependent chromatin remodeling complexes, such as ISWI or Mi-2, are unable to substitute for BRG1 activity including chromatin remodeling, transcription factor binding and transcriptional activation of the stably integrated promoter. In this cell model, a select number of endogenous GR-responsive promoters were analyzed including p21, HSD-11β-type 2 and PLZF, each of which displays a similar transcriptional dependence for BRG1 activity [Trotter and Archer, 2004].
Further insight into the molecular mechanism of BRG1 action was provided through the generation and characterization of chimeric proteins constructed from two of the best studied ATP-dependent chromatin remodeling proteins, BRG1 and SNF2h [Fan et al., 2005]. Both BRG1 and SNF2h represent the central catalytic subunit of the SWI/SNF- and ISWI-based chromatin remodeling complexes, respectively, and genetic analyses indicate these ATPase proteins can not complement each other (27.8% homology between the two remodeling proteins; 44.3% homology over ATPase domain). To determine if ATPase activity provided from another remodeling protein could function within the context of SWI/SNF, a chimeric protein was generated by replacing the BRG1 ATPase region with the ATPase domain derived from SNF2h. The chimeric protein, designated B-S-B, was evaluated using in vitro chromatin remodeling assays which revealed the mechanism of remodeling is specific to the ATPase present. In vivo, the BRG1/SNF2h chimera induced expression from a subset of BRG1-dependent genes when introduced into SW-13 cells. Interestingly, the chimeric protein was unable to stimulate GR-mediated transcriptional activation or chromatin remodeling of integrated MMTV, indicating the ATPase activities of BRG1 and SNF2h are not functionally interchangeable [Fan et al., 2005]. Together, these data suggest BRG1 activity is a critical component for hormone-dependent transcriptional activation.
Numerous studies have utilized promoter reporter models to analyze the role of BRG1 in NR-mediated transcription regulation. Results from these reports reveal BRG1 is a critical component required for transcriptional activation involving NRs including PR, AR and ER [Chen et al., 2006]. Studies using chromatin MMTV show PR competes with GR for available BRG1 upon glucocorticoid treatment, indicating the remodeling protein is a required component of PR-mediated promoter activation [Fryer and Archer, 1998; Li et al., 2003]. Examination of AR-mediated transcriptional regulation suggests recruitment of SWI/SNF has been established for AR-dependent gene expression [Marshall et al., 2003]. Estrogen receptor has also been reported to require BRG1 activity for activation of ER-responsive genes. Interestingly, transcriptional activation from ER-dependent promoters, within SW-13 cells, was significantly reduced in the absence of BRG1; conversely, ER-mediated transcription was restored upon expression of the remodeling protein [Chen et al., 2006]. Additionally, BRG1 can associate with ligand-bound VDR within the WINAC complex which is recruited to VDR-responsive promoters activating transcription. Defects in this VDR-signaling complex lead to the autosomal genetic disorder Williams syndrome [Kitagawa et al., 2003].
In summary, chromatin remodeling is an essential regulatory step where BRG1-containing complexes can directly and indirectly support ligand-mediated transcriptional activation by NRs. Given the functional diversity of NR, it is thought that coactivators differentially affect chromatin remodeling based on target gene and its biological context.
Transcriptional repression
The role of BRG1 has been widely associated with transcriptional activation; however, studies indicate the remodeling protein can play critical roles in gene silencing through interactions with a variety of transcriptional corepressors [Gaston and Jayaraman, 2003; Underhill et al., 2000]. BRG1 has been shown to interact with retinoblastoma tumor suppressor [Zhang et al., 2000] to form a repressor complex which inhibits cell cycle proteins such as cyclin A, D1 and E [Giacinti and Giordano, 2006; Zhang et al., 2000]. Complexes purified from mammalian extracts demonstrate BRG1 associates and is a member of the mSin3A/HDAC complex that is implicated in transcriptional repression of a variety of genes [Sif et al., 2001]. The chromatin remodeling protein was also found in association with the NCoR corepressor complex, which displays HDAC activity and includes the heterochromatin protein-1 (HP1) interacting protein TIF1β [Underhill et al., 2000].
Several studies have reported multisubunit complexes possessing ATPase activity exist in combination with histone deacetylases (HDACs), methyl CpG-binding proteins (MBDs) and histone methyltransferases (HMTs) in transcriptional repression complexes coupling ATP-dependent chromatin remodeling with a wide variety of mechanisms involved in gene silencing [Bilodeau et al., 2006; Dacwag et al., 2007; Datta et al., 2005; Pal et al., 2003; Xu et al., 2006]. Interestingly, subunits of the mSin3A/HDAC corepressor complex associate with BRG1, suggesting the SWI/SNF complex may be involved in transcriptional repression and gene silencing [Sif, 2004]. Recent studies have shown the SWI/SNF complex and their associated histone-modifying enzymes are involved in the transcriptional repression of genes important for various cellular processes such as cycle regulation [Giacinti and Giordano, 2006]. BRG1 has shown to associate with Rb proteins to induce cell cycle arrest and this Rb-mediated transcriptional repression is dependent on HDAC activity. Additionally, BRG1/HDAC-containing complexes have been shown to repress expression of genes involved in cell cycle regulation including cdc2, cyclin A, D1, E and the Myc/Max/Mad target cad [Pal et al., 2003; Zhang et al., 2000]. This repression is thought to occur through direct interaction and recruitment of PRMT5 and the mSin3A/HDAC2 corepressor complex to target promoters for transcriptional inhibition [Pal et al., 2003].
BRG1 also associates with the SYT oncoprotein assembly, which has been reported to display corepressor activity through interactions with mSin3A/HDAC transcriptional regulatory complexes [Ito et al., 2004]. Taken together, these studies suggest BRG1 activity is a critical component of the mSin3A/HDAC repression complex, which acts on chromatin through transcriptional regulators, corepressors and histone-modifying enzymes and leads to gene silencing.
Promoter inhibition by BRG1 has been reported for a number of genes using either the HDAC-dependent or -independent pathways. The repressor element 1-silencing transcription factor (REST) is a transcriptional regulator that has been shown to repress expression of genes involved in neuronal function [Battaglioli et al., 2002; Schoenherr and Anderson, 1995]. Interestingly, REST function depends on its associations with a wide variety of proteins including HDAC1, HDAC2 and core SWI/SNF subunits BRG1, BAF170 and BAF57, as well as histone demethylase LSD1 and histone methyltransferase G9a [Battaglioli et al., 2002; Gu et al., 2005; Roopra et al., 2004; Shi et al., 2004]. Although the precise mechanism regarding BRG1-dependent REST-mediated repression remains unclear, BRG1 is thought to be involved in the establishment of an altered promoter state which allows histone acetylation-dependent binding of REST and results in transcriptional inhibition [Ooi et al., 2006].
Nuclear hormone receptors have been shown to mediate gene repression through association with BRG1. Ligand-bound GR is critical for the expression of a variety of proteins including tryptophan oxygenase (TO) and tyrosine amino transferase [Tamkun et al., 1992] during liver development [Danesch et al., 1987; Grange et al., 2001]. Interestingly, SWI/SNF complexes containing BRG1 were shown to significantly downregulate the GR-mediated transcription of both genes in a HDAC-independent manner. This observation suggests BRG1 activity may induce a closed conformation, thus restricting transcription factor binding and resulting in attenuated gene expression [Inayoshi et al., 2005].
BRG1 has also been characterized as a coactivator and corepressor at the same promoter, switching modes of action by ligand-directed differential coordination with various transcriptional regulators. For example, BRG1 is recruited to the same ER-responsive promoters by different cofactors in response to hormone or antagonists to transcriptionally activate or repress gene expression, respectively. SWI/SNF complexes recruited by ligand-bound ER result in active gene transcription; conversely, when BRG1 is recruited via HDAC1, p300 and prohibitin, in response to ER-bound antagonist, the promoter activation was significantly diminished [Zhang et al., 2007]. Although the mechanism underlying this molecular switching remains unclear, it does provide interesting insight into the role of BRG1 as a transcriptional coregulator. Taken together, these studies demonstrate the BRG1 chromatin remodeling protein can associate with corepressor complexes and be recruited by a variety of transcription factors to specific promoters to inhibit transcription and induce gene silencing.
Biological role of BRG1 – deregulated expression
BRG1 and hBrm are highly conserved, mutually exclusive, catalytic proteins found incorporated within the context of numerous enzymatic complexes that modify chromatin structure and alter gene expression patterns. In vitro, these ATPase-containing proteins display similar chromatin remodeling activities and studies indicate that BRG1 and hBrm can regulate transcriptional activity at distinct genes [Kadam and Emerson, 2003]. However, in vivo studies suggest these proteins exhibit distinct non-redundant biological functions. Interestingly, genetic inactivation of BRG1 or hBrm in mice resulted in moderate to severe deregulation of cellular processes such as proliferation. BRG1-deficient mice die at the pre-implantation stage, suggesting that BRG1 is essential for early development. Moreover, BRG1 heterozygotes were found to be predisposed to tumor development. Conversely, hBrm null mice were found to be viable, fertile and dispensable for cell growth, although these mice showed increased expression of BRG1 in certain tissues and displayed aberrant cell cycle regulation [Reyes et al., 1998]. Together, these knock-out studies show BRG1- and hBrm-containing complexes are regulated and targeted differently to alter gene expression patterns during critical stages of cell proliferation and differentiation.
The SWI/SNF chromatin remodeling complex has been described as a key regulator of skeletal muscle differentiation, where the ATPase of BRG1 is required for gene expression at different stages of skeletal myogenesis. Recent studies have shown that expression of myogenic late genes requires BRG1 ATPase activity in conjunction with myogenin and Mef2D [Ohkawa et al., 2007]. Interestingly, BRG1 associates with both MyoD and Mef2 which targets BRG1-based chromatin remodeling activity to myogenic-specific gene promoters to regulate protein expression at different stages of skeletal muscle differentiation [Ohkawa et al., 2006].
Genetic and molecular evidence indicate BRG1 can act as tumor suppressors in a variety of human cancers or cell lines due to mutations or downregulation of protein expression [Wong et al., 2000]. In BRG1-null cell lines, reintroduction of exogenous BRG1 induces a growth arrest phenotype by repression of E2F-genes and activation of cyclin-dependent kinase (CDK) inhibitors p21 and p15 [Hendricks et al., 2004]. BRG1 has also been reported to interact with other tumor suppressors including pRb, BRCA1, c-Myc, c-Fos and member proteins of the Wnt signaling pathway [Barker et al., 2001; Bochar et al., 2000; Eroglu et al., 2006; Murphy et al., 1999; Pal et al., 2003]. Therefore, due to the multitude of transcriptional events which require SWI/SNF activity, loss of BRG1 would tend to alter normal gene expression patterns and possibly lead to oncogenic phenotypes.
Conclusions
The role of BRG1 in transcriptional activation by numerous Type-I and Type-II NRs or transcriptional repression through association with mSin3A/HDAC complexes has been firmly established. Among the various BRG1-based complexes, the SWI/SNF family has been the best characterized regarding function and enzymatic activity in transcriptional activation with chromatin remodeling. Activated receptors bound to target DNA sequences recruit transcriptional coactivators, such as BRG1, which are essential to modulate chromatin structure, and initiate gene expression. Studies suggest NRs recruit SWI/SNF activity to gene-specific promoters, in a ligand-dependent manner, where BRG1 functions to remodel the chromatin architecture. This remodeled nucleosomal state allows access to and binding of members of the preinitiation complex (PIC) [Trotter and Archer, 2007]. Conversely, BRG1-mediated gene silencing is also thought to involve manipulation of the promoter structure, resulting in increased binding of chromatin modifying enzymes and/or repressor complexes, such as mSin3A/HDAC1/2, to inhibit transcription. Targeting BRG1 activity is thought to be mediated through interactions with various members of the enzymatic complex, including BAF subunits, which have been shown to associate with numerous transcriptional coactivator or corepressor proteins. Taken together, these observations imply BRG1 is an essential component of a diverse group of chromatin-modifying complexes which alter chromatin structure to regulate transcription. To this end, clear mechanistic insights gained from the functional interactions between BRG1 and nuclear receptors may provide a better understanding regarding the precise role the remodeling protein plays in molecular events within the cell.
Abbreviations
- AR
androgen receptor
- ARID
AT-rich interaction domain
- BAF
BRG1/hBrm-associated factors
- BRG1
brahma-related gene-1
- BRK
BRM and KIS
- CARM1
coactivator-associated arginine methyltransferase-1
- CDK
cyclin-dependent kinase
- DBD
DNA-binding domain
- ER
estrogen receptor
- GR
glucocorticoid receptor
- hBrm
human brahma
- HDAC
histone deacetylase
- HDAC3
histone deacetyltransferase 3
- HMT
histone methyltransferase
- HP-1
heterochromatin protein-1
- HRE
hormone response element
- HSA
helicase/SANT-associated
- KAP-1
Krab associated protein 1
- LTR
long terminal repeat
- MBD
methyl CpG-binding protein
- MMTV
mouse mammary tumor virus
- N-CoR-1
nuclear receptor corepressors-1
- NF1
nuclear factor-1
- NLS
nuclear localization signal
- NR
nuclear receptor
- NUMAC
nucleosomal methylation activation complex
- OTF
octamer transcription factor
- PBAF
polybromo-associated BAF
- PIC
preinitiation complex
- PPARγ
peroxisome proliferator-activated receptor γ
- PR
progesterone receptor
- QLQ
Glutamine-Leucine-Glutamine
- RAR
retinoic acid receptor
- Rb
retinoblastoma tumor suppressor
- REST
repressor element 1-silencing transcription factor
- RSC
remodeling the structure of chromatin
- SMARCA4
SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily a, member 4
- SWI/SNF
mating-type switching and sucrose non-fermenting
- TAT
tyrosine aminotransferase
- TBP
TATA binding protein
- TO
tryptophan oxygenase
- VDR
vitamin-D receptor
- WINAC
WSTF including nucleosome assembly complex
- WSTF
Williams syndrome transcription factor
- ZFP
zinc finger proteins
References
- Allen M. D., Religa T. L., Freund S. M., Bycroft M. Solution structure of the BRK domains from CHD7. J Mol Biol. 2007;371:1135–40. doi: 10.1016/j.jmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Aoyagi S., Trotter K. W., Archer T. K. ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm. 2005;70:281–307. doi: 10.1016/S0083-6729(05)70009-1. [DOI] [PubMed] [Google Scholar]
- Archer T. K., Cordingley M. G., Wolford R. G., Hager G. L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–98. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. Embo J. 2001;20:4935–43. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli E., Andres M. E., Rose D. W., Chenoweth J. G., Rosenfeld M. G., Anderson M. E., Mandel G. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277:41038–45. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- Belandia B., Parker M. G. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell. 2003;114:277–80. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- Belandia B., Orford R. L., Hurst H. C., Parker M. G. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. Embo J. 2002;21:4094–103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S., Vallette-Kasic S., Gauthier Y., Figarella-Branger D., Brue T., Berthelet F., Lacroix A., Batista D., Stratakis C., Hanson J., Meij B., Drouin J. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20:2871–86. doi: 10.1101/gad.1444606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar D. A., Wang L., Beniya H., Kinev A., Xue Y., Lane W. S., Wang W., Kashanchi F., Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–65. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cairns B. R., Kim Y. J., Sayre M. H., Laurent B. C., Kornberg R. D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci U S A. 1994;91:1950–4. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., Kornberg R. D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–60. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran R., Thompson M. Polybromo-1-bromodomains bind histone H3 at specific acetyl-lysine positions. Biochem Biophys Res Commun. 2007;355:661–6. doi: 10.1016/j.bbrc.2007.01.193. [DOI] [PubMed] [Google Scholar]
- Chen J., Kinyamu H. K., Archer T. K. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384–94. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesch U., Gloss B., Schmid W., Schutz G., Schule R., Renkawitz R. Glucocorticoid induction of the rat tryptophan oxygenase gene is mediated by two widely separated glucocorticoid-responsive elements. Embo J. 1987;6:625–30. doi: 10.1002/j.1460-2075.1987.tb04800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J., Majumder S., Bai S., Ghoshal K., Kutay H., Smith D. S., Crabb J. W., Jacob S. T. Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer Res. 2005;65:10891–900. doi: 10.1158/0008-5472.CAN-05-1455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Debril M. B., Gelman L., Fayard E., Annicotte J. S., Rocchi S., Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–86. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Dilworth F. J., Fromental-Ramain C., Yamamoto K., Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol Cell. 2000;6:1049–58. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- Dingwall A. K., Beek S. J., McCallum C. M., Tamkun J. W., Kalpana G. V., Goff S. P., Scott M. P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–91. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Copley R. R., Schultz J., Ponting C. P., Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., Becker P. B. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117:3707–11. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- Eroglu B., Wang G., Tu N., Sun X., Mivechi N. F. Critical role of Brg1 member of the SWI/SNF chromatin remodeling complex during neurogenesis and neural crest induction in zebrafish. Dev Dyn. 2006;235:2722–35. doi: 10.1002/dvdy.20911. [DOI] [PubMed] [Google Scholar]
- Fan H. Y., Trotter K. W., Archer T. K., Kingston R. E. Swapping function of two chromatin remodeling complexes. Mol Cell. 2005;17:805–15. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–53. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fletcher T. M., Xiao N., Mautino G., Baumann C. T., Wolford R., Warren B. S., Hager G. L. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–63. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J., Archer T. K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Garcia-Pedrero J. M., Kiskinis E., Parker M. G., Belandia B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281:22656–64. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- Gaston K., Jayaraman P. S. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol Life Sci. 2003;60:721–41. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P. T., Fletcher T. M., Hager G. L., Hansen J. C. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev. 2003;17:1617–29. doi: 10.1101/gad.1097603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–7. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Grange T., Cappabianca L., Flavin M., Sassi H., Thomassin H. In vivo analysis of the model tyrosine aminotransferase gene reveals multiple sequential steps in glucocorticoid receptor action. Oncogene. 2001;20:3028–38. doi: 10.1038/sj.onc.1204327. [DOI] [PubMed] [Google Scholar]
- Gu H., Liang Y., Mandel G., Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci U S A. 2005;102:7571–6. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–92. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Hansen J. C. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev. 2001;11:124–9. doi: 10.1016/s0959-437x(00)00168-4. [DOI] [PubMed] [Google Scholar]
- Hendricks K. B., Shanahan F., Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–76. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn P. J., Peterson C. L. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- Hsiao P. W., Fryer C. J., Trotter K. W., Wang W., Archer T. K. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–20. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H., Garnier J. M., Chambon P., Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- Inayoshi Y., Kaneoka H., Machida Y., Terajima M., Dohda T., Miyake K., Iijima S. Repression of GR-mediated expression of the tryptophan oxygenase gene by the SWI/SNF complex during liver development. J Biochem (Tokyo) 2005;138:457–65. doi: 10.1093/jb/mvi147. [DOI] [PubMed] [Google Scholar]
- Inoue H., Furukawa T., Giannakopoulos S., Zhou S., King D. S., Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277:41674–85. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- Ito T., Ouchida M., Ito S., Jitsumori Y., Morimoto Y., Ozaki T., Kawai A., Inoue H., Shimizu K. SYT, a partner of SYT-SSX oncoprotein in synovial sarcomas, interacts with mSin3A, a component of histone deacetylase complex. Lab Invest. 2004;84:1484–90. doi: 10.1038/labinvest.3700174. [DOI] [PubMed] [Google Scholar]
- Johnson C. N., Adkins N. L., Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005;83:405–17. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- Kadam S., Emerson B. M. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–4. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Choi D., Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kinyamu H. K., Archer T. K. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim Biophys Acta. 2004;1677:30–45. doi: 10.1016/j.bbaexp.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Kitagawa H., Fujiki R., Yoshimura K., Mezaki Y., Uematsu Y., Matsui D., Ogawa S., Unno K., Okubo M., Tokita A., Nakagawa T., Ito T., Ishimi Y., Nagasawa H., Matsumoto T., Yanagisawa J., Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–17. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E., Green M. R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–81. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treich I., Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–91. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Lemon B., Inouye C., King D. S., Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–8. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Link K. A., Burd C. J., Williams E., Marshall T., Rosson G., Henry E., Weissman B., Knudsen K. E. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–15. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol. 2003;23:3763–73. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., Zhang M., Kornberg R. D. RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- Marshall T. W., Link K. A., Petre-Draviam C. E., Knudsen K. E. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem. 2003;278:30605–13. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- McNally J. G., Muller W. G., Walker D., Wolford R., Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Mohrmann L., Verrijzer C. P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–90. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. J., Hardy S., Engel D. A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–33. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich A. K., Walker D. A., Wolford R., Hager G. L. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–74. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–58. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Xue Y., Yang D., Zhou S., Deroo B. J., Archer T. K., Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18:6385–95. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y., Yoshimura S., Higashi C., Marfella C. G., Dacwag C. S., Tachibana T., Imbalzano A. N. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007;282:6564–70. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y., Marfella C. G., Imbalzano A. N. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. Embo J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L., Belyaev N. D., Miyake K., Wood I. C., Buckley N. J. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281:38974–80. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–87. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin M. J., Kadonaga J. T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–40. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Dingwall A., Scott M. P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A. 1994;91:2905–8. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Workman J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–92. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Tamkun J. W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–6. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- Phelan M. L., Sif S., Narlikar G. J., Kingston R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Randazzo F. M., Khavari P., Crabtree G., Tamkun J., Rossant J. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994;161:229–42. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- Reyes J. C., Barra J., Muchardt C., Camus A., Babinet C., Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17:6979–91. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A., Qazi R., Schoenike B., Daley T. J., Morrison J. F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–38. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–78. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C. J., Anderson D. J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–3. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Shen W., Xu C., Huang W., Zhang J., Carlson J. E., Tu X., Wu J., Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–10. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–98. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- Sif S., Saurin A. J., Imbalzano A. N., Kingston R. E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–18. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., D'Silva L., Holak T. A. DNA-binding properties of the recombinant high-mobility-group-like AT-hook-containing region from human BRG1 protein. Biol Chem. 2006;387:1469–78. doi: 10.1515/BC.2006.184. [DOI] [PubMed] [Google Scholar]
- Singh M., Popowicz G. M., Krajewski M., Holak T. A. Structural Ramification for Acetyl-Lysine Recognition by the Bromodomain of Human BRG1 Protein, a Central ATPase of the SWI/SNF Remodeling Complex. Chembiochem. 2007;8:1308–1316. doi: 10.1002/cbic.200600562. [DOI] [PubMed] [Google Scholar]
- Stern M., Jensen R., Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–68. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–72. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tremethick D. J. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–4. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Trotter K. W., Archer T. K. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265-266:162–7. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter K. W., Archer T. K. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol. 2004;24:3347–58. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C., Qutob M. S., Yee S. P., Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–70. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., Kuo A., Cairns B. R., Crabtree G. R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996b;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang W., Cote J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M., Workman J. L., Crabtree G. R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996a;15:5370–82. [PMC free article] [PubMed] [Google Scholar]
- Williamson M. P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297 ( Pt 2):249–60. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–91. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wixon Jo. Website Review: Protein-protein interactions on the web. Comparative and Functional Genomics. 2001;2:338–343. doi: 10.1002/cfg.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Chromatin structure and function. Academic Press; 1998. 447 pp. [Google Scholar]
- Wolffe A. P. Transcriptional regulation in the context of chromatin structure. Essays Biochem. 2001;37:45–57. doi: 10.1042/bse0370045. [DOI] [PubMed] [Google Scholar]
- Wong A. K., Shanahan F., Chen Y., Lian L., Ha P., Hendricks K., Ghaffari S., Iliev D., Penn B., Woodland A. M., Smith R., Salada G., Carillo A., Laity K., Gupte J., Swedlund B., Tavtigian S. V., Teng D. H., Lees E. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–7. [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Xu W., Cho H., Kadam S., Banayo E. M., Anderson S., Yates J. R., 3rd, Emerson B. M., Evans R. M. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–56. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Canman J. C., Lee C. S., Nie Z., Yang D., Moreno G. T., Young M. K., Salmon E. D., Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–20. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Meng X., Cai Y., Koury M. J., Brandt S. J. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J. 2006;399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Cui K., Murray D. M., Ling C., Xue Y., Gerstein A., Parsons R., Zhao K., Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zhang H. S., Gavin M., Dahiya A., Postigo A. A., Ma D., Luo R. X., Harbour J. W., Dean D. C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- Zhang B., Chambers K. J., Faller D. V., Wang S. Reprogramming of the SWI/SNF complex for co-activation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene. 2007;26:7153–7. doi: 10.1038/sj.onc.1210509. [DOI] [PubMed] [Google Scholar]